Abstract
RNA folding landscapes have been described alternately as simple and as complex. The limited diversity of RNA residues and the ability of RNA to form stable secondary structures prior to adoption of a tertiary structure would appear to simplify folding relative to proteins. Nevertheless, there is considerable evidence for long-lived misfolded RNA states, and these observations have suggested rugged energy landscapes. Recently, single molecule fluorescence resonance energy transfer (smFRET) studies have exposed heterogeneity in many RNAs, consistent with deeply furrowed rugged landscapes. We turned to an RNA of intermediate complexity, the P4-P6 domain from the Tetrahymena group I intron, to address basic questions in RNA folding. P4-P6 exhibited long-lived heterogeneity in smFRET experiments, but the inability to observe exchange in the behavior of individual molecules led us to probe whether there was a non-conformational origin to this heterogeneity. We determined that routine protocols in RNA preparation and purification, including UV shadowing and heat annealing, cause covalent modifications that alter folding behavior. By taking measures to avoid these treatments and by purifying away damaged P4-P6 molecules, we obtained a population of P4-P6 that gave near-uniform behavior in single molecule studies. Thus, the folding landscape of P4-P6 lacks multiple deep furrows that would trap different P4-P6 molecules in different conformations and contrasts with the molecular heterogeneity that has been seen in many smFRET studies of structured RNAs. The simplicity of P4-P6 allowed us to reliably determine the thermodynamic and kinetic effects of metal ions on folding and to now begin to build more detailed models for RNA folding behavior.
Keywords: Biophysics, RNA, RNA Folding, RNA Modification, Single Molecule Biophysics, Heterogeneity
Introduction
Highly structured RNAs such as the ribosome, spliceosome, and riboswitches must fold to function and undergo conformational changes in the course of their function (1, 2). With the majority of the transcriptome largely unexplored, it seems likely that additional roles of RNA structure will emerge (3). Yet the folding and conformational changes of structured RNAs remain poorly understood.
Two general models for how RNAs fold have been widely discussed. In one view, RNA folding is a simple process, particularly in comparison to protein folding (4, 5). In a contrasting view, RNA folding is complex, replete with numerous deep kinetic traps on a rugged energy landscape (6, 7). The simple view is derived from the limited diversity of RNA structural components and the hierarchical nature of RNA folding. The isolated RNA secondary structure is highly stable in contrast to the marginal stability of isolated protein α-helices or β-sheets. In transitioning from a secondary to a tertiary structure, junctions that link RNA helices can favor particular orientations of the helical elements that are then enforced by modular tertiary motifs (8, 9). In contrast, globular proteins appear to have more extensive and interconnected packing arrangements with less distinct separation in the formation of the secondary and tertiary structure.
Despite this apparent simplicity, many RNAs fold slowly, on the timescale of seconds, minutes, and even longer, and many RNAs form long-lived misfolded states (10–18). The slow folding and propensity to fall into kinetic traps has led to the view that RNA folding involves a complex, rugged energy landscape. Indeed, single molecule fluorescence resonance energy transfer (smFRET)2 studies have strongly reinforced the idea of rugged RNA folding landscapes (19–26). In these experiments, individual molecules often show distinct folding behaviors that persist over observation times of seconds to minutes.
Nevertheless, a critical limitation of the smFRET experiments has been the difficulty in probing and even observing the exchange between distinct folding behaviors. Indeed, we are aware of only one RNA for which extensive exchange between distinct folding states has been observed, and this RNA, the Tetrahymena group I ribozyme (14), is a complex RNA that renders further in-depth dissection difficult. Here we turn to the 160-nucleotide P4-P6 domain of this ribozyme (27) (Fig. 1, A and B). P4-P6 was the first RNA with a side-by-side arrangement of helices to have its structure elucidated by x-ray crystallography and has been the subject of numerous folding studies (27–44). The knowledge provided by these prior studies along with its relative simplicity, with only two long-range tertiary interactions, render P4-P6 an attractive target for addressing the general issue of complexity in RNA folding.
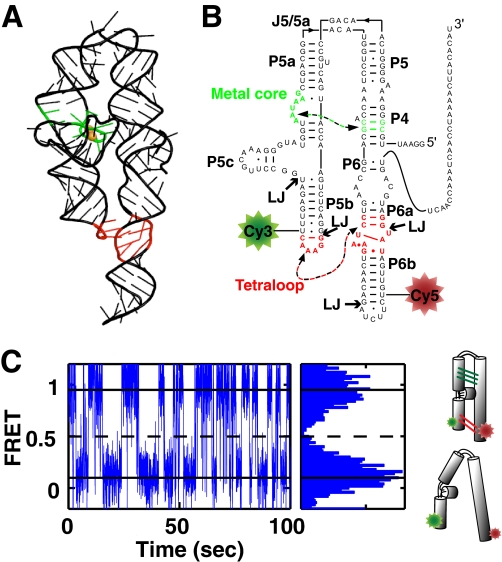
FIGURE 1.
smP4-P6 for smFRET studies. A, rendering of the P4-P6 tertiary structure (PDB code 1GID). The TL/TLR interaction is shown in red, and the MC/MCR is shown in green. B, the secondary structure of smP4-P6 used herein. Ligation junctions are denoted LJ, and the tertiary contacts are colored as in A. C, smFRET trace of smP4-P6 in 2.5 mm Mg2+ (left panel) and cumulative FRET histogram for this trace (right panel). The histogram shows distinct high and low FRET states, as indicated by the solid horizontal lines. The dashed line indicates the position of the FRET threshold. The schematics on the right represent the folded high FRET and unfolded low FRET states.
EXPERIMENTAL PROCEDURES
A full description of the methods is provided in the supplemental information. Initial smP4-P6 samples were prepared as described previously using standard laboratory techniques (45–49). Length homogeneity of in vitro transcribed pieces was ensured by cleaving at the 3′ end with a trans-acting DNAzyme. For the two synthetic cy3- and cy5-labeled oligonucleotides, single-nucleotide resolution was obtained via denaturing PAGE purification. Prior to ligation, the five RNA pieces and two DNA splints were annealed at 95 °C for 5 min in initial preparations and for 1 min in non-UV-shadowed preparations. Non-UV shadowed smP4-P6 RNA was made as in the initial preparation, except the in vitro transcribed pieces were DNAzyme treated immediately after transcription and were purified using Sephadex PD-10 desalting columns (GE Healthcare), thereby eliminating two PAGE purification and UV shadowing steps. α-32P-labeled P4-P6 was transcribed from a PCR-derived DNA template using standard in vitro transcription conditions. Denaturing gels were 8–20% polyacrylamide gel, with 7 m urea and TBE (100 mm Tris, 83 mm boric acid, 1 mm EDTA) running buffer. Native gels were 10% polyacrylamide, 1 mm MgCl2 and TB buffer (100 mm Tris, 83 mm boric acid) and were run at 4 °C. Spin column purification was completed with Microcontm YM-30 (Millipore) ultrafiltration columns. UV shadowing was performed with a 6-watt, 252 nm UV light (UVP). When autoradiography was used, ∼10,000 cpm of analyte was loaded in each gel lane, and gels were dried and exposed to storage phosphor screens (GE Healthcare).
smFRET measurements were made on a custom-built prism-based total internal reflection microscope (14, 37). Buffers used in smFRET measurements contained 1–10 mm MgCl2, 5–50 mm BaCl2, or 700 mm additional NaCl, 50 mm sodium MOPS (pH 7.0), 100 mm NaCl, 2 mg/ml glucose, 1.8 mm Trolox, 100 units/ml glucose oxidase, and 1000 units/ml catalase. smFRET data were collected with 40-ms time resolution and analyzed using software written in-house.
Simulated distributions of thermodynamic and kinetic parameters were extracted from simulated single-molecule time traces. Traces were simulated assuming a two-state kinetic model. Each simulated trace has a length and donor/acceptor channel signal with a mean and standard deviation corresponding to that obtained from fits of actual traces.
RESULTS AND DISCUSSION
Construction of Dye-labeled P4-P6 and Initial Single Molecule FRET Assays
P4-P6 for single molecule FRET studies, referred to as smP4-P6 herein, was made with cy3 and cy5 fluorescent dyes positioned to provide a large FRET signal upon folding and with a 26-nucleotide 3′ tail for surface immobilization (Fig. 1B). smP4-P6 was assembled via splinted ligation from five RNA pieces, two of which were synthetic and modified with cy3 or cy5 and three of which were prepared via in vitro transcription. DNA splints were used to position the ends to be ligated, and the ligation was catalyzed by T4 DNA ligase. This methodology is routinely used in RNA studies (45, 46, 49) and was used previously to make smP4-P6 to study tertiary folding (37, 38) (see “Experimental Procedures”).
smFRET traces for individual smP4-P6 molecules were stereotyped by well defined high and low FRET states with fast transitions between these states, as observed previously (37). With the use of additives that stabilize the dyes (50), it is possible to routinely observe single RNA molecules for 100 s and longer with high signal to noise and millisecond time resolution (Fig. 1C). These long traces allow the precise determination of equilibrium and rate parameters for individual molecules. To obtain folding equilibrium constants (Kfold) and free energies (ΔG°fold) for each smP4-P6 molecule, we assigned threshold FRET values for the unfolded and folded states and determined the time spent in each state (Fig. 1C). The kinetics of each trace were determined with a two-state hidden Markov model (51–53).
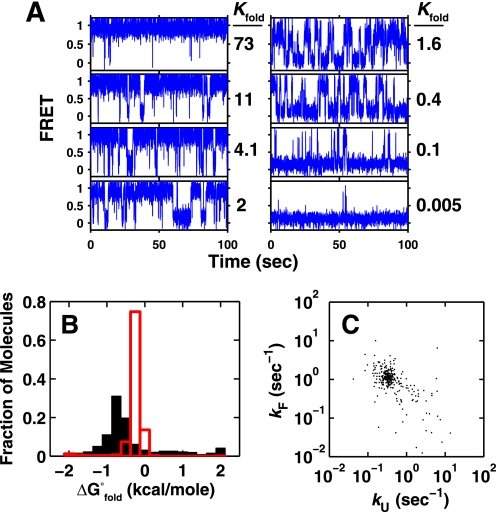
Inspection of individual FRET traces revealed molecules that appeared to exhibit a wide range of thermodynamic and kinetic behaviors (Fig. 2A). The folding free energies for the > 500 individual molecules plotted in Fig. 2B exhibit values that span a range of over 4 kcal/mol. The rate constants underlying these individual equilibria spanned a similarly large range of 3 orders of magnitude (Fig. 2C). Stochastic simulations indicate that these distributions are much broader than expected from experimental factors alone (Fig. 2B, red lines, and “Experimental Procedures”).
FIGURE 2.
smP4-P6 shows heterogeneity. A, anecdotal traces of smP4-P6 showing a broad range of equilibrium constants. B, histogram of the free energies of folding of individual P4-P6 molecules (n = 566) in 2.5 mm Mg2+. The predicted width of the distribution because of the inherent noise in the measurement is overlaid in red. The peak center of the simulated distribution is shifted to the right of the observed peak because of the large number of mostly unfolded molecules on the right side of the observed distribution. C, the folding and unfolding rate constants for 300 randomly chosen molecules from the distribution in B. (See supplemental Fig. S7 for all data points.)
The spread in the data from individual smP4-P6 molecules strongly suggests that there are differences between molecules that result in a range of kinetic and equilibrium behaviors. Such heterogeneity is commonly observed in single molecule experiments and has typically been interpreted in terms of long-lived conformational differences between molecules (14, 19–26). Below, we first describe probes of this potential origin of P4-P6 heterogeneity. We then describe tests of the alternative model that considers if the differential behaviors arise from covalent differences between smP4-P6 molecules within the population.
Testing the Conformational Exchange between Thermodynamic States
Molecules on the same energy landscape can, in principle, adopt all other conformations present on the landscape, given sufficient time. To test whether smP4-P6 molecules that appear different on the 1–2 min time scale of a typical smFRET measurement can sample the other states, a discontinuous assay was used. This approach established that heterogeneity observed in the docking behavior of the P1 helix of the Tetrahymena group I ribozyme arises from conformational differences among the ribozyme molecules (14). We carried out these smP4-P6 experiments in Ba2+, as folding transitions are more frequent in Ba2+ than in Mg2+ at the folding midpoint, and more frequent transitions allow accurate determination of equilibrium constants with shorter observation times.
Fig. 3A shows the initial ΔG°fold distribution (determined from 1 min of observation) of smP4-P6 in 15 mm Ba2+, and Fig. 3B shows the distribution for these same molecules after waiting 60 min to provide time for reequilibration. A consistent color coding is used in Fig. 3 to follow the fate of molecules originally in the center and in the extremes of the free energy distribution. The overwhelming majority of smP4-P6 molecules return to their initial ΔG°fold value, with only 9 of 175 molecules separated by more than 0.5 kcal/mole from the initially determined folding free energy. Experiments conducted with a 10-min wait or with unfolding via Ba2+ removal to temporarily unfold smP4-P6 showed similarly low levels of apparent exchange (supplemental Fig. S1).
FIGURE 3.
smP4-P6 states persist for more than 60 min. A, histogram of folding free energies of smP4-P6 (n = 175) in 15 mm Ba2+. B, the same molecules as in A, imaged after the laser was turned off for 60 min. Colors correspond to the initial free energies of the molecules in A, and the black outline demarcates the initial distribution.
If the occasional examples of exchange were due to conformational rearrangement, a reproducibly higher exchange would be expected with increased observation time and with unfolding. Our results suggest that we were at the sensitivity limit of this assay and thus provide no evidence for the model in which the heterogeneity of smP4-P6 molecules arises from conformationally distinct molecules. The remaining possibilities were the presence of very deep energy wells that persist even in unfolded states or covalent differences between the molecules. As single molecule assays cannot readily be used to directly probe covalent differences, we turned to bulk assays for these tests.
Testing Whether P4-P6 Heterogeneity Originates from Covalent Differences
P4-P6 folding has been studied extensively with bulk assays (27–36, 39, 40). Among the most widely used techniques with this and other RNAs is native gel electrophoresis, which allows separation of molecules based on conformational differences. Native gels have been used to monitor the thermodynamic effects of P4-P6 mutations by measuring the relative mobility of a P4-P6 mutant compared with the wild type (28–31). In a simple extension of this approach, we used native gels to monitor for multiple distinct species in P4-P6 (32–34).
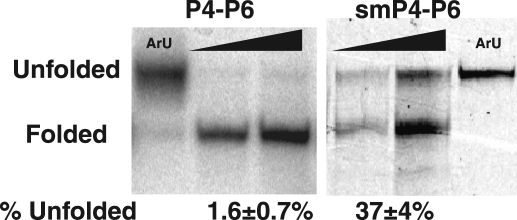
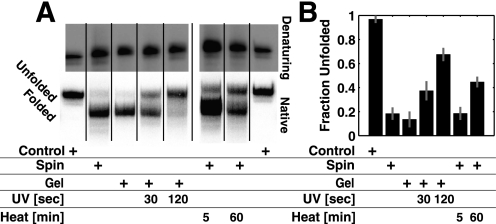
Freshly transcribed P4-P6 runs predominantly as a single band on a 1 mm Mg2+ native gel, but a small amount of a slower migrating band is also present (Fig. 4). This slower band migrates the same as an unfolded mutant control (Fig. 4, ArU). The mutant has one of the tertiary contacts ablated, resulting in a [Mg2+]1/2 of 18 mm (37), 10-fold higher than WT P4-P6 (29, 30, 37) and thus is in a predominantly unfolded state in 1 mm Mg2+. In contrast to the small amount of transcribed P4-P6 that runs with the unfolded control, a sample of purified smP4-P6 gave more than one-third of unfolded material. Side-by-side comparisons of P4-P6 in bulk (determined using hydroxyl radical footprinting) and smP4-P6 revealed no significant difference in overall folding stability (37). Nevertheless, the differences observed in native gel assays introduce the possibility that damaged molecules within the smP4-P6 population might be responsible for the heterogeneity observed in single molecule experiments.
FIGURE 4.
P4-P6 migrates as two bands on a native gel (1 mm Mg2+). ArU is an unfolded control (see text). P4-P6 gel, RNA freshly transcribed with [α-32P]ATP visualized using autoradiography; smP4-P6 gel, RNA visualized using fluorescence from the incorporated cy3 dye. The amount of P4-P6 loaded per lane was either 1× or 3×.
To further test the origin of the two populations observed by native gel we first determined whether these populations were exchangeable. Exchange was not observed in experiments using native gel assays (supplemental Fig. S2), consistent with covalent differences between the two populations. In the following experiments, to directly test for covalent differences, we built upon the observation that freshly prepared and unpurified P4-P6 behaved as a nearly homogenous population, whereas smP4-P6, which had been subject to several purification and assembly steps, exhibited a large amount of unfolded material.
P4-P6 was transcribed with α-32P-labeled XTP (where X = A, G, C, or U) and then separated from low molecular weight material by spin column treatment or purified by denaturing PAGE or denaturing PAGE with UV shadowing. UV shadowing was used to simulate purification conditions for non-radiolabeled RNA (47–49). We also treated a sample of spin column purified RNA at 95 °C, as heat treatments were used in our sample preparation and are commonly used in RNA studies (45, 46, 49).
Each sample had the same mobility on a denaturing polyacrylamide gel (Fig. 5A, top panel). This assay verified that the P4-P6 RNA was of uniform length and suggested that if covalent differences are present, they do not affect the RNA migration under denaturing conditions. In contrast, when these samples were run in parallel on a native gel, differences in electrophoretic mobility arising from the purification treatments became apparent (Fig. 5A, bottom panel). The amount of unfolded P4-P6 increased in a dose-dependent manner with UV and heat treatment, as quantified for a series of experiments in Fig. 5B.
FIGURE 5.
UV and heat treatment change P4-P6 RNA. A, 1 mm Mg2+ native gel of [α-32P]UTP-labeled P4-P6 prepared with different purifications and UV and heat treatments. The control lanes have unfolded ArU RNA (see text). B, quantification of experiments carried out as in A. Experiments were carried out in duplicate for each of the four nucleoside triphosphates. This set of experiments gave an average of 15–20% unfolded P4-P6 in the transcribed and spin-column-treated material. In additional independent experiments (Fig. 4 and data not shown), this amount varied from 1–20%. Most experiments gave 1–3% unfolded material, and the origin of this variability was not identified. Data from eight gels were combined to obtain the average results in B. See supplemental Fig. S4 for the non-averaged results.
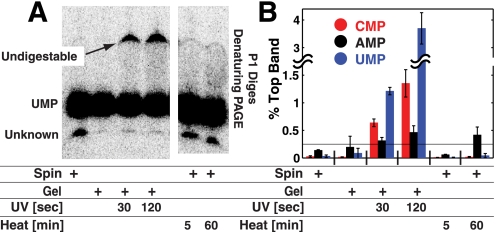
These results indicated that UV and heat treatment alter P4-P6 RNA. To more directly assay covalent damage, P4-P6 RNA subjected to the different treatments was digested to completion with P1 nuclease, and the digested samples were separated using denaturing PAGE. P1 nuclease is a single-stranded endonuclease that produces 5′-nucleotide monophosphate products (54). Fig. 6A shows the result of P1 digestions of P4-P6 made by transcription with α-32P-labeled UTP. All samples gave an identical major band, which was assigned as [α-32P]UMP. The UV-treated samples showed a slower-running band, which increased with increased UV treatment and was insensitive to increased nuclease P1 concentration and digestion time (supplemental Fig. S3). This slower-running band was absent in the heat-treated samples, suggesting that the heat-induced modifications responsible for preventing P4-P6 folding do not prevent P1 digestion. Reverse-phase HPLC following P1 nuclease and shrimp alkaline phosphatase treatment of a heat-treated 22-nucleotide RNA (data not shown) revealed additional peaks not present in control samples, suggesting heat-induced damage to nucleotide bases.
FIGURE 6.
Testing for covalent damage in P4-P6 RNA. A, [α-32P]UTP-labeled P4-P6 prepared with different purifications and UV and heat treatments were digested using nuclease P1 to nucleoside monophosphates. For UV-treated samples, indigestible RNA appears in significant quantities, indicating the presence of covalently damaged bases. B, quantification of experiments carried out as in A. Each experiment was carried out in duplicate for each of the four labeled NTPs. The percent of total RNA running as the top (indigestible) band is plotted. Significant indigestible RNA was not observed for GTP-labeled samples. The horizontal line at 0.25% represents background error, which can arise because of background subtraction.
The results of Figs. 5 and 6 indicate that UV and heat treatment can damage RNA and affect its folding behavior. Although the 30-s UV exposure and 5-min heat treatment are longer than needed or as used in some protocols, the amount of damage observed herein was extensive. For example, 30-s UV exposure led to 1% indigestible residues when assayed just with labeled uridine. Thus, essentially every P4-P6 RNA, which is 160 nucleotides in length, would be covalently modified, and many would contain multiple modifications.
It is not surprising that UV light could produce indigestible RNA products, as UV cross-linking is a commonly used structure mapping technique, and UV-induced DNA damage is well known. Nevertheless, some standard laboratory protocols involve UV light in the preparation of RNA and could be affecting the experimental results. In addition, heat treatment is typically used to renature RNAs but, as shown herein, can lead to RNA damage even in the absence of strand cleavage events. Although it is tempting to speculate on the chemical identity of this damage, the evidence points to multiple and nonspecific sources, arising from the UV and heat treatments, none of which are intrinsic to P4-P6 RNA and hence did not warrant further study. Rather, as our initial preparations of smP4-P6 included UV shadowing and heat annealing steps, we turned to alternative protocols to remove these problems.
smFRET with smP4-P6 Prepared via a New Purification Procedure
We prepared RNA pieces for smP4-P6 assembly without UV shadowing and then carried out ligations following a 1-min heat-annealing step. Shortening this annealing time from the 5-min step used in previous protocols reduced the yield but gave enough ligated material for the experiments herein. After purification of the ligated material by denaturing gel electrophoresis, the full-length RNA was further purified by native gel. On a 1 mm Mg2+ native gel, 20–40% of the RNA comigrated with the unfolded control, a higher percentage than that observed directly following transcription of P4-P6 (supplemental Fig. S5). Thus, this unfolded RNA likely resulted because of the ligation procedure or the 1-min heat-annealing step. To separately assess the folded and unfolded smP4-P6 populations, we excised the bands from a native gel, eluted the RNA, and used these purified fractions in subsequent smFRET experiments. This purification method does not guarantee a covalently pure population of smP4-P6. A single band on a native gel could still be composed of multiple covalently distinct species.
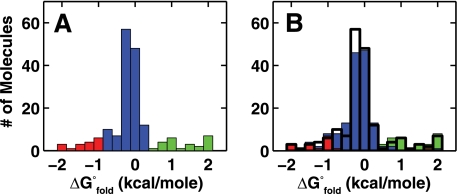
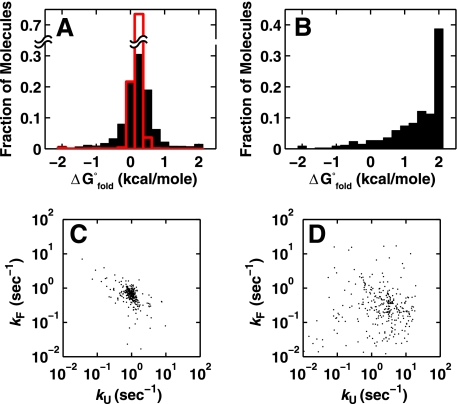
We refer to these purified populations as “smP4-P6”” and “damaged smP4-P6” below. Individual smP4-P6 molecules gave a narrow distribution of folding behaviors, as judged by their folding free energies (Fig. 7A) and folding kinetics (C). In contrast, damaged smP4-P6 molecules exhibited less favorable folding and much broader equilibrium and kinetic behaviors (Fig. 7, B and D). Thus, purification of smP4-P6 eliminates many molecules with disparate folding behaviors.
FIGURE 7.
Thermodynamic and kinetic analysis of individual smP4-P6 molecules prepared without UV shadowing and purified by native gel. A, histogram of the free energies of folding for smP4-P6 in 2.5 mm Mg2+ (n = 1953). The predicted width of the distribution because of the inherent noise in the measurement is shown in red. B, same experiment as in A but with damaged smP4-P6 (n = 338). Shown are a scatter plot of the folding and unfolding rates of smP4-P6 (C) and damaged smP4-P6 (D). For C and D, 300 randomly selected data points were plotted. All data for C are shown in supplemental Fig. S11.
Although purified smP4-P6 molecules were highly homogeneous, a fraction of molecules behaved as outliers: 20% had folding free energies differing by > 0.5 kcal/mol from the mean folding free energy and folding or unfolding rate constants differing by > 2-fold from their respective means (< 10% had both rate constants beyond the 2-fold range) (Figs. 7, A and C). These distributions are considerably narrower than those for unpurified ligated material (e.g. supplemental Fig. S6) but are still broader than predicted from simulations using estimates of the experimental noise (see Fig. 7A, red bars, and supplemental Fig. S8).
The extensive heterogeneity observed in previous RNA folding studies and prior to purification of P4-P6 implied complex kinetic behavior. The inability to observe the exchange prevented the development of unifying kinetic models that could account for this behavior or be used as the starting points for further in-depth investigation. The much simpler behavior of our purified P4-P6, with the majority of folding rate and equilibrium constants of individual molecules falling within a 2-fold range, now allows such fundamental studies to commence.
The Folding Properties of P4-P6 RNA
Simple versus Complex Folding Behavior for Structured RNAs
Several single molecule studies with RNAs that form a tertiary structure have revealed heterogeneous behavior of individual molecules, consistent with rugged and complex landscapes for folding (19–26). Our results with smP4-P6 RNA indicate that much of its observed molecular heterogeneity arises from covalent modifications introduced by standard preparative protocols. The use of alternative protocols and native gel purification revealed that smP4-P6 behaves in a highly uniform fashion. There remains a modest amount of heterogeneity, which could arise from unresolved covalent modifications, from an underestimate of the intrinsic uncertainty in our measurements and/or from a limited degree of ruggedness of the folding landscape of this RNA.
We showed previously that the full-length Tetrahymena group I ribozyme, which includes the P4-P6 independently folding domain studied herein, does indeed exhibit a rugged landscape, with folding occurring via parallel pathways and with the formation of multiple native conformers that do not exchange over many minutes and require unfolding to efficiently equilibrate (12, 14, 55). The simple folding behavior of smP4-P6 suggests that the ruggedness of the Tetrahymena group I ribozyme folding landscape arises when the components of the molecule are put together and is not an inherent property of each structural unit of the more complex molecule.
Analysis of P4-P6 Folding Kinetics and Thermodynamics
Folded P4-P6 has two tertiary interactions: the metal core/metal core receptor (MC/MCR) interaction and tetraloop/tetraloop receptor (TL/TLR) interaction (Fig. 1, A and B). Both tertiary interactions can form in Mg2+, but as Ba2+ cannot occupy the metal ion binding sites in the metal core, only the TL/TLR interaction forms in Ba2+ (35). The TL/TLR interaction can form even in the absence of divalent metal ions, provided that sufficiently high concentrations of monovalent cations are present (39). To explore the effect of different molecular features on folding and to begin to develop folding models for P4-P6, we quantitatively compared the folding of smP4-P6 as a function of Mg2+, Ba2+, and Na+.
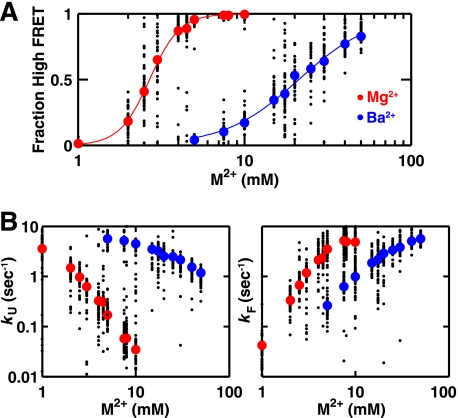
Fig. 8 shows the dependence of the folding equilibrium and folding and unfolding rate constants on the concentration of Mg2+ and Ba2+. The midpoint for folding in Ba2+ is 10-fold higher than that for folding in Mg2+, consistent with an additional stabilizing contribution from formation of the MC/MCR interaction in Mg2+ but not Ba2+. A difference in free energy for folding in Mg2+ versus Ba2+ of 3.6–4.1 kcal/mol can be directly determined by comparing the folding equilibrium at 5–10 mm metal ion (supplemental Fig. S10). This free energy value is almost the same as the metal ion core contribution of 3.2 ± 0.2 kcal/mol determined previously from mutant cycle analysis in Mg2+ (37) and indicates that folding stability is the same or nearly the same in Mg2+ and Ba2+ if the Mg2+-dependent metal ion core and its tertiary interaction are prevented from forming (35). This result implies that there are no other Mg2+ sites that significantly contribute to the folding stability.
FIGURE 8.
Comparison of smP4-P6 folding thermodynamics and kinetics in Mg2+ and Ba2+. Folding thermodynamics (A) and kinetics (B) of smP4-P6 as a function of Mg2+ (red) and Ba2+ (blue). All buffers contained 100 mm Na+. Large circles represent the median of all molecules analyzed at the indicated ion concentration, and smaller black circles represent 50 randomly chosen molecules. (See supplemental information for all single molecule data.) Lines in A are empirical Hill fits to the median data points, giving nHill = 4.1 for Mg2+ and 1.9 for Ba2+.
The above comparisons require the high sensitivity of single molecule data to allow an accurate folding equilibrium and free energies to be determined when nearly all of the RNA is either folded or unfolded (Fig. 8A). In the absence of single molecule data, most free energy comparisons for RNA folding involve extrapolations to common metal ion concentrations using a Hill model for metal ion binding, an approach that is problematic because of the non-two-state nature of ion atmosphere effects and can give incorrect thermodynamic values (40, 56).
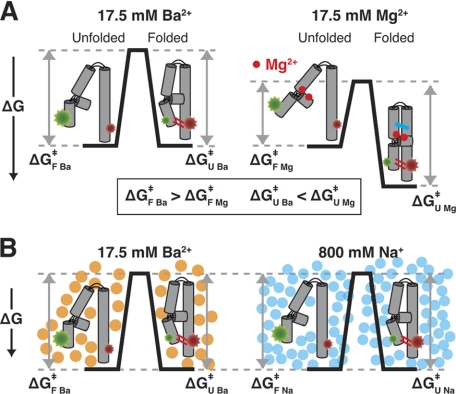
The slower unfolding rate constant for a given metal ion concentration in Mg2+ relative to Ba2+ (Fig. 8B, kU) suggests that the MC/MCR tertiary interaction fully or partially breaks prior to the rate-limiting transition state for unfolding (Fig. 9A, ΔGU Mg‡ > ΔGU Ba‡. In the model of Fig. 9A, the bound Mg2+ ions are depicted by the red dots, and the MC/MCR tertiary interaction that can form in Mg2+ but not Ba2+ is depicted by the blue lines. These interactions would be broken prior to the unfolding transition state, according to the model.
FIGURE 9.
Models for effects of Mg2+, Ba2+, and Na+ on the folding of P4-P6 RNA. A, Mg2+ (red dots) but not Ba2+ can bind to the metal ion core (35), and this binding can occur prior to tertiary folding (44). According to this model, Mg2+ binding lowers the barrier for folding (ΔGF Ba‡ > ΔGF Mg‡) by increasing the probability of formation of the tetraloop/tetraloop receptor interaction (red lines in folded state) via a conformational change or restriction of available conformations in the unfolded state. In addition, unfolding is slowed in Mg2+ (ΔGU Ba‡ < ΔGU Mg‡) because an additional tertiary interaction is formed (blue lines in folded state) that must be broken prior to the transition state in the unfolding process. The free energy profiles are shown for 17.5 mm metal ion, as this is near the folding midpoint in Ba2+. B, near the folding midpoint in Ba2+ (orange circles) and Na+ (blue circles) (i.e. the fraction folded was 0.39 and 0.45 in 17.5 mm Ba2+ and 800 mm Na+ respectively), the rate constants for folding and unfolding are nearly identical (ΔGF Ba‡ = ΔGF Na‡ and ΔGU Ba‡ = ΔGU Na‡), consistent with a simple model of ion effects via electrostatic screening.
The folding is faster in Mg2+ relative to Ba2+ (Fig. 8B, kF), suggesting that Mg2+ also affects the unfolded state. We propose that Mg2+ binds to the metal core in unfolded P4-P6 (Fig. 9A, red dots) and alters the conformation of the P5abc subdomain of P4-P6 so that formation of the TL/TLR interaction is more probable, as depicted in the free energy profile of Fig. 9A (ΔGF Mg‡ < ΔGF Ba‡). This model is consistent with data suggesting that Mg2+ ions bind the P5abc subdomain of P4-P6 at low concentrations and in the absence of tertiary structure formation (33, 44).
As Ba2+ does not bind to the metal core, the simplest model for its effects on folding invokes charge screening via its increasing presence in the ion atmosphere surrounding unfolded and folded P4-P6 as the Ba2+ concentration increases. To provide a first test of this model, we turned to monovalent cation-dependent folding, as previous work has shown that P4-P6 can fold via formation of the TL/TLR tertiary interaction in Na+ in the absence of any divalent metal ions (39). Much more Na+ than Ba2+ is required to achieve the folded state, with a [Na+]1/2 value of ∼700 mm (39). Although ion atmosphere theoretical treatments are not accurate enough to quantitatively predict the differences between the screening effects of monovalent and divalent cations, divalent cations are expected to be much more efficient in charge screening, and numerous empirical studies support this expectation (39, 57–59).
The simplest prediction from the ion atmosphere model is as follows. If charge screening dominates the effects of Ba2+ and Na+, then charge screening would be the same in Ba2+ and Na+ at their respective folding midpoints, and the folding kinetics would also be the same. We therefore determined smP4-P6 folding kinetics at the Na+ midpoint. At 800 mm Na+, which gave 0.45 folded smP4-P6, values of kF and kU of 2.9 and 3.8 s−1 were obtained (supplemental Fig. S13). These values are nearly the same as the values of kF = 2.2 s−1 and kU = 3.0 s−1 at 17.5 mm Ba2+, which gives a similar fraction folded of 0.39 (Fig. 8, A and B), supporting the view that simple charge screening effects dominate both the kinetic and thermodynamic behavior of P4-P6 in Ba2+ and Na+. Thus, at the folding midpoint in Ba2+ and Na+, the barriers for folding and unfolding are virtually the same, as depicted in the free energy profile of Fig. 9B (ΔGF Ba‡ = ΔGF Na‡ and ΔGU Ba‡ = ΔGU Na‡). These observations also suggest that there are no significant contributions to the kinetic barriers for folding and unfolding from the larger number of monovalent than divalent cations that must reposition themselves as the RNA compacts to its folded state or expands to its unfolded state.
Overall, the P4-P6 kinetic and thermodynamic constants obtained by smFRET measurements agree well with those obtained by multiple bulk approaches, including hydroxyl radical footprinting, stopped-flow fluorescence with a pyrene label, and small angle x-ray scattering (41–44). Nevertheless, although some of these approaches provide additional structural information, none can match smFRET for the range of conditions that can be investigated, and the ease at which reversible folding can be followed.
Further Implications
In the course of investigating the origins of complex folding behavior of P4-P6 RNA revealed by single molecule experiments, we discovered that standard RNA preparative protocols damage this RNA. Although there is extensive precedent for UV-induced damage, popular protocols nevertheless advocate visualization by UV, typically without caution. Further, heat steps, whereas recognized as potentially leading to strand scission, have been nearly universally employed to renature RNAs; thus, our finding that heat treatment can lead to damage in the absence of cleavage that is undetectable by denaturing gel electrophoresis suggests that additional cautions and controls will be needed.
Moreover, this damage results in heterogeneous behavior at the level of single molecules. Thus, the folding of P4-P6 is simpler than it at first appeared, and is in contrast with the complex kinetics and multiple long-lived alternative states for the full-length Tetrahymena group I ribozyme (14, 55).
Evidence for kinetic traps in group I intron folding included slower folding at higher concentrations of Mg2+, a result attributable to stabilization of a long-lived kinetic trap (12, 13, 15, 16). In contrast, smP4-P6 folding rates increase as Mg2+ or Ba2+ concentrations increase, giving no indication of long-lived kinetic traps and such complex folding behavior (Fig. 8B).
Although our smP4-P6 data provide no indication of extensive ruggedness and kinetic traps, the fastest observed rate constants for P4-P6 folding are on the order of 10 s−1 (Fig. 8B), much slower than formation of basic structures such as RNA hairpins, which form on the microsecond timescale (60, 61). Understanding the nature of the barriers for RNA folding and the transition states that define these barriers remain important challenges to be met by combining smFRET with other experimental and computational approaches.
Supplementary Material
Acknowledgments
We thank H. Mabuchi (Department of Applied Physics, Stanford University) for technical support and D. Pavlichin for helping to create data analysis software. We also thank R. Das for helpful discussions and the Herschlag laboratory for critical advice throughout this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM49243 (to D. H.) and Training Grant in Biotechnology 5T32GM008412 (to M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S13, Table S1, and experimental procedures.
- smFRET
- single molecule fluorescence resonance energy transfer
- P4–P6
- P4–P6 domain of the Tetrahymena group I ribozyme
- smP4–P6
- single molecule P4–P6
- MC/MCR
- metal core/metal core receptor
- TL/TLR
- tetraloop/tetraloop receptor.
REFERENCES
- 1. Noller H. F. (2005) Science 309, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 2. Al-Hashimi H. M., Walter N. G. (2008) Curr. Opin. Chem. Biol. 18, 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Birney E., Stamatoyannopoulos J. A., Dutta A., Guigó R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., Kuehn M. S., Taylor C. M., Neph S., Koch C. M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J. A., Andrews R. M., Flicek P., Boyle P. J., Cao H., Carter N. P., Clelland G. K., Davis S., Day N., Dhami P., Dillon S. C., Dorschner M. O., Fiegler H., Giresi P. G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K. D., Johnson B. E., Johnson E. M., Frum T. T., Rosenzweig E. R., Karnani N., Lee K., Lefebvre G. C., Navas P. A., Neri F., Parker S. C., Sabo P. J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F. S., Dekker J., Lieb J. D., Tullius T. D., Crawford G. E., Sunyaev S., Noble W. S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I. L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H. A., Sekinger E. A., Lagarde J., Abril J. F., Shahab A., Flamm C., Fried C., Hackermüller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J. S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M. C., Thomas D. J., Weirauch M. T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K. G., Sung W. K., Ooi H. S., Chiu K. P., Foissac S., Alioto T., Brent M., Pachter L., Tress M. L., Valencia A., Choo S. W., Choo C. Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T. G., Brown J. B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C. N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J. S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R. M., Rogers J., Stadler P. F., Lowe T. M., Wei C. L., Ruan Y., Struhl K., Gerstein M., Antonarakis S. E., Fu Y., Green E. D., Karaöz U., Siepel A., Taylor J., Liefer L. A., Wetterstrand K. A., Good P. J., Feingold E. A., Guyer M. S., Cooper G. M., Asimenos G., Dewey C. N., Hou M., Nikolaev S., Montoya-Burgos J. I., Löytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N. R., Holmes I., Mullikin J. C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W. J., Stone E. A., Batzoglou S., Goldman N., Hardison R. C., Haussler D., Miller W., Sidow A., Trinklein N. D., Zhang Z. D., Barrera L., Stuart R., King D. C., Ameur A., Enroth S., Bieda M. C., Kim J., Bhinge A. A., Jiang N., Liu J., Yao F., Vega V. B., Lee C. W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M. J., Inman D., Singer M. A., Richmond T. A., Munn K. J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J. C., Couttet P., Bruce A. W., Dovey O. M., Ellis P. D., Langford C. F., Nix D. A., Euskirchen G., Hartman S., Urban A. E., Kraus P., Van Calcar S., Heintzman N., Kim T. H., Wang K., Qu C., Hon G., Luna R., Glass C. K., Rosenfeld M. G., Aldred S. F., Cooper S. J., Halees A., Lin J. M., Shulha H. P., Zhang X., Xu M., Haidar J. N., Yu Y., Ruan Y., Iyer V. R., Green R. D., Wadelius C., Farnham P. J., Ren B., Harte R. A., Hinrichs A. S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A. S., Smith K., Thakkapallayil A., Barber G., Kuhn R. M., Karolchik D., Armengol L., Bird C. P., de Bakker P. I., Kern A. D., Lopez-Bigas N., Martin J. D., Stranger B. E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrímsdóttir I. B., Huppert J., Zody M. C., Abecasis G. R., Estivill X., Bouffard G. G., Guan X., Hansen N. F., Idol J. R., Maduro V. V., Maskeri B., McDowell J. C., Park M., Thomas P. J., Young A. C., Blakesley R. W., Muzny D. M., Sodergren E., Wheeler D. A., Worley K. C., Jiang H., Weinstock G. M., Gibbs R. A., Graves T., Fulton R., Mardis E. R., Wilson R. K., Clamp M., Cuff J., Gnerre S., Jaffe D. B., Chang J. L., Lindblad-Toh K., Lander E. S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P. J., Consortium E. P. (2007) Nature 447, 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tinoco I., Bustamante C. (1999) J. Mol. Biol. 293, 271–281 [DOI] [PubMed] [Google Scholar]
- 5. Brion P., Westhof E. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 113–137 [DOI] [PubMed] [Google Scholar]
- 6. Treiber D. K., Williamson J. R. (1999) Curr. Opin. Struct. Biol. 9, 339–345 [DOI] [PubMed] [Google Scholar]
- 7. Woodson S. A. (2000) Cell Mol. Life Sci. 57, 796–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu V. B., Lipfert J., Bai Y., Pande V. S., Doniach S., Herschlag D. (2009) RNA 15, 2195–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al-Hashimi H. M., Pitt S. W., Majumdar A., Xu W., Patel D. J. (2003) J. Mol. Biol. 329, 867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan T., Sosnick T. R. (1997) Nat. Struct. Biol. 4, 931–938 [DOI] [PubMed] [Google Scholar]
- 11. Elenko M. P., Szostak J. W., van Oijen A. M. (2009) J. Am. Chem. Soc. 131, 9866–9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell R., Herschlag D. (2001) J. Mol. Biol. 308, 839–851 [DOI] [PubMed] [Google Scholar]
- 13. Zarrinkar P. P., Williamson J. R. (1994) Science 265, 918–924 [DOI] [PubMed] [Google Scholar]
- 14. Solomatin S. V., Greenfeld M., Chu S., Herschlag D. (2010) Nature 463, 681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan J., Woodson S. A. (1998) J. Mol. Biol. 280, 597–609 [DOI] [PubMed] [Google Scholar]
- 16. Sclavi B., Sullivan M., Chance M. R., Brenowitz M., Woodson S. A. (1998) Science 279, 1940–1943 [DOI] [PubMed] [Google Scholar]
- 17. Lindahl T., Adams A., Fresco J. R. (1966) Proc. Natl. Acad. Sci. U.S.A. 55, 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gartland W. J., Sueoka N. (1966) Proc. Natl. Acad. Sci. U.S.A. 55, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhuang X., Kim H., Pereira M. J., Babcock H. P., Walter N. G., Chu S. (2002) Science 296, 1473–1476 [DOI] [PubMed] [Google Scholar]
- 20. Steiner M., Karunatilaka K. S., Sigel R. K., Rueda D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemay J. F., Penedo J. C., Tremblay R., Lilley D. M., Lafontaine D. A. (2006) Chem. Biol. 13, 857–868 [DOI] [PubMed] [Google Scholar]
- 22. Rueda D., Bokinsky G., Rhodes M. M., Rust M. J., Zhuang X., Walter N. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10066–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie Z., Srividya N., Sosnick T. R., Pan T., Scherer N. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan E., Wilson T. J., Nahas M. K., Clegg R. M., Lilley D. M., Ha T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9308–9313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kobitski A. Y., Nierth A., Helm M., Jäschke A., Nienhaus G. U. (2007) Nucleic Acids Res. 35, 2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ditzler M. A., Rueda D., Mo J., Håkansson K., Walter N. G. (2008) Nucleic Acids Res. 36, 7088–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murphy F. L., Cech T. R. (1993) Biochemistry 32, 5291–5300 [DOI] [PubMed] [Google Scholar]
- 28. Szewczak A. A., Cech T. R. (1997) RNA 3, 838–849 [PMC free article] [PubMed] [Google Scholar]
- 29. Silverman S. K., Cech T. R. (1999) Biochemistry 38, 8691–8702 [DOI] [PubMed] [Google Scholar]
- 30. Battle D. J., Doudna J. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Young B. T., Silverman S. K. (2002) Biochemistry 41, 12271–12276 [DOI] [PubMed] [Google Scholar]
- 32. Juneau K., Cech T. R. (1999) RNA 5, 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cate J. H., Hanna R. L., Doudna J. A. (1997) Nat. Struct. Biol. 4, 553–558 [DOI] [PubMed] [Google Scholar]
- 34. Basu S., Rambo R. P., Strauss-Soukup J., Cate J. H., Ferre-D'Amare A. R., Strobel S. A., Doudna J. A. (1998) Nat. Struct. Biol. 5, 986–992 [DOI] [PubMed] [Google Scholar]
- 35. Travers K. J., Boyd N., Herschlag D. (2007) RNA 13, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cate J. H., Gooding A. R., Podell E., Zhou K., Golden B. L., Kundrot C. E., Cech T. R., Doudna J. A. (1996) Science 273, 1678–1685 [DOI] [PubMed] [Google Scholar]
- 37. Sattin B. D., Zhao W., Travers K., Chu S., Herschlag D. (2008) J. Am. Chem. Soc. 130, 6085–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee T. H., Lapidus L. J., Zhao W., Travers K. J., Herschlag D., Chu S. (2007) Biophys. J. 92, 3275–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takamoto K., Das R., He Q., Doniach S., Brenowitz M., Herschlag D., Chance M. R. (2004) J. Mol. Biol. 343, 1195–1206 [DOI] [PubMed] [Google Scholar]
- 40. Das R., Travers K. J., Bai Y., Herschlag D. (2005) J. Am. Chem. Soc. 127, 8272–8273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlatterer J. C., Kwok L. W., Lamb J. S., Park H. Y., Andresen K., Brenowitz M., Pollack L. (2008) J. Mol. Biol. 379, 859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silverman S. K., Cech T. R. (2001) RNA 7, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silverman S. K., Deras M. L., Woodson S. A., Scaringe S. A., Cech T. R. (2000) Biochemistry 39, 12465–12475 [DOI] [PubMed] [Google Scholar]
- 44. Deras M. L., Brenowitz M., Ralston C. Y., Chance M. R., Woodson S. A. (2000) Biochemistry 39, 10975–10985 [DOI] [PubMed] [Google Scholar]
- 45. Akiyama B. M., Stone M. D. (2009) Methods Enzymol. 469, 27–46 [DOI] [PubMed] [Google Scholar]
- 46. Solomatin S., Herschlag D. (2009) Methods Enzymol. 469, 47–68 [DOI] [PubMed] [Google Scholar]
- 47. Andrus A., Kuimelis R. G. (2001) Curr. Protoc. Nucleic Acid Chem. Unit 10.14 [DOI] [PubMed] [Google Scholar]
- 48. Robert F. E. (2009) RNA Methodologies: Laboratory Guide for Isolation and Characterization, 4th ed., Elsevier, Amsterdam [Google Scholar]
- 49. Frilander M. J., Turunen J. J. (2005) RNA Ligation Using T4 DNA Ligase, Wiley-VCH, Hoboken, New Jersey [Google Scholar]
- 50. Rasnik I., McKinney S. A., Ha T. (2006) Nat. Meth. 3, 891–893 [DOI] [PubMed] [Google Scholar]
- 51. McKinney S. A., Joo C., Ha T. (2006) Biophys. J. 91, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cappé O., Moulines E., Rydén T. (2005) Inference in Hidden Markov Models, Springer Science and Business Media, New York [Google Scholar]
- 53. Welch L. R. (2003) IEEE Information Theory Society Newsletter 53 [Google Scholar]
- 54. Volbeda A., Lahm A., Sakiyama F., Suck D. (1991) EMBO J. 10, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Russell R., Zhuang X., Babcock H. P., Millett I. S., Doniach S., Chu S., Herschlag D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Draper D. E. (2004) RNA 10, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bai Y., Greenfeld M., Travers K. J., Chu V. B., Lipfert J., Doniach S., Herschlag D. (2007) J. Am. Chem. Soc. 129, 14981–14988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lambert D., Leipply D., Shiman R., Draper D. E. (2009) J. Mol. Biol. 390, 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takamoto K., He Q., Morris S., Chance M. R., Brenowitz M. (2002) Nat. Struct. Biol. 9, 928–933 [DOI] [PubMed] [Google Scholar]
- 60. Van Orden A. V., Jung J. (2008) Biopolymers 89, 1–16 [DOI] [PubMed] [Google Scholar]
- 61. Ma H., Proctor D. J., Kierzek E., Kierzek R., Bevilacqua P. C., Gruebele M. (2006) J. Am. Chem. Soc. 128, 1523–1530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.