Abstract
A large body of evidence supports the involvement of heparan sulfate (HS) proteoglycans in physiological processes such as development and diseases including cancer and neurodegenerative disorders. The role of HS emerges from its ability to interact and regulate the activity of a vast number of extracellular proteins including growth factors and extracellular matrix components. A global view on how protein-HS interactions influence the extracellular proteome and, consequently, cell function is currently lacking. Here, we systematically investigate the functional and structural properties that characterize HS-interacting proteins and the network they form. We collected 435 human proteins interacting with HS or the structurally related heparin by integrating literature-derived and affinity proteomics data. We used this data set to identify the topological features that distinguish the heparin/HS-interacting network from the rest of the extracellular proteome and to analyze the enrichment of gene ontology terms, pathways, and domain families in heparin/HS-binding proteins. Our analysis revealed that heparin/HS-binding proteins form a highly interconnected network, which is functionally linked to physiological and pathological processes that are characteristic of higher organisms. Therefore, we then investigated the existence of a correlation between the expansion of domain families characteristic of the heparin/HS interactome and the increase in biological complexity in the metazoan lineage. A strong positive correlation between the expansion of the heparin/HS interactome and biosynthetic machinery and organism complexity emerged. The evolutionary role of HS was reinforced by the presence of a rudimentary HS biosynthetic machinery in a unicellular organism at the root of the metazoan lineage.
Keywords: Cell Surface Protein, Evolution, Extracellular Matrix, Extracellular Matrix Proteins, Glycosaminoglycan, Heparan Sulfate, Heparin, Heparin-binding Protein, Protein-Protein Interactions, Network
Introduction
A major challenge for the postgenomics era is to establish functional and structural relationships between the components of biological systems. In the last decade, the development of high throughput methods for the study of genetic interactions (1) and protein-protein interactions (2–4) has enabled the collection of large data sets describing binary relationships between primary gene products. The accumulation of these large data sets required innovative ways to represent and analyze molecular networks, thus stimulating the development of a new discipline known as network biology (5–8). This new approach has been successfully used to integrate data from different experimental platforms (9), infer properties of interaction networks by applying statistical theories (6), assign protein function (10), identify network signatures characteristic of diseases such as cancer (8, 10), and investigate the evolution of interaction networks (11, 12). However, the chemical complexity of secondary gene products such as glycans and lipids and the technical challenges associated with the study of their interactions have generated a gap in our current models of interaction networks, and as a consequence, the interactions of proteins with secondary gene products such as glycosaminoglycans (GAGs)4 have been excluded from the above systematic analyses.
The GAGs are linear polysaccharides whose synthesis is not template-driven. As the most complex of biological polymers, they provide access to a vast chemical information space. This has been exploited in eumetazoans to provide structural frameworks and active mediation of cell-cell communication, both absolute requirements for multicellularity. The sulfated GAGs such as heparin/heparan sulfate (HS) are synthesized and serine-linked to the core proteins of proteoglycans (HSPGs) and are located on the plasma membrane and in the extracellular matrix (ECM). The chemical complexity of heparin/HS arises from the initially synthesized monotonous polymer being extensively modified (epimerization and sulfation at various positions in the sugar rings). These modifications are substoichiometric and grouped to produce characteristic domains, which vary in size and number within each chain (13, 14). Analysis of functional structures in vivo demonstrates that there is specific regulation of the structures of heparin/HS that are expressed at the cellular level (15–18), and thus, it seems that biology exploits a substantial amount of the chemical information space of heparin/HS. The functions of heparin/HS are exerted through their capacity to engage protein ligands. The consequences of these interactions range from elaborating large scale structures to regulating the gradient formation and signaling activities of growth factors, cytokines, and morphogens and the localization and activity of extracellular enzymes (Refs. 16, 19, and 20; for a review, see Ref. 17). The scope of these functions is evidenced by the size of the human heparin/HS interactome: 216 proteins in a review published in 2008 (17). Many pathogens express proteins that interact with heparin/HS as part of their molecular adaptation to infection of mammals (21). Thus, HSPGs are key players in molecular networks driving biological phenomena such as development (22), inflammation and immune response (23, 24), and disease (21, 25).
The first aim of this study was to integrate and rationalize available data on heparin/HS-protein interactions. The current coverage of heparin/HS-protein interactions in public databases is largely incomplete (Table 1). Therefore, a literature mining effort (17) was combined with an affinity proteomics approach for the identification of heparin/HS-binding proteins (HBPs) (supplemental Results and supplemental Figs. 1 and 2), and data were retrieved from public databases to generate a comprehensive list of the interactions between heparin/HS and proteins described so far. The term “HBP” is used because heparin is commonly used as an experimental proxy for the sulfated domains of HS, and many interactions have not been validated with HS. This data set then enabled a new systematic way of analyzing heparin/HS-protein interactions using tools widely applied in genomics and proteomics studies. The system-level analysis allowed the investigation of how HBPs interact with each other by computing the topological properties of the network they form and the identification of functional and structural features that are associated with the heparin/HS binding activity. Finally, to generate insights into the role of HSPGs in the evolution of multicellular organisms, the presence of orthologs of HS biosynthetic enzymes in the genome of the choanoflagellate Monosiga brevicollis was investigated. Choanoflagellates are unicellular and colony-forming organisms found in marine and freshwater environments. They use a single apical flagellum surrounded by a collar of actin-filled microvilli to swim and capture bacterial prey (26). Because choanoflagellates are not metazoans and did not evolve from sponges or more recently derived metazoan phyla, they are indicated as the last unicellular organisms that evolved before the origin and diversification of metazoans (27). Previous works indicate the presence in the genome of M. brevicollis of protein families that were thought to be exclusive to multicellular organisms (26–28). The presence of functional signaling cascades based on tyrosine phosphorylation has been also demonstrated (28). For these reasons, the study of M. brevicollis is considered to be crucial for the identification of the molecular networks that were present in the last common ancestor of choanoflagellates and metazoans and that likely contributed to the emergence of multicellularity and the development of animals.
TABLE 1.
Current coverage of human HBPs in publicly available databases
| Source | Search criteria | Output | Ref. |
|---|---|---|---|
| GO consortium (April 2010) | GO:0008201 heparin binding | 109 human genes | 29 |
| GO consortium (April 2010) | GO:0043395 heparan sulfate binding | 4 human genes | 29 |
| MatrixDB (April 2010) | Heparin + heparan sulfate | 90 human entries | 31 |
| UniProtKB (Release 2010_04) | KW-0358 heparin binding | 66 human genes | 30 |
| Literature-based review | 216 human genes | 17 |
EXPERIMENTAL PROCEDURES
Construction of Heparin/HS Interactome
The heparin/HS interactome was built using a combination of literature curation, data retrieval from public databases, and experimental data obtained by the affinity proteomics approach described in the supplemental information. An initial version of the literature-curated data set was first published in 2008 (17) and originally included 216 HBPs. The original data set was expanded, and its current version (December 2009) includes 280 interactors. Additional data were retrieved from publicly available databases and gene ontology (GO) classifications using the search criteria listed in Table 1. For GO (29) and UniProtKB (30), the search was restricted to human genes, whereas this was not necessary for MatrixDB because all the interactions described in it are associated by default to human protein identifiers (31). Finally, the 147 HBPs identified by the heparin affinity proteomics approach described in the supplemental Results and supplemental Methods were included as a set of experimentally derived, non-literature-based data (supplemental Figs. 1–3 and 5 and supplemental Tables 1 and 7). The integration of these data sets resulted in a non-redundant list of 435 human proteins. This list, referred to as the heparin/HS interactome, was used for all the subsequent analysis presented, and it is available in supplemental Table 1.
Construction and Analysis of Heparin/HS-interacting Network
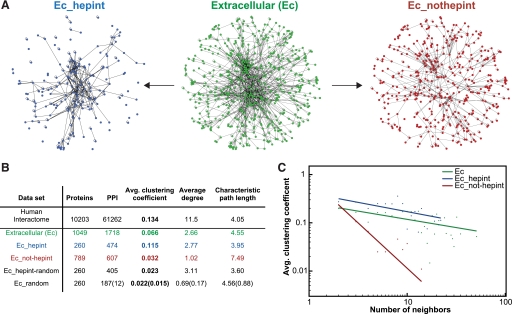
A protein-protein interaction network based on the heparin/HS interactome was built using Cytoscape v.2.6.3 (32, 33). The protein-protein interaction resource was the Cytoscape Human Interactome Dataset (2007), which was obtained by merging molecular interaction data from a variety of sources including IntAct (34), Database of Interacting Proteins (35), and Human Protein Reference Database (36). The NCBI Entrez GeneID for each HBP was obtained from their UniProt accession number using DAVID 6.7 (37) and used to extract HBPs and their interactions from the human interactome data set. Topological parameters of the heparin/HS-interacting network, treated as an undirected network, were computed using the Cytoscape plugin Network Analyzer v.2.6 (38). For a context-relevant analysis, the topological parameters of the extracellular human interactome were compared with those of the network formed by extracellular HBPs. Thus, the extracellular interactome was extracted from the human data set by applying filters based on GO cellular component terms. The terms used were GO:0005576 (extracellular region), GO:0005615 (extracellular space), GO:0031012 (ECM), and GO:0005604 (basement membrane). Extracellular HBPs and their interactions were then selected from the extracellular proteome using the NCBI Entrez GeneIDs from the heparin/HS interactome. The properties and topological parameters of the networks are summarized in Fig. 1B.
FIGURE 1.
Topological and functional analysis of heparin/HS-interacting network. A, the extracellular heparin/HS-interacting network (“Ec_hepint”; blue) was extracted from a data set of extracellular protein-protein interactions, and it was compared with the non-heparin/HS-interacting network (“Ec_not-hepint”; red) and the whole extracellular interactome (“Ec”; green) (see “Experimental Procedures”). The position of the nodes in the networks and the length of edges are arbitrary and only have a graphical purpose. The properties and topological parameters of the network analyzed are summarized in B. “Proteins” indicate the number of proteins (nodes) that form each network, and “PPI” (protein-protein interactions) indicates the number of interactions (edges) connecting them. The “Average degree” indicates the mean number of neighbors per node in the network. The “Characteristic path length” is the average over the shorter distances (number of links) separating all pairs of nodes in the network and offers a measure of the overall navigability of a network. The clustering coefficient is defined as the number of links connecting the first neighbors of a given node divided by the total possible number of connections between them. It is a measure of the tendency of nodes to form highly interconnected modules. The “Avg. clustering coefficient” for a network is calculated as the mean of the clustering coefficients for each node having a degree ≥2. The “Ec_hepint-random” network was generated from the extracellular heparin/HS-interacting network by applying a degree-preserving random shuffle of the edges (1320 shuffles). The “Ec_random” network was generated by randomly selecting a network of the same size as the extracellular heparin/HS-interacting network from the total extracellular interactome. The procedure was iterated 50 times, and mean network parameters are shown with S.D. in parentheses. The average clustering coefficient of the extracellular heparin/HS-interacting network is six standard deviations higher than the average value calculated for randomly picked networks. In C, nodes are binned according to their degree, and the average (Avg.) clustering coefficient for each bin is plotted applying the same color code used in A. The distribution of the clustering coefficients is characterized by the typical slope of protein-protein interaction networks, which indicates the presence of hierarchical modularity.
Functional and Structural Analysis of Heparin/HS Interactome
The over-representation (enrichment) of GO terms (29), Kyoto encyclopedia of genes and genomes (KEGG) pathways (39), and Pfam domain families (40) in the heparin/HS interactome was analyzed using the web-accessible program DAVID 6.7 (37). The list of UniProt accession numbers of the HBPs was used as the input list, and the default human proteome was used as the background list. The significance of the enrichments was statistically evaluated with a modified Fisher's exact test (Expression Analysis Systematic Explorer score), and a p value for each term was calculated by applying a Benjamini-Hochberg false discovery rate correction (37). Cutoff values were 0.01 for GO biological process terms enrichment and 0.05 for KEGG pathways and Pfam domains. Furthermore, Pfam domains significantly enriched in the heparin/HS interactome were associated with the corresponding structural classification of proteins (SCOP) superfamilies (41) to reduce redundancy and perform the analysis described in Fig. 4. For GO term enrichments, the GO FAT annotation available in DAVID was used. The GO FAT is a subset of the GO term set created by filtering out the broadest ontology terms to avoid overshadowing more specific terms. The enrichment of GO biological process terms was also analyzed using the Cytoscape plugin BiNGO v2.3 (42) using the complete GO term set and a hypergeometric statistical test with Benjamini-Hochberg false discovery rate correction.
FIGURE 4.
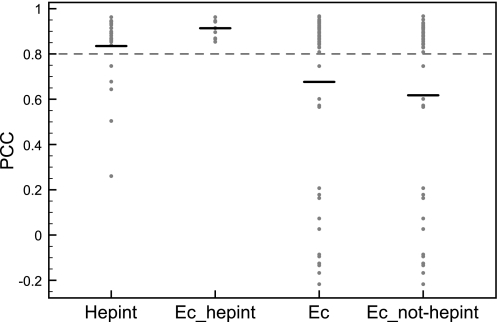
Correlation between abundance of superfamilies associated with heparin/HS interactome and organism complexity. The PCCs describing the association between superfamily abundance and organism complexity were extracted from Vogel and Chothia (50). The PCCs for superfamilies enriched in the heparin/HS interactome (“Hepint”), enriched in the heparin/HS interactome and annotated as extracellular in Vogel and Chothia (50) (“Ec_hepint”), annotated as extracellular (“Ec”), and annotated as extracellular but not enriched in the heparin/HS interactome (“Ec_not-hepint”) are plotted along the y axis. For each group, a horizontal bar indicates the mean PCC. The dashed line indicates the threshold for strong correlation between superfamily abundance and organism complexity.
Identification of HS Biosynthetic Enzymes in M. brevicollis
The orthologs of human biosynthetic enzymes responsible for synthesis of the protein-GAG linker tetrasaccharide and for the polymerization and modification of HS chains were identified using a reciprocal Blast best hit approach (43). Thus, the protein sequences of human enzymes were searched against the non-redundant protein sequences databases of M. brevicollis (taxid 81824), fungi (taxid 4751), Dictyostelium discoideum (taxid 44689), and plants (taxid 3193) using Blastp with default settings. The best hits for each group were then searched against the human non-redundant protein sequences database (taxid 9606) and selected as orthologs only in the case where the reciprocal best hit criterion was satisfied. In the cases where more than one paralog enzyme was present in the human genome, a paralog was arbitrarily chosen from the family and used for the Blast search. Only for these cases was the reciprocal best hit criterion considered satisfied if the best hit of the reciprocal Blast search was a member of the same enzyme family but not necessarily the paralog used for the first search.
RESULTS
Topological Analysis of Heparin/HS-interacting Network
The first step toward a systematic analysis of the heparin/HS interactome was to build and analyze the topological parameters of the network of protein-protein interaction formed by HBPs. This analysis is based on the representation of protein-protein interaction networks as graphs where proteins are represented as nodes connected by edges that indicate the presence of an interaction between them (Fig. 1A). By applying statistical tools used for graph theory, it is then possible to compute topological parameters describing the properties of the network and use such parameters to compare different networks (for reviews, see Refs. 6, 38, and 44) (Fig. 1, B and C). Thus, HBPs and their interactions were extracted from a data set of human protein interactions obtained by merging data retrieved from different repositories (see “Experimental Procedures”). Because the predominant location of HSPGs is extracellular, the extracellular heparin/HS-interacting network was compared with the network formed by extracellular non-heparin/HS-interacting proteins and the total extracellular interactome (Fig. 1A). These networks were extracted from the human interactome data set using filters based on GO cellular component terms associated with extracellular protein localization (see “Experimental Procedures”). The topological parameters of the networks analyzed are summarized in Fig. 1B. By comparing the properties of these three extracellular networks, the main topological parameter associated with the heparin/HS binding function was the high average clustering coefficient displayed by the heparin/HS-interacting network (Fig. 1, B and C). The clustering coefficient is a measure of the modularity of a network (i.e. the tendency of nodes to form groups or clusters). Therefore, a high average clustering coefficient indicates the presence of highly interconnected groups of nodes (modules) within the network. Clustering coefficients of the heparin/HS-interacting network are distributed at higher values when compared with the non-heparin/HS-interacting network and the total extracellular interactome (Fig. 1C). The high average clustering coefficient of the extracellular heparin/HS-interacting network indicates a stronger tendency of HBPs to form highly interconnected modules than other extracellular proteins. In Fig. 2, selected examples of highly clustered modules extracted from the heparin/HS interactome are shown. These examples indicate how the tendency to form highly connected modules is independent of the nature of the HBP. The clusters shown are in fact formed by secreted growth factors such as VEGFB and transforming growth factor-β2 (TGFβ2) and their transmembrane receptors (Fig. 2, A and D) as well as by structural components of the ECM such as fibrillins (Fig. 2B) and plasma proteins such as coagulation factors (Fig. 2C). Furthermore, the architecture of the heparin/HS-interacting network also has functional implications. These clusters represent examples of functional modules responsible for the regulation of complex biological processes such as angiogenesis (45), morphogenesis (46), ECM assembly (47), and regulation of the coagulation cascade (48). These data support the view of HSPGs as key mediators of the assembly of molecular complexes at the cell surface and in the extracellular space (49).
FIGURE 2.
Examples of highly clustered modules of heparin/HS-interacting network. HBPs with a high clustering coefficient were extracted together with their first neighbors from the extracellular heparin/HS-interacting network. The node color indicates the clustering coefficient of each node according to the legend. The node label indicates the UniProt short name of the HBP. Protein-protein interactions are represented as green edges. The highest clustering coefficients represented were 1.0 for VEGFB and vascular endothelial growth factor receptor-1 (FLT1) (A), 0.67 for fibrillin-2 (FBN2) (B), 0.60 for coagulation factor IX (F9) (C), and 0.40 for TGFβ2 (D). The graphs were generated using Cytoscape v.2.6.3 (32). VWF, von Willebrand factor; CTGF, connective tissue growth factor; ELN, elastin; VTN, vitronectin; APP, amyloid precursor protein. NRP1, neuropilin-1; LTBP-1, latent-transforming growth factor beta-binding protein-1; BMP2, bone morphogenetic protein-2.
Functional Analysis of Heparin/HS Interactome
To gain insights into functional roles of HBPs, the over-representation (enrichment) of ontology terms and components of molecular pathways in the heparin/HS interactome was analyzed in comparison with their occurrence in the human proteome. The GO biological process terms describe biological objectives to which the gene or gene product contributes (29), and they can be either broad, generic terms such as “response to stimulus” or a more specific term such as “fibroblast growth factor receptor signaling pathway.” Therefore, the identification of biological process terms associated with a particular set of genes, in this case the HBPs, can be useful to highlight their functional roles at a network level. Ninety-four percent of the HBPs were annotated with at least one biological process term (Table 2 and supplemental Table 2). The remaining 6% were not annotated due to the incompleteness of the current GO annotation. From this analysis, a strong correlation between the heparin/HS interactome and biological functions characteristic of multicellular and higher organisms emerged. Significantly enriched terms are associated with fundamental processes common to all multicellular organisms such as cell-cell signaling but also with more complex processes such as wound healing and the immune response that are characteristic of higher organisms (Table 2). As with other ontologies, the biological process terms can be visualized as a graph where directed links describe the hierarchy and relationships between terms (29). This kind of visualization helps to group highly related/redundant terms typical of ontology classifications and identify relevant functional modules. The graph shown in Fig. 3 highlights the existence of four main functional groups strongly enriched in the heparin/HS interactome. These include two clusters of biological processes involved in the control of the immune system and of developmental processes and two other clusters related to the regulation of cellular processes such as cell proliferation and cell-cell signaling (Fig. 3). It has to be highlighted that the graph in Fig. 3 is based for clarity only on the subsets of terms with the highest p value; therefore, other significant and perhaps less investigated biological functions (such as, for example, “cation homeostasis”; p value, 2.1e−16) were not included in this analysis.
TABLE 2.
GO biological process terms enriched in heparin/HS interactome
“Count” indicates the number of HBPs, and “Percent” indicates the percentage of the mapped proteins associated to each term. For clarity, only the 20 most significant terms are listed. A complete list can be found in supplemental Table 2.
| Term | Name | Count | Percent | Corrected p value |
|---|---|---|---|---|
| GO:0009611 | Response to wounding | 120 | 27.8 | 6.6e−69 |
| GO:0042330 | Taxis | 55 | 12.8 | 2.6e−39 |
| GO:0006935 | Chemotaxis | 55 | 12.8 | 2.6e−39 |
| GO:0006954 | Inflammatory response | 73 | 16.9 | 3.4e−39 |
| GO:0006952 | Defense response | 91 | 21.1 | 8.1e−35 |
| GO:0007626 | Locomotory behavior | 62 | 14.4 | 3.3e−33 |
| GO:0006955 | Immune response | 91 | 21.1 | 5.7e−31 |
| GO:0042060 | Wound healing | 51 | 11.8 | 1.5e−30 |
| GO:0016477 | Cell migration | 57 | 13.2 | 3.1e−28 |
| GO:0007610 | Behavior | 71 | 16.5 | 3.7e−27 |
| GO:0051674 | Localization of cell | 58 | 13.5 | 9.7e−27 |
| GO:0048870 | Cell motility | 58 | 13.5 | 9.7e−27 |
| GO:0042127 | Regulation of cell proliferation | 90 | 20.9 | 4.0e−26 |
| GO:0006928 | Cell motion | 70 | 16.2 | 4.0e−26 |
| GO:0032101 | Regulation of response to external stimulus | 43 | 10.0 | 8.7e−26 |
| GO:0001568 | Blood vessel development | 51 | 11.8 | 2.4e−25 |
| GO:0001944 | Vasculature development | 51 | 11.8 | 7.3e−25 |
| GO:0051605 | Protein maturation by peptide bond cleavage | 33 | 7.7 | 1.2e−24 |
| GO:0007267 | Cell-cell signaling | 76 | 17.6 | 1.8e−24 |
| GO:0016485 | Protein processing | 36 | 8.4 | 3.9e−24 |
FIGURE 3.
GO biological process terms enriched in heparin/HS interactome are shown as nodes connected by directed edges that indicate hierarchies and relationships between terms. The node size is proportional to the number of HBPs belonging to the functional category. The node color indicates the corrected p value (Benjamini-Hochberg false discovery rate correction) for the enrichment of the term according to the legend. For clarity, only highly significant terms are displayed (p < 1e−21). The graphs were generated using Cytoscape v.2.6.3 (32) and its plugin BiNGO v2.3 (42).
Next, a similar approach to identify those pathways that have a statistically significant over-representation of HBPs was applied by projecting the heparin/HS interactome on the KEGG collection of pathways. KEGG pathways are manually drawn graphs that are based on the current knowledge of molecular interactions and reaction networks (39). In Table 3 and supplemental Table 3, the pathways enriched in HBPs are summarized. HBPs are involved in pathways responsible for the control of key physiological and pathological processes characteristic of multicellular organisms. In particular, the enriched pathways highlight the role of the heparin/HS interactome in key mechanisms implicated in the regulation of the cellular response to external stimuli. These include interactions between soluble ligands and their cell surface receptors (“cytokine-cytokine receptor interaction”) as well as cross-talk with components of the ECM (“ECM-receptor interaction”). These mechanisms are directly linked to the control of cell behavior via the regulation of processes such as cytoskeleton reorganization (“regulation of actin cytoskeleton” and “focal adhesion”) and activation of intracellular signaling cascades (e.g. “TGFβ signaling pathway”). The deregulation of these pathways can lead to the establishment of pathological conditions such as cancer (e.g. “melanoma”) and immunological disorders (e.g. “systemic lupus erythematosus”). Furthermore, pathways linked to other pathologies caused by structural alteration of extracellular proteins and accumulation of amyloid plaques (e.g. “prion diseases”) are significantly correlated with the heparin/HS binding activity because most of these proteins directly interact with GAGs.
TABLE 3.
KEGG pathways enriched in heparin/HS interactome
“Count” indicates the number of HBPs, and “Percent” indicates the percentage of the mapped proteins associated to each term. NOD, nucleotide binding and oligomerization domain.
| Term | Name | Count | Percent | Corrected p value |
|---|---|---|---|---|
| hsa04610 | Complement and coagulation cascades | 42 | 9.7 | 1.4e−33 |
| hsa04060 | Cytokine-cytokine receptor interaction | 63 | 14.6 | 7.3e−24 |
| hsa04512 | ECM-receptor interaction | 35 | 8.1 | 7.6e−21 |
| hsa04510 | Focal adhesion | 43 | 10.0 | 9.3e−14 |
| hsa05200 | Pathways in cancer | 52 | 12.1 | 1.7e−11 |
| hsa05218 | Melanoma | 22 | 5.1 | 7.8e−10 |
| hsa04062 | Chemokine signaling pathway | 34 | 7.9 | 7.9e−09 |
| hsa05020 | Prion diseases | 15 | 3.5 | 1.3e−08 |
| hsa04810 | Regulation of actin cytoskeleton | 33 | 7.7 | 8.7e−07 |
| hsa04350 | TGF-β signaling pathway | 18 | 4.2 | 2.6e−05 |
| hsa04672 | Intestinal immune network for IgA production | 13 | 3.0 | 6.8e−05 |
| hsa05322 | Systemic lupus erythematosus | 18 | 4.2 | 1.4e−04 |
| hsa04010 | MAPK signaling pathway | 30 | 7.0 | 1.4e−03 |
| hsa04640 | Hematopoietic cell lineage | 14 | 3.2 | 4.5e−03 |
| hsa04621 | NOD-like receptor signaling pathway | 11 | 2.6 | 1.0e−02 |
| hsa05219 | Bladder cancer | 9 | 2.1 | 1.1e−02 |
| hsa05310 | Asthma | 7 | 1.6 | 2.5e−02 |
| hsa05222 | Small cell lung cancer | 12 | 2.8 | 3.0e−02 |
In summary, the functional analysis of the heparin/HS interactome highlighted the following. (i) HBPs are functionally enriched in biological processes that are characteristic of multicellular and higher organisms. (ii) HBPs are candidates for a potential key role in mediating the information flow between the extracellular space and intracellular signaling pathways. (iii) This role could imply a direct involvement of the heparin/HS interactome in complex physiological and pathological systems such as organismal development, inflammation, cancer, and neurodegenerative disorders.
Structural Analysis of Heparin/HS Interactome
The same strategy used for the functional analysis of the heparin/HS interactome was performed at a structural level by investigating the enrichment of protein domains in HBPs. Ninety-eight percent of the HBPs were annotated to at least one Pfam domain, and the domain families significantly enriched in the heparin/HS interactome are listed in Table 4. Pfam domain families associated with heparin/HS binding activity are typically extracellular, and the majority of them appear to be characteristic of the metazoan lineage (65% of the top 20 most enriched families (Table 4)). Highlighting the structural diversity of the heparin/HS interactome, the list includes domains that are characteristic of small soluble, single domain proteins such as cytokines and growth factors (e.g. “small cytokines, interleukin-8-like,” “fibroblast growth factor,” and “transforming growth factor-β-like domain”), domains that assemble in large multi-domain proteins (e.g. “thrombospondin type 1 domain,” “laminin G domain,” and “fibronectin type III domain”) and domains associated with enzymatic activity (e.g. “trypsin family,” associated with proteolytic activity). The enrichment of some domain families is the result of the presence of families of HBP paralogs that expanded during evolution (e.g. the chemokine and FGF family). Whereas in other cases, non-homologous proteins contribute to one domain family by the arrangement of the same structural unit in various multidomain architectures (e.g. thrombospondin type 1 domain). Furthermore, some domain families are enriched in the heparin/HS interactome without being directly responsible for the interaction with the carbohydrate (e.g. EGF-like domain) because of their co-occurrence with heparin/HS-binding domains in multidomain proteins.
TABLE 4.
Pfam domain families enriched in heparin/HS interactome
“Count” indicates the number of HBPs associated to each domain family. For clarity, only the 20 most significant Pfam families are listed. A complete list can be found in supplemental Table 4.
| Term | Name | Count | Corrected p value | Metazoan-specific |
|---|---|---|---|---|
| PF00048 | Small cytokines, interleukin-8-like | 32 | 1.23e−38 | √ |
| PF00167 | Fibroblast growth factor | 19 | 5.08e−22 | √ |
| PF01391 | Collagen triple helix repeat | 24 | 1.09e−16 | |
| PF00090 | Thrombospondin type 1 domain | 21 | 1.17e−14 | |
| PF00008 | EGF-like domain | 22 | 1.03e−09 | |
| PF00089 | Subtilase family | 21 | 1.29e−09 | |
| PF01410 | Fibrillar collagen C-terminal domain | 8 | 5.98e−08 | √a |
| PF00079 | Serpin | 12 | 2.14e−07 | |
| PF02210 | Laminin G domain | 12 | 3.49e−07 | |
| PF00019 | Transforming growth factor-β-like domain | 11 | 1.15e−06 | √ |
| PF00093 | von Willebrand factor type C domain | 11 | 2.44e−06 | √ |
| PF00052 | Laminin B (Domain IV) | 6 | 2.27e−05 | √b |
| PF00053 | Laminin EGF-like | 10 | 2.88e−05 | |
| PF00084 | Sushi domain | 11 | 5.07e−05 | |
| PF02412 | Thrombospondin type 3 repeat | 5 | 7.16e−05 | √b |
| PF05735 | Thrombospondin C-terminal region | 5 | 7.16e−05 | √b |
| PF06008 | Laminin Domain I | 5 | 7.16e−05 | √b |
| PF06009 | Laminin Domain II | 5 | 7.16e−05 | √b |
| PF00219 | Insulin-like growth factor-binding protein | 7 | 2.60e−04 | √ |
| PF00688 | TGF-β propeptide | 7 | 3.27e−04 | √b |
a Domain family also present in the choanoflagellate M. brevicollis.
b Domain family also present in bacteria but not in non-metazoan eukaryotes.
Next, the Pfam domain families enriched in the heparin/HS interactome were mapped to the corresponding SCOP superfamilies. The SCOP classification uses structural information derived from the Protein Data Bank instead of sequence alignments to establish evolutionary relationships between proteins and domains (41). Seventy-one percent of the Pfam domain families significantly enriched in the heparin/HS interactome were mapped to a SCOP superfamily (see supplemental Table 4). This approach allowed the reduction of the redundancy of the Pfam classification by grouping domains of related structure into 25 superfamilies (for example the entries “EGF-like domain” and “laminin EGF-like” of Pfam are grouped in the SCOP superfamily “EGF/laminin”) (Table 5). As mentioned above, not all the structures are directly associated with heparin/HS binding activity. Literature curation of the heparin/HS-binding sites (HBSs) identified so far revealed that 11 of the 25 superfamilies have never been described as mediating the interaction with the carbohydrate (Table 5). Although the lack of evidence in the current literature does not necessarily exclude a role of these structures in heparin/HS binding, this analysis highlights those structures that are predominantly mediating the interaction with GAGs. For each of these structures, a referenced example and, when available, a three-dimensional structure of the domain in complex with heparin or a heparin-related molecule are reported in Table 5.
TABLE 5.
Superfamilies associated with Pfam domain families enriched in heparin/HS interactome
Not all the superfamilies are directly associated with heparin binding activity. In the cases where an HBS was located in a domain belonging to a superfamily, the columns “HBS: UniProt AC” and “HBS: Ref.” report the UniProt accession number of the HBP and the publication describing the interaction, respectively. The column “PDB” reports, when available, the Protein Data Bank code of a three-dimensional structure including a domain belonging to a superfamily in complex with heparin or a heparin derivative. TSP, thrombospondin; FnI, fibronectin type I; TIMP, tissue inhibitor of metalloproteinases; vWA, von Willebrand factor type A; SCR, short consensus repeat; BPTI, bovine pancreatic trypsin inhibitor; GLA, γ-carboxyglutamic acid.
| Superfamily | Name | HBS |
||
|---|---|---|---|---|
| UniProt AC | Ref. | PDB | ||
| 54117 | Interleukin-8-like chemokines | P02776 | 70 | 1U4L |
| 50353 | Cytokine | P09038 | 70 | 1AXM |
| 82895 | TSP-1 type 1 repeat | Q9UHI8 | 71 | |
| 57196 | EGF/laminin | |||
| 50494 | Trypsin-like serine proteases | P04070 | 72 | 1A7S |
| 56574 | Serpins | P01008 | 73 | 1AZX |
| 57501 | Cystine knot cytokines | P01137 | 74 | |
| 57603 | FnI-like domain | P02751 | 75 | |
| 57535 | Complement control module/SCR domain | P04003 | 76 | |
| 103647 | TSP type 3 repeat | |||
| 49899 | Concanavalin A-like lectins/glucanases | |||
| 57184 | Growth factor receptor domain | |||
| 50242 | TIMP-like | O95631 | 77 | |
| 53300 | vWA-like | P04275 | 78 | |
| 57610 | Thyroglobulin type-1 domain | P18065 | 79 | |
| 57440 | Kringle-like | P00749 | 80 | 1GMN |
| 49265 | Fibronectin type III | P02751 | 81 | |
| 47874 | Annexin | P07355 | 82 | 2HYU, 2HYV |
| 57424 | LDL receptor-like module | |||
| 49410 | α-Macroglobulin receptor domain | |||
| 48239 | Terpenoid cyclases | |||
| 57362 | BPTI-like | P10646 | 83 | |
| 56491 | A heparin-binding domain | P05067 | 84 | |
| 50755 | Phosphotyrosine-binding domain | |||
| 57630 | GLA domain | |||
| 69179 | Integrin domains | |||
| 58010 | Fibrinogen coiled coil and central regions | |||
In summary, the structural analysis of the heparin/HS interactome revealed a high level of diversity between structures associated with heparin/HS binding function possibly with a direct link to the structural complexity of GAGs. These structures are typically found in extracellular proteins, and the majority of them are specific to metazoans. They can either occur in single domain proteins, which are in general part of large families of paralogs, or as units mediating the heparin/HS binding activity that have been arranged during evolution in different multidomain architectures. Finally, a fraction of the structures enriched in the heparin/HS interactome does not seem to be directly involved in the interaction with the carbohydrate, and they might be over-represented because they are functionally and structurally linked with the heparin/HS-interacting modules.
Evolutionary Aspects of Heparin/HS Interactome
Because a strong association emerged between HBPs and biological functions and pathways characteristic of higher organisms, the evolution of the protein domains enriched in the heparin/HS interactome was analyzed in correlation with organism complexity across the tree of life. In a recent publication, Vogel and Chothia (50) investigated the expansion of domain superfamilies across the genomes of 38 uni- and multicellular eukaryotes and correlated it to the increase in organism complexity as measured by the number of different cell types of which the organisms are composed. They mapped the occurrence of different superfamilies using a database of 1219 hidden Markov models based on the superfamily classification of domains (51). For each genome, they annotated single domain proteins and the individual domains of multidomain proteins to their respective superfamily and then calculated the abundance of each superfamily as the number of proteins that contain at least one domain belonging to that particular superfamily. The normalized abundance profiles were then used to calculate a Pearson correlation coefficient (PCC), r, describing the correlation between superfamily abundance and the estimated number of cell types per genome (50). The PCCs for the 27 superfamilies enriched in the heparin/HS interactome were extracted from the data set of Vogel and Chothia (50), and they are plotted in Fig. 4. The distribution of PCCs of the heparin/HS interactome superfamilies indicates a strong correlation between their abundance and organism complexity (mean r = 0.83 with r ≥ 0.8 indicating a strong positive correlation). To establish a link between domain functions and their correlation with organism complexity, the authors extended the domain annotations by manually assigning functional categories to each superfamily (50). They observed that only 15% of the superfamilies have a strong positive correlation (r ≥ 0.8) with organism complexity. Moreover, just two functional categories contributed nearly half of these positively correlated superfamilies. These two functional categories are superfamilies associated with extracellular processes (20%) and regulation (29%) (50). Therefore, because most of the superfamilies characteristic of the heparin/HS interactome are also associated with extracellular processes, their relative contribution to the extracellular functional category was investigated. Thus, the heparin/HS interactome superfamilies were annotated using the functional classification described by Vogel and Chothia (50), and the distribution of the correlation coefficients of extracellular superfamilies enriched in the heparin/HS interactome was compared with that of superfamilies annotated as extracellular but not enriched in the heparin/HS interactome and of the whole extracellular category (Fig. 4). The mean correlation coefficient of the heparin/HS interactome superfamilies annotated as extracellular is even higher than the whole heparin/HS interactome set (mean r = 0.89), and most importantly, the distribution of their PCCs is significantly different from that of the whole extracellular category (p = 9.2e−3; one-sided Wilcoxon rank sum test) and other extracellular superfamilies (p = 1.9e−3; one-sided Wilcoxon rank sum test).
Next, the analysis of the correlation between HSPGs and organism evolution was extended to their biosynthetic enzymes. In this case, the analysis was not performed using the SCOP superfamily classification because functional specificity cannot be entirely recapitulated by the structural annotation (i.e. structurally related enzymes can be grouped in the same superfamily despite having different substrate specificity). Only the HS co-polymerases EXT1 and EXT2 are currently annotated to a superfamily (“nucleotide-diphospho-sugar transferases”; 53448), which also includes other sugar transferases such as galactosyltransferases. However, this superfamily also shows a positive correlation with organism complexity (r = 0.77). Therefore, the occurrence of HS biosynthetic enzymes across the tree of life was investigated by multiple sequence alignment across 638 sequenced genomes using STRING (52). The results are schematically summarized in Fig. 5. HS biosynthetic enzymes appear to be characteristic of the eumetazoan lineage and therefore strongly associated with the emergence of multicellularity. No homology is detectable in unicellular eukaryotes such as fungi and in plants (Fig. 5). Interestingly, some level of homology is detected in certain species of bacteria. The occurrence of enzymes able to synthesize GAG-related carbohydrates has been described already in some vertebrate pathogens such as Streptococci (53). The emergence of these microbial enzymes may have occurred by either convergent evolution or horizontal gene transfer under the selective pressure of the vertebrate host defense as a mechanism to enhance the pathogen virulence (53). Interestingly, some HS postsynthesis editing mechanisms appear to have evolved at a later stage than the core biosynthetic machinery. No homology is detectable for the extracellular HS-cleaving heparanases in the nematode Caenorhabditis elegans, although homology, albeit low, is observed for the extracellular HS sulfatases (Fig. 5). The association between an increase in organism complexity and HSPGs is further corroborated by the expansion of the HS biosynthetic enzymes in particular in the vertebrate lineage. Thus, although C. elegans possesses only one isoform for each of the key enzymes involved in HS biosynthesis (HS co-polymerase exostosin (EXT), N-deacetylase/N-sulfotransferase (NDST), glucuronic acid C5-epimerase (GLCE), and the sulfotransferases HS2ST, HS6ST, and HS3ST), in humans, all these enzymes have expanded by gene duplication with the exception of GLCE and HS2ST. Two EXT, four NDST, three HS6ST, and six HS3ST isoforms together with one isoform each for GLCE and HS2ST form the HS biosynthetic machinery in humans. A similar pattern is observed for the HS core proteins. Only one syndecan and two glypican genes are found in C. elegans, whereas humans possess four syndecan and six glypican isoforms (54). Interestingly, a similar expansion is not observed for the ECM core proteins perlecan and agrin (54).
FIGURE 5.
Occurrence of heparin/HS biosynthetic enzymes across tree of life. The occurrence of heparin/HS biosynthetic enzymes across 630 organisms was analyzed using STRING 8.2 (52). The UniProt Accession numbers of the human HS biosynthetic enzymes were used as input. The conservation of each gene across different species is indicated by squares colored according to the sequence homology detected by STRING. For clarity, some species (e.g. bacteria) were grouped in collapsed nodes colored in gray, and the number indicated in parentheses reports the number of species grouped in each node. In these cases, split squares report the highest and lowest score for the given gene within the grouped species.
In summary, a strong correlation emerged between the abundance in a genome of domain superfamilies associated with the heparin/HS interactome and organism complexity. This correlation has already been observed for extracellular domains (50), but it is statistically more pronounced for the heparin/HS interactome superfamilies than others. A similar correlation is also suggested for HS biosynthetic enzymes and core proteins based on the occurrence of these proteins across the tree of life and their expansion during evolution, especially in the vertebrate lineage.
Choanoflagellate M. brevicollis Possesses Rudimental HS Biosynthetic Machinery
Because GAGs are commonly considered a hallmark of eumetazoans and the expansion of HS biosynthetic enzymes and interacting domains is associated with an increase in organism complexity, the presence of orthologs of HS biosynthetic enzymes in the genome of M. brevicollis was investigated. Thus, orthologs of human HS biosynthetic enzymes were established using a reciprocal Blast best hit procedure (43) against the non-redundant protein sequences database of M. brevicollis. The Blast search was also performed against the protein sequence databases of the nematode C. elegans, known to produce GAGs, of the colony-forming slime mold D. discoideum, and of Fungi and Plantae.
The biosynthesis of HS and heparin requires the formation of a linker tetrasaccharide by sequential transfer of a xylose, two galactose units, and a glucuronic acid (GlcUA) unit to serine residues located in a serine-glycine repeat consensus on the core protein. This linker region is common to other GAGs such as chondroitin sulfate and dermatan sulfate. High score orthologs of the four enzymes required for the assembly of the protein-GAG linker tetrasaccharide are present in the genome of M. brevicollis, whereas clear orthology cannot be established for the full set in lower eukaryotes and plants (Fig. 6). The addition to the linker region of an N-acetylglucosamine (GlcNAc) or an N-acetylgalactosamine (GalNAc) residue commits the biosynthesis toward heparin/HS or chondroitin sulfate/dermatan sulfate, respectively (55). The biosynthetic reaction then proceeds with the polymerization of the sugar backbone by alternate addition of GlcUA and GlcNAc units in the case of heparin and HS. In human, the addition of the first GlcNAc residue is performed by the enzyme exostosin-like 3 (EXTL3) that possesses only GlcNAc-transferase activity, whereas the sugar polymerization is carried out by the enzymes EXTs that present both GlcUA- and GlcNAc-transferase activities (56). The two activities can be localized in the EXT enzymes in two separate domains with the N-terminal domain (EXT(N)) responsible for GlcUA-transferase activity and the C-terminal domain (EXT(C)) responsible for the GlcNAc-transferase activity (57). The EXT enzymes are likely to be the result of a gene fusion event between two functionally interacting enzymes carrying the GlcUA- and GlcNAc-transferase activities (55). The more rudimentary biosynthetic machinery of the nematode C. elegans comprises just a single enzyme (rib-2) with GlcNAc-transferase activity that is responsible for both chain initiation and polymerization and a separate enzyme (rib-1) that presents high sequence homology with EXT(N) and possesses GlcUA-transferase activity but only when in complex with rib-2 (58) (Fig. 6). Similarly, in M. brevicollis, two genes are present that possess homology with the C-terminal portion of EXTL3 and EXT(N) (Fig. 6). These two genes present conserved features that have been associated with GlcUA- and GlcNAc-transferase activity in human genes such as a conserved DXD motif in the C-terminal portion of EXTL3 (55, 57) (supplemental Fig. 4), and they indicate the possibility of the production of heparosan-like chains by choanoflagellates. The heparin/HS polymerizing chains undergo a series of modification by four classes of sulfotransferases (NDST, HS2ST, HS6ST, and HS3ST) that introduce sulfate groups at different positions on both the GlcUA and GluNAc units and by an epimerase (GLCE) that converts some GlcUA residues to iduronic acid (55, 59). These modifications largely contribute to generate sequence diversity in HS chains. M. brevicollis possesses two genes homologous to members of two of the four families of HS sulfotransferases: HS2ST and HS3ST (Fig. 6). Also in this case, the choanoflagellate genes present conservation of key residues involved in sulfotransferase catalytic activity such as binding sites for the sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (supplemental Fig. 4). These data suggest that M. brevicollis potentially possesses a complete, albeit rudimentary, HS biosynthetic machinery able to synthesize sulfated forms of HS-like polysaccharides. There is no evidence supporting the presence of a GLCE enzyme in M. brevicollis, suggesting that this kind of postsynthesis modification might have been a more recent innovation in the metazoan lineage.
FIGURE 6.
Choanoflagellate M. brevicollis possesses rudimentary HS biosynthetic machinery. Orthologs of the human HS biosynthetic enzymes were established by applying the reciprocal Blast best hit criterion (43) against the non-redundant protein sequences databases of C. elegans, M. brevicollis, Fungi, D. discoideum, and Plantae. The figure reports the Blastp score of the best hits only in the cases when the reciprocal best hit criterion was satisfied. The “Linker” enzymes are responsible for the synthesis of the protein-GAG linker tetrasaccharide, which is shared between HS and other GAGs. The “HS” group includes enzymes that are specific for the HS-specific biosynthetic pathway that follow the formation of the linker tetrasaccharide. The full report of the Blastp search including M. brevicollis hits is provided in supplemental Table 6.
In summary, the identification of orthologs of key GAG biosynthetic enzymes in the genome of a unicellular choanoflagellate suggests for the first time the presence of HS-like sulfated polysaccharides in non-metazoans. Intriguingly, the presence of a rudimentary HS biosynthetic machinery at the root of the metazoan lineage but not in other eukaryotes could support the role of GAGs as key molecules for the emergence of metazoan multicellularity as it has been suggested for other cell signaling, adhesion, and ECM molecules exclusively shared by M. brevicollis and metazoans (27, 28). Further studies including the biochemical characterization of the enzymes identified in this study and isolation of GAGs from M. brevicollis cells are required to validate this hypothesis.
DISCUSSION
The analysis undertaken of the heparin/HS interactome allowed the investigation of the properties of the heparin/HS-interacting network of protein-protein interaction and the analysis at a system level of the functional and structural features characterizing HBPs. The heparin/HS interactome was built combining literature mining, data retrieval from public databases, and proteomics experimental data. The affinity proteomics strategy described in the supplementary information led to the identification of 147 extracellular proteins of which 32 were previously described HBPs (supplemental Table 1). The remaining 115 newly discovered HBPs were also included for the analysis of the heparin/HS interactome, although these interactions will require independent experimental validation. The resulting data set included 435 human proteins of which most are extracellular (supplemental Table 5). The analysis of the heparin/HS-interacting network revealed that HBPs tend to form more highly clustered modules than other extracellular proteins. These clusters can assemble both at the cell surface and in the ECM, and they often also represent functional modules. From a functional point of view, the heparin/HS interactome is strongly associated with biological processes characteristic of multicellular organisms and with pathways that are crucial for the conversion of extracellular cues into intracellular signaling events and finally into a phenotypic response. These processes and pathways are central to complex biological phenomena particular to higher organisms such as development and the immune response, and they are consequently linked to pathological conditions such as cancer and neurodegenerative disorders. In this perspective, potential intracellular roles of HSPGs could provide additional mechanisms for GAG-mediated regulation of intracellular signaling (60). The structural analysis of the heparin/HS interactome revealed the existence of two main categories of domains associated with heparin/HS binding activity: domains that are characteristic of soluble single domain proteins and domains that occur mainly in multidomain proteins and that have been assembled during evolution in different architectures. From an evolutionary perspective, the expansion of domains associated with the heparin/HS interactome strongly correlates with an increase in organism complexity that is independent of their nature. A similar correlation was already described for domains and proteins generically associated with extracellular processes (50, 61); however, also within this set, the heparin/HS interactome-associated domains display a statistically significant higher correlation than other extracellular domains. It has been shown that the evolutionary rate of the extracellular proteome is faster then the intracellular proteome probably due to the less chemically constrained environment faced by extracellular proteins (62). This evolutionary plasticity of the extracellular proteome could have been a driving force for the organization of more complex systems of intercellular communication and organization. The functional and structural link between the heparin/HS interactome and biological processes characteristic of complex organisms and the fact that HBPs are more correlated than other extracellular proteins with an increase in organism complexity strongly suggest a pivotal role of HSPGs in driving the evolution of multicellular and higher organisms. In fact, the expansion of enzymes and core proteins responsible for the synthesis and localization of HS chains also correlates with an increase in organism complexity in the metazoan lineage. The core HSPG biosynthetic machinery was thought to have evolved in early eumetazoans concomitantly with the emergence of multicellularity (63). Biochemical data describing the presence of heparin/HS-like GAGs in phyla at the root of the eumetazoan lineage such as Cnidaria and Ctenophora (64, 65) supported this view. However, the presence of orthologs of key HS biosynthetic enzymes in the choanoflagellate M. brevicollis suggests that GAGs are likely to be a molecular innovation that predates the origin of metazoans. HSPGs might have been a critical step for the assembly of an extracellular network of proteins required for the structural organization of the extracellular space and for the establishment of cell-cell communication and cell differentiation. Later on, gene duplication events, in particular the whole genome duplications that occurred at the root of the vertebrate lineage (66), contributed to expand the repertoire of both HSPG biosynthetic enzymes and their interacting partners on which evolution could act. The fact that domains characteristic of the heparin/HS interactome are strongly correlated with organism complexity could indicate that their expansion has been one of the driving forces toward the organization of the more sophisticated and tunable extracellular networks that are required for the development of higher organisms and for the control of organism-level biological processes such as the establishment of an immune system. In support of this, others have already proposed HSPGs as key molecules for the emergence of neural connectivity (54, 67). The authors collected a series of experimental evidences obtained in different model organisms describing the specific involvement of HSPGs in all the key processes required for the establishment of neural connectivity including axon guidance, neuron-target interaction, and synapse development (54). Similar to what has been proposed here, the authors suggested a role for HSPGs as versatile extracellular scaffolds that modulate extracellular cues influencing the response of neurons to their environment (54). In the future, heparin affinity proteomics strategies similar to that described in the supplemental information could be implemented in combination with quantitative mass spectrometry techniques such as targeted proteomics (68) and staple isotope labeling with amino acids in cell culture (69) to investigate dynamic changes of the heparin/HS interactome associated, for example, with different developmental stages or pathological conditions. Such data, complemented by the characterization of the dynamic changes in HS structure, could be extremely valuable to elucidate HSPG-mediated mechanisms involved in the control of physiological and pathological processes. This opens the door to the design of new, network-based therapeutic strategies targeting multiple protein-glycan interactions that are associated with multifactorial diseases. Finally, the structural and functional characterization of the proteome and glycome of organisms at the root of the metazoan lineage could illuminate the role of protein and glycan co-evolution in the assembly of the extracellular molecular networks necessary for the development of complex forms of life.
Supplementary Material
Acknowledgments
We thank Dr. Martin Beck and the European Molecular Biology Laboratory Proteomic Core Facility for help with mass spectrometry data acquisition and analysis, Dr. Olga Vasieva and Dr. Krzysztof Wicher for many helpful discussions and bioinformatics support, and Hassanul Choudhury for technical assistance.
This work was supported by the European Commission Marie Curie Early Stage Training Program MolFun (to A. O.), the Cancer and Polio Research Fund Laboratories, and the North West Cancer Research Fund (to D. G. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Results, Figs. 1–5, and Tables 1–7.
- GAG
- glycosaminoglycan
- B3GA3
- galactosylgalactosylxylosylprotein 3-β-glucuronosyltransferase 3
- B3GT6
- β-1,3-galactosyltransferase 6
- B4GT7
- xylosylprotein 4-β-galactosyltransferase 7
- ECM
- extracellular matrix
- EXT
- exostosin/heparan sulfate polymerase
- EXTL3
- exostosin-like 3/glucosamine transferase
- GlcUA
- glucuronic acid
- GlcNAc
- N-acetylglucosamine
- GLCE
- glucuronic acid C5-epimerase
- GO
- gene ontology
- HBP
- heparin-binding protein
- HBS
- heparin-binding site
- HS
- heparan sulfate
- HS2ST
- heparan sulfate 2-O-sulfotranferase
- HS3ST
- heparan sulfate 3-O-sulfotranferase
- HS6ST
- heparan sulfate 6-O-sulfotranferase
- HSPG
- heparan sulfate proteoglycan
- KEGG
- Kyoto encyclopedia of genes and genomes
- NDST
- heparan sulfate N-deacetylase/N-sulfotransferase
- PCC
- Pearson correlation coefficient
- SCOP
- structural classification of proteins
- taxid
- NCBI taxonomy identifier.
REFERENCES
- 1. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 2. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 3. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 4. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 5. Jeong H., Mason S. P., Barabási A. L., Oltvai Z. N. (2001) Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 6. Barabási A. L., Oltvai Z. N. (2004) Nat. Rev. Genet. 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 7. Grove C. A., De Masi F., Barrasa M. I., Newburger D. E., Alkema M. J., Bulyk M. L., Walhout A. J. (2009) Cell 138, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taylor I. W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J. L. (2009) Nat. Biotechnol. 27, 199–204 [DOI] [PubMed] [Google Scholar]
- 9. Ideker T., Thorsson V., Ranish J. A., Christmas R., Buhler J., Eng J. K., Bumgarner R., Goodlett D. R., Aebersold R., Hood L. (2001) Science 292, 929–934 [DOI] [PubMed] [Google Scholar]
- 10. Bandyopadhyay S., Sharan R., Ideker T. (2006) Genome Res. 16, 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fraser H. B. (2006) Curr. Opin. Genet. Dev. 16, 637–644 [DOI] [PubMed] [Google Scholar]
- 12. Beltrao P., Serrano L. (2007) PLoS Comput. Biol. 3, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turnbull J. E., Gallagher J. T. (1991) Biochem. J. 273, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy K. J., Merry C. L., Lyon M., Thompson J. E., Roberts I. S., Gallagher J. T. (2004) J. Biol. Chem. 279, 27239–27245 [DOI] [PubMed] [Google Scholar]
- 15. Allen B. L., Rapraeger A. C. (2003) J. Cell Biol. 163, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bornemann D. J., Park S., Phin S., Warrior R. (2008) Development 135, 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ori A., Wilkinson M. C., Fernig D. G. (2008) Front. Biosci. 13, 4309–4338 [DOI] [PubMed] [Google Scholar]
- 18. Thompson S. M., Jesudason E. C., Turnbull J. E., Fernig D. G. (2010) Birth Defects Res. C Embryo Today 90, 32–44 [DOI] [PubMed] [Google Scholar]
- 19. Vyas N., Goswami D., Manonmani A., Sharma P., Ranganath H. A., VijayRaghavan K., Shashidhara L. S., Sowdhamini R., Mayor S. (2008) Cell 133, 1214–1227 [DOI] [PubMed] [Google Scholar]
- 20. Yu S. R., Burkhardt M., Nowak M., Ries J., Petrásek Z., Scholpp S., Schwille P., Brand M. (2009) Nature 461, 533–536 [DOI] [PubMed] [Google Scholar]
- 21. Chen Y., Götte M., Liu J., Park P. W. (2008) Mol. Cells 26, 415–426 [PMC free article] [PubMed] [Google Scholar]
- 22. Lin X. (2004) Development 131, 6009–6021 [DOI] [PubMed] [Google Scholar]
- 23. Parish C. R. (2006) Nat. Rev. Immunol. 6, 633–643 [DOI] [PubMed] [Google Scholar]
- 24. Handel T. M., Johnson Z., Crown S. E., Lau E. K., Proudfoot A. E. (2005) Annu. Rev. Biochem. 74, 385–410 [DOI] [PubMed] [Google Scholar]
- 25. Fuster M. M., Esko J. D. (2005) Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 26. Abedin M., King N. (2008) Science 319, 946–948 [DOI] [PubMed] [Google Scholar]
- 27. King N., Westbrook M. J., Young S. L., Kuo A., Abedin M., Chapman J., Fairclough S., Hellsten U., Isogai Y., Letunic I., Marr M., Pincus D., Putnam N., Rokas A., Wright K. J., Zuzow R., Dirks W., Good M., Goodstein D., Lemons D., Li W., Lyons J. B., Morris A., Nichols S., Richter D. J., Salamov A., Sequencing J. G., Bork P., Lim W. A., Manning G., Miller W. T., McGinnis W., Shapiro H., Tjian R., Grigoriev I. V., Rokhsar D. (2008) Nature 451, 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. King N., Hittinger C. T., Carroll S. B. (2003) Science 301, 361–363 [DOI] [PubMed] [Google Scholar]
- 29. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. UniProt Consortium (2010) Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chautard E., Ballut L., Thierry-Mieg N., Ricard-Blum S. (2009) Bioinformatics 25, 690–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C., Dimmer E., Feuermann M., Friedrichsen A., Huntley R., Kohler C., Khadake J., Leroy C., Liban A., Lieftink C., Montecchi-Palazzi L., Orchard S., Risse J., Robbe K., Roechert B., Thorneycroft D., Zhang Y., Apweiler R., Hermjakob H. (2007) Nucleic Acids Res. 35, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salwinski L., Miller C. S., Smith A. J., Pettit F. K., Bowie J. U., Eisenberg D. (2004) Nucleic Acids Res. 32, D449–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keshava Prasad T. S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Harrys Kishore C. J., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., Pandey A. (2009) Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 38. Assenov Y., Ramírez F., Schelhorn S. E., Lengauer T., Albrecht M. (2008) Bioinformatics 24, 282–284 [DOI] [PubMed] [Google Scholar]
- 39. Kanehisa M., Goto S. (2000) Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 42. Maere S., Heymans K., Kuiper M. (2005) Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 43. Koonin E. V. (2005) Annu. Rev. Genet. 39, 309–338 [DOI] [PubMed] [Google Scholar]
- 44. Tucker C. L., Gera J. F., Uetz P. (2001) Trends Cell Biol. 11, 102–106 [DOI] [PubMed] [Google Scholar]
- 45. Uniewicz K. A., Fernig D. G. (2008) Front. Biosci. 13, 4339–4360 [DOI] [PubMed] [Google Scholar]
- 46. Belenkaya T. Y., Han C., Yan D., Opoka R. J., Khodoun M., Liu H., Lin X. (2004) Cell 119, 231–244 [DOI] [PubMed] [Google Scholar]
- 47. Ritty T. M., Broekelmann T. J., Werneck C. C., Mecham R. P. (2003) Biochem. J. 375, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu H., Muñoz E. M., Edens R. E., Linhardt R. J. (2005) Biochim. Biophys. Acta 1726, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turnbull J., Powell A., Guimond S. (2001) Trends Cell Biol. 11, 75–82 [DOI] [PubMed] [Google Scholar]
- 50. Vogel C., Chothia C. (2006) PLoS Comput. Biol. 2, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gough J., Karplus K., Hughey R., Chothia C. (2001) J. Mol. Biol. 313, 903–919 [DOI] [PubMed] [Google Scholar]
- 52. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DeAngelis P. L. (2002) Anat. Rec. 268, 317–326 [DOI] [PubMed] [Google Scholar]
- 54. Van Vactor D., Wall D. P., Johnson K. G. (2006) Curr. Opin. Neurobiol. 16, 40–51 [DOI] [PubMed] [Google Scholar]
- 55. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 56. Kim B. T., Kitagawa H., Tamura J., Saito T., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7176–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei G., Bai X., Gabb M. M., Bame K. J., Koshy T. I., Spear P. G., Esko J. D. (2000) J. Biol. Chem. 275, 27733–27740 [DOI] [PubMed] [Google Scholar]
- 58. Kitagawa H., Izumikawa T., Mizuguchi S., Dejima K., Nomura K. H., Egusa N., Taniguchi F., Tamura J., Gengyo-Ando K., Mitani S., Nomura K., Sugahara K. (2007) J. Biol. Chem. 282, 8533–8544 [DOI] [PubMed] [Google Scholar]
- 59. Kusche-Gullberg M., Kjellén L. (2003) Curr. Opin. Struct. Biol. 13, 605–611 [DOI] [PubMed] [Google Scholar]
- 60. Chen L., Sanderson R. D. (2009) PLoS One 4, e4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huxley-Jones J., Pinney J. W., Archer J., Robertson D. L., Boot-Handford R. P. (2009) Int. J. Exp. Pathol. 90, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Julenius K., Pedersen A. G. (2006) Mol. Biol. Evol. 23, 2039–2048 [DOI] [PubMed] [Google Scholar]
- 63. Freilich S., Goldovsky L., Ouzounis C. A., Thornton J. M. (2008) BMC Evol. Biol. 8, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Medeiros G. F., Mendes A., Castro R. A., Baú E. C., Nader H. B., Dietrich C. P. (2000) Biochim. Biophys. Acta 1475, 287–294 [DOI] [PubMed] [Google Scholar]
- 65. Yamada S., Morimoto H., Fujisawa T., Sugahara K. (2007) Glycobiology 17, 886–894 [DOI] [PubMed] [Google Scholar]
- 66. Panopoulou G., Poustka A. J. (2005) Trends Genet. 21, 559–567 [DOI] [PubMed] [Google Scholar]
- 67. Lee J. S., Chien C. B. (2004) Nat. Rev. Genet. 5, 923–935 [DOI] [PubMed] [Google Scholar]
- 68. Domon B., Aebersold R. (2010) Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 69. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 70. Ori A., Free P., Courty J., Wilkinson M. C., Fernig D. G. (2009) Mol. Cell. Proteomics 8, 2256–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuno K., Matsushima K. (1998) J. Biol. Chem. 273, 13912–13917 [DOI] [PubMed] [Google Scholar]
- 72. Shen L., Villoutreix B. O., Dahlbäck B. (1999) Thromb. Haemost. 82, 72–79 [PubMed] [Google Scholar]
- 73. Ersdal-Badju E., Lu A., Zuo Y., Picard V., Bock S. C. (1997) J. Biol. Chem. 272, 19393–19400 [DOI] [PubMed] [Google Scholar]
- 74. Lyon M., Rushton G., Gallagher J. T. (1997) J. Biol. Chem. 272, 18000–18006 [DOI] [PubMed] [Google Scholar]
- 75. Ingham K. C., Brew S. A., Atha D. H. (1990) Biochem. J. 272, 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blom A. M., Kask L., Dahlbäck B. (2001) J. Biol. Chem. 276, 27136–27144 [DOI] [PubMed] [Google Scholar]
- 77. Kappler J., Franken S., Junghans U., Hoffmann R., Linke T., Müller H. W., Koch K. W. (2000) Biochem. Biophys. Res. Commun. 271, 287–291 [DOI] [PubMed] [Google Scholar]
- 78. Rastegar-Lari G., Villoutreix B. O., Ribba A. S., Legendre P., Meyer D., Baruch D. (2002) Biochemistry 41, 6668–6678 [DOI] [PubMed] [Google Scholar]
- 79. Kuang Z., Yao S., Keizer D. W., Wang C. C., Bach L. A., Forbes B. E., Wallace J. C., Norton R. S. (2006) J. Mol. Biol. 364, 690–704 [DOI] [PubMed] [Google Scholar]
- 80. Stephens R. W., Bokman A. M., Myöhänen H. T., Reisberg T., Tapiovaara H., Pedersen N., Grøndahl-Hansen J., Llinás M., Vaheri A. (1992) Biochemistry 31, 7572–7579 [DOI] [PubMed] [Google Scholar]
- 81. Sachchidanand, Lequin O., Staunton D., Mulloy B., Forster M. J., Yoshida K., Campbell I. D. (2002) J. Biol. Chem. 277, 50629–50635 [DOI] [PubMed] [Google Scholar]
- 82. Shao C., Zhang F., Kemp M. M., Linhardt R. J., Waisman D. M., Head J. F., Seaton B. A. (2006) J. Biol. Chem. 281, 31689–31695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mine S., Yamazaki T., Miyata T., Hara S., Kato H. (2002) Biochemistry 41, 78–85 [DOI] [PubMed] [Google Scholar]
- 84. Clarris H. J., Cappai R., Heffernan D., Beyreuther K., Masters C. L., Small D. H. (1997) J. Neurochem. 68, 1164–1172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.