Abstract
Metastasis is the deadliest and most poorly understood feature of malignant diseases. Recent work has shown that Metadherin (MTDH) is overexpressed in over 40% of breast cancer patients and promotes metastasis and chemoresistance in experimental models of breast cancer progression. Here we applied mass spectrometry-based screen to identify staphylococcal nuclease domain-containing 1 (SND1) as a candidate MTDH-interacting protein. After confirming the interaction between SND1 and MTDH, we tested the role of SND1 in breast cancer and found that it strongly promotes lung metastasis. SND1 was further shown to promote resistance to apoptosis and to regulate the expression of genes associated with metastasis and chemoresistance. Analyses of breast cancer clinical microarray data indicated that high expression of SND1 in primary tumors is strongly associated with reduced metastasis-free survival in multiple large scale data sets. Thus, we have uncovered SND1 as a novel MTDH-interacting protein and shown that it is a functionally and clinically significant mediator of metastasis.
Keywords: Apoptosis, Breast Cancer, Lung, Mass Spectrometry (MS), Mouse, SND-1, Metadherin, Metastasis
Introduction
Metastasis is responsible for the majority of cancer-related deaths (1). An increasing number of genes have been implicated in mediating different steps of metastasis, but relatively few are mechanistically well characterized (2). One recently verified mediator of distant metastasis is Metadherin (MTDH3; also called Lyric and AEG1 (3–5)). MTDH has been shown to be a key functional target of the 8q22 genomic gain that is frequently observed in poor prognosis breast cancer patients (6). Beyond promoting experimental lung metastasis (3, 6), multiple studies have implicated MTDH as a mediator of several cancer-related processes, such as oncogenesis and angiogenesis (7, 8), invasion (9), chemoresistance (6, 10–12), apoptosis resistance (13), and autophagy (14). Despite the abundance of MTDH phenotypes, a consensus understanding of its underlying molecular mechanisms has not yet been reached. MTDH has been shown to influence several oncogenic signaling pathways and transcription factors, such as Ha-Ras (15), PI3K/AKT (13), ERK, Wnt/β-catenin (8), NF-κB (16), c-Myc (15), and FOXO1/FOXO3a (17, 18). However, MTDH regulation of signaling pathways appears to be context-dependent with affected pathways varying by tumor type and cell line (19, 20). Furthermore, prior work on MTDH molecular mechanisms has largely focused on hypothesis-driven investigations into the ability of MTDH to influence classical oncogenic pathways with little exploration to date on protein-level interactions (16, 21).
In this study, we aimed to expand the knowledge of MTDH molecular functionality and also uncover novel genes involved in promoting distant metastasis. Our approach utilized an unbiased, mass spectrometry-based screen for MTDH-interacting partners. We identified and characterized the interaction between MTDH and one such protein, SND1. SND1 is a multifunctional protein with reported roles in transcriptional activation (22), RNA editing (23, 24), formation of the RNA-induced silencing complex (25), and regulation of spliceosome activity (26) and the apoptotic cascade (27). However, its functional role in tumorigenesis or metastasis has not yet been evaluated. Here we report experimental and clinical data indicating that, in addition to interacting with MTDH, SND1 is in its own right a powerful novel mediator of breast cancer lung metastasis.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
SCP28 and LM2 cells were derived from the parental cell line MDA-MB-231 (American Type Culture Collection) (28, 29). These sublines, their genetically modified variants, the retroviral packaging cell line H29, and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, Fungizone, and antibiotics. H29 cells were cultured with doxycycline when not actively producing virus.
Construction of MTDH and SND1 Expression Constructs
Either full-length or deletion mutant human SND1 coding sequences were inserted into the pCMV5-N-HA expression plasmid using the restriction sites 5′ BamHI and 3′ Xba1. For deletion mutants, the N-terminal staphylococcal nuclease (SN) repeat domain (amino acids 1–639) and the C-terminal Tudor-SN domain (amino acids 640–885) were used. For MTDH expression constructs, either full-length or deletion mutant human MTDH coding sequences were inserted into the pCMV-N-Myc expression plasmid using the restriction sites 5′ SalI and 3′ BamHI. N-terminal MTDH deletions mutants ND1–ND5 consisted of MTDH amino acids 95–582, 146–582, 260–582, 364–582, and 471–582, respectively. C-terminal MTDH deletion mutants CD1–CD5 consisted of MTDH amino acids 1–470, 1–363, 1–259, 1–145, and 1–94, respectively. A C-terminally eGFP-tagged MTDH expression plasmid was constructed by inserting the MTDH coding sequence in-frame to the N terminus of eGFP in the pEGFP-N1 expression plasmid (Clontech) using 5′ EcoRI and 3′ Xho1 restriction sites. The MTDH-eGFP sequence was also subcloned into the retroviral pMSCVpuro vector (Clontech) using 5′ EcoRI and 3′ Xho1 restriction sites.
Mass Spectrometry Analysis
Affinity purification of MTDH-GFP and subsequent mass spectrometry analysis were performed as described previously (30–32). LC-MS/MS-based identification of proteins isolated from bands sliced from silver-stained SDS-PAGE gels was performed by ProtTech, Inc. (Norristown, PA).
Immunoprecipitation, Western Blot, and Immunofluorescence Analyses
For immunoprecipitation (IP) experiments, 80% confluent cells from 15-cm dishes were washed in cold PBS and lysed in 800 μl of lysis buffer (same as used for mass spectrometry protein preparation) (30) with an EDTA-free protease inhibitor mixture (Roche Applied Science) and PMSF. Cell lysates were then sonicated and centrifuged. For IP bead preparation, 10 μl of protein A/G beads (Santa Cruz Biotechnology) were washed with PBS, incubated with 10 μl of antibodies for 1 h at room temperature, washed again, and then cross-linked with 20 mm dimethyl pimelimidate dihydrochloride prior to addition to 800 μl of cell lysate for IP. IPs were carried out for 4–12 h at 4 °C. Beads were subsequently boiled for 5 min in SDS protein loading buffer to elute bound protein.
For Western blot analysis, either IP or whole cell lysate samples were subjected to SDS-PAGE and subsequently transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked in milk prior to primary antibody incubation (all antibodies were diluted 1:1,000 in milk).
For immunofluorescence, cells were grown on ethanol-sterilized glass coverslips in 24-well plates, washed with PBS, and fixed in 4% paraformaldehyde. 0.2% Triton X-100 was used for membrane permeabilization. Primary antibodies used for IPs, Western blots, and immunofluorescence were as follows: anti-SND1 (AbCam ab70422), anti-Metadherin (Zymed Laboratories Inc., Invitrogen), anti-β-actin (AbCam ab6726), anti-Myc 9E10 (Santa Cruz Biotechnology Inc. sc-40), and anti-HA (Santa Cruz Biotechnology Inc. sc-7392).
Generation of Stable Cell Lines
Stable short hairpin RNA (shRNA)-mediated knockdown was achieved with the pSuper-Retro system (OligoEngine) targeting the sequences 5′-AAGGAGCGATCTGCTAGCTAC-3′ (KD1) and 5′-GGAACGGTTCACATACTA-3′ (KD2) for SND1. Stable overexpression of MTDH-eGFP was achieved using pMSCVpuro retroviruses. Retroviral vectors were transfected into the packaging cell line H29. After 48 h, viruses were collected, filtered, and used to infect target cells in the presence of 5 μg/ml Polybrene. The infected cells were selected with 0.8 μg/ml puromycin.
Reverse Transcription and Quantitative PCR
Total RNA was isolated using the RNeasy kit (Qiagen) and reverse transcribed with the Superscript III kit (Invitrogen) following the manufacturer's instructions. Quantitative PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) with the ABI Prism 7900HT thermocycler (Applied Biosystems). Primers used for quantitative PCR were as follows: human SND1, 5′-GTGGACAGCGTAGTTCGGGA-3′ (forward) and 5′-CCCACGAGACATTTCCACACAC-3′ (reverse); human KiSS1, 5′-CACTTTGGGGAGCCATTAGA-3′ (forward) and 5′-CAGTAGCAGCTGGCTTCCTC-3′ (reverse); and human GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse). SND1 and KiSS1 mRNA levels were normalized to GAPDH levels.
Microarray Analysis
To identify genes regulated by SND1 knockdown, RNA samples of SCP28 control and SND1-KD cells were analyzed with Agilent Whole Human Genome 4x44K microarrays. RNA samples were labeled with Cy5 using the Agilent Low RNA Input Linear Amplification kit and hybridized with the Cy3-labeled Human Universal Reference RNA (Stratagene). Duplicate arrays were performed for each sample. Arrays were scanned with an Agilent G2565BA scanner and analyzed initially with Agilent Feature Extraction v9.5 software and subsequently with GeneSpring GX. The Cy5/Cy3 ratios were calculated using the feature medium signal and normalized by the array median. Probes with an average of >2.0-fold changes were identified as SND1-regulated genes.
Tumor Xenografts and Bioluminescence Analyses
All animal work was performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Princeton University under approved protocols. 2 × 105 cells were washed in PBS and injected intravenously into female athymic Ncr-nu/nu mice to study lung metastasis activity as described previously (29). To study primary tumor growth, cancer cells harvested from culture were resuspended in PBS at a concentration of 1 × 107 cells/ml. An incision was made in the abdomen, and the skin was recessed to locate the number 4 mammary fat pad into which 1 × 105 cells (10 μl) were injected. Noninvasive bioluminescence imaging was performed to quantify the metastasis burden using an IVIS 200 Imaging System (Caliper Life Sciences). Analysis was performed with Living Image software (Xenogen) by measuring photon flux of the region of interest. Data were normalized to the signal obtained immediately after injection (day 1).
Histological Analysis
Mice were sacrificed, and lungs were harvested followed by fixation in 10% neutral buffered formalin for 24 h, washing with PBS, and dehydration in 70% ethanol. Lungs were then paraffin-embedded, sectioned, and subjected to hematoxylin and eosin (H&E) staining.
In Vitro Growth Curve and Invasion Assays
To establish growth curves, 1 × 106 cells were seeded into 10-cm dishes in triplicate at each time point and counted every 3 days over the course of 12 days. For invasion assays, 1 × 105 cells suspended in serum-free DMEM were seeded in triplicate into the upper chambers of the insert membranes with 8-μm pore size (BD Biosciences) in a 24-well plate. The upper chambers were coated with 40 μl of 1.0 mg/ml Matrigel (BD Biosciences) and incubated to allow the Matrigel to solidify for 1 h before adding cells. Serum-containing growth medium was used in the bottom chamber as an attractant. After 24 h in culture, medium was aspirated from the top and bottom chambers, and the bottom chambers were washed with PBS prior to addition of 300 μl of trypsin. After full trypsinization, cells were collected from the bottom chamber and counted.
M30 CytoDEATH Assays
Cells were grown to 70% confluence and treated with either 0.1 μm Adriamycin or ethanol for 24 h. Conditioned medium was collected, and cells were trypsinized and then added back to their respective conditioned medium. Cells were washed with PBS and then either fixed with methanol for 30 min at −20 °C (experimental samples) or incubated unfixed in PBS at 4 °C (negative control samples). All samples were then washed with PBS-Tween 20, blocked for 1 h with PBS-Tween 20 plus 1% BSA, and incubated with either M30 CytoDEATH or M30 CytoDEATH fluorescein (Roche Applied Science) at a 1:250 dilution in PBS-Tween 20. Cells were then either incubated with rhodamine red-conjugated secondary antibody or analyzed directly (if using M30 CytoDEATH fluorescein). For flow cytometry, the background signal was determined using unfixed (non-permeabilized) samples and used to set gating thresholds.
Transfection and Reporter Assays
For reporter assays, 90% confluent LM2 or 293T cells in 12-well plates were transfected with KiSS1-luciferase (a gift from Dr. Marc Montminy), a Renilla luciferase expression plasmid, and (for 293T cells) either HA-SND1-pCMV5 or pCMV5. Lipofectamine 2000 was used for transfections at concentrations designated by the manufacturer's instructions. 24 h post-transfection cells were lysed, and both firefly and Renilla luciferase activity was measured. All firefly luciferase readings were normalized to their corresponding Renilla luciferase readings. All experiments were performed in triplicate and repeated at least once for a minimum sample size of six wells per experimental group.
Gene Set Enrichment Analysis and Gene Ontology Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) v2.0 (33, 34) was used to analyze normalized microarray data of control and SND1-KD SCP28 cells. Multiple probe matches for the same gene were collapsed into one value, using the highest probe reading in each case. Only probes with matches to gene symbols were used. Genes were ranked using the provided signal-to-noise ranking statistic, and GSEA was run using a weighted statistic and evaluated for statistical significance by comparison with results obtained using 1,000 random permutations of each gene set. For all other GSEA parameters, default settings were used. Gene sets tested for enrichment consisted of all gene sets from the C2 collection provided in the Molecular Signatures Database v3.0 as well as several gene sets collected manually from published microarray studies.
For gene ontology enrichment analysis, the signature of SND1-regulated genes was loaded into the DAVID v6.7 (35, 36) website and submitted to the functional annotation tool. Ontologies from the “cellular component” database were considered in enrichment analyses.
Clinical Data Set Analyses
Microarray and patient survival data from the MSK-82 data set were downloaded from the on-line supplemental data provided in the Minn et al. (29) study. For Kaplan-Meier Plotter database (37) analysis, distant metastasis-free survival was assessed in lymph node-positive patients stratified by median SND1 expression. All other parameters were left at default settings.
Statistical Analyses
Results were reported as average ± S.E. Two-sided independent Student's t tests without equal variance assumption or nonparametric Mann-Whitney U tests were performed to analyze the data with p < 0.05 considered as statistically significant. Multiple hypothesis testing was controlled as indicated using either Bonferroni-corrected p values or by considering the false discovery rate in addition to p values. Kaplan-Meier analyses were performed using Intercooled Stata 7.0 and evaluated for statistical significance using log rank tests.
RESULTS
Identification of SND1 as an MTDH-interacting Protein
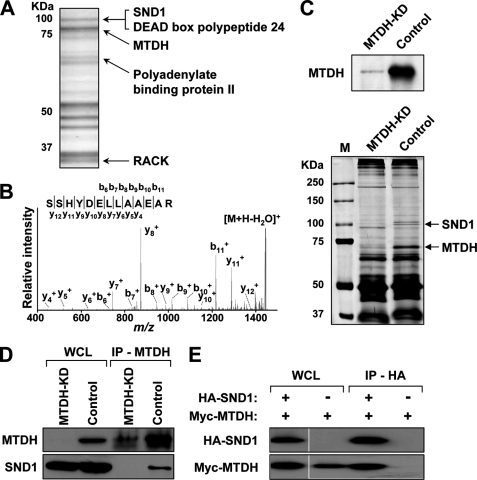
To search for novel proteins in MTDH-associated signaling pathways, we used two approaches to identify candidate MTDH-interacting proteins. In the first method, an MTDH-eGFP fusion protein was created and stably overexpressed in LM2 cells. Whole cell lysates of LM2-MTDH-eGFP were subjected to immunoprecipitation with a highly specific anti-eGFP antibody (30) and separated by SDS-PAGE. Coomassie-stained bands (Fig. 1A) were subjected to MALDI-TOF MS/MS, and multiple candidate MTDH-interacting proteins were determined, including the 100-kDa staphylococcal nuclease and Tudor domain-containing protein 1 (p100/SND1/TSN), receptor of activated protein kinase C (RACK1), and several DEAD box-containing proteins (Fig. 1, A and B). In the second approach, to more specifically determine candidate MTDH-interacting partners, whole cell lysates of either constitutive MTDH knockdown (KD) or vector control LM2 cells (an aggressively lung-metastatic subline of MDA-MD-231) (29) were subjected to anti-MTDH immunoprecipitation followed by SDS-PAGE separation. Silver staining of the SDS-PAGE gel revealed two bands clearly present in the vector control IP but absent in the MTDH-KD IP, suggesting that their pulldown was specific to the presence of MTDH (Fig. 1C). These two bands were cut, digested, and subjected to LC-MS/MS analysis for protein identification. One of the proteins was again determined to be SND1.
FIGURE 1.
Identification of SND1 as MTDH-interacting partner. A, lysates from LM2-MTDH-eGFP cells were immunoprecipitated with anti-eGFP and subjected to SDS-PAGE and Coomassie staining. Bands were sequenced by MALDI MS, and the indicated proteins were confirmed by MS/MS. B, MS/MS confirmation of an SND1 peptide. C, lysates from control or MTDH-KD LM2 cells (top) were immunoprecipitated with anti-MTDH and subjected to SDS-PAGE and silver staining (bottom). Bands specific to control cells (arrows) were identified by LC-MS/MS. D, lysates from control or MTDH-KD LM2 cells were immunoprecipitated with anti-MTDH and immunoblotted with the indicated antibodies. E, 293T cells were transfected with Myc-MTDH and either HA-SND1 or eGFP, and lysates were immunoprecipitated with anti-HA and immunoblotted with the indicated antibodies. WCL, whole cell lysate; RACK, receptor of activated protein kinase C; M, molecular mass standards.
To confirm the interaction between MTDH and SND1, we performed co-immunoprecipitation experiments. Anti-MTDH IPs of either MTDH-KD or control LM2 cells were immunoblotted with anti-SND1, and SND1 was detected only in the IPs of control (MTDH-expressing) cells (Fig. 1D). To perform the reciprocal co-IP experiment, HA-SND1 and Myc-MTDH overexpression constructs were transiently transfected in 293T cells and subjected to anti-HA IP for SND1 pulldown. Anti-Myc immunoblotting for MTDH indicated that Myc-MTDH was pulled down along with HA-SND1 but not with eGFP-transfected control IPs (Fig. 1E).
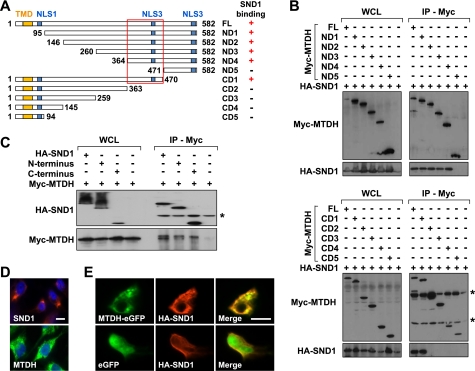
MTDH has no predicted conserved structural domains and does not have significant homology to any known proteins. Thus, an unbiased series of 10 Myc-tagged MTDH deletion mutant constructs was created and used for domain mapping experiments (Fig. 2A). 293T cells were co-transfected with HA-SND1 and either a Myc-MTDH construct (full length or deletion mutant) or eGFP (negative control) and immunoprecipitated with anti-Myc. HA-SND1 was observed to co-immunoprecipitate with full-length Myc-MTDH, with N-terminal MTDH deletion mutants 1–4 (Fig. 2B, upper panel), and with the first C-terminal MTDH deletion mutant (Fig. 2B, lower panel). Thus, the region required for interaction of MTDH and SND1 maps to a 107-amino acid stretch (amino acids 364–470) in the middle of the MTDH coding sequence (Fig. 2A, indicated by the red box).
FIGURE 2.
Mapping of MTDH and SND1 domains of interaction. A, schematic of MTDH deletion mutant coding sequences and region required for MTDH-SND1 interaction (red box). +/− indicate whether the denoted sequence interacts with SND1. The locations of the transmembrane domain (TMD; amino acids 52–74) and three putative nuclear localization signals (NLS1–3; amino acids 79–91 for NLS1, amino acids 432–451 for NLS2, and amino acids 561–580 for NLS3) are marked by yellow and blue boxes, respectively. B, 293T cells were transfected with HA-SND1 and the indicated full-length (FL) or deletion mutant Myc-MTDH construct. Lysates were immunoprecipitated with anti-Myc and immunoblotted with the indicated antibodies. * indicates nonspecific bands. C, 293T cells were transfected with Myc-MTDH and the indicated full-length or deletion mutant HA-SND1 construct. Lysates were immunoprecipitated with anti-HA and immunoblotted with the indicated antibodies. D, LM2 cells were subjected to immunofluorescence analyses with the indicated antibodies to detect endogenous MTDH or SND1. E, for co-localization, LM2 cells were co-transfected with HA-SND1 and either eGFP-MTDH (top row) or eGFP (bottom row) and subjected to anti-HA immunofluorescence. Scale bar in D and E, 10 μm. WCL, whole cell lysate.
SND1 is a multifunctional protein that contains four N-terminal SN domain repeats and a C-terminal Tudor-SN hybrid domain. The SN domains are suggested to have nuclease activity (38), whereas the Tudor-SN domain is suggested to bind methylated protein substrates (39). To test whether MTDH could interact with the SN domains, Tudor-SN domain, or both, two HA-tagged SND1 deletion mutant constructs were created, one containing the four SN repeat domains and the other containing the Tudor-SN domain. 293T cells co-transfected with Myc-MTDH and either an HA-SND1 construct (full length or deletion mutant) or eGFP were immunoprecipitated with anti-HA and immunoblotted with anti-Myc. Myc-MTDH was found to co-immunoprecipitate with full-length HA-SND1 as well as with both major domains (SN repeat and Tudor-SN) of HA-SND1 (Fig. 2C).
To further explore the potential interaction between MTDH and SND1, their subcellular localizations were investigated. MTDH has been localized most commonly to the endoplasmic reticulum/perinuclear region but has also been observed in the nucleus, nucleolus, and plasma membrane (19, 20). SND1 has been localized to both the nucleus and the endoplasmic reticulum (22, 23, 25–27). Using immunofluorescence analyses in LM2 cells, endogenous MTDH and SND1 were each observed to abundantly localize to the endoplasmic reticulum region with a more diffuse signal observed in the cytoplasm (Fig. 2D). For co-localization analyses, HA-SND1 and either MTDH-eGFP or eGFP were co-transfected into LM2 cells and imaged. Significant co-localization was observed for HA-SND1 and MTDH-eGFP with the overexpressed proteins localizing in punctate patterns to the endoplasmic reticulum/cytoplasm (Fig. 2E).
SND1 Globally Regulates Metastatic and Oncogenic Gene Signatures
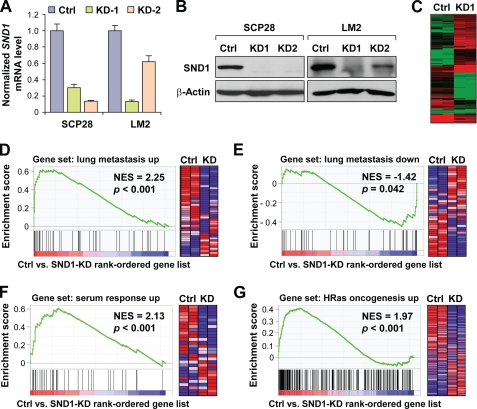
SND1 has been reported to have several different molecular roles. To explore the potential functionality of SND1 in the current context as a novel MTDH-interacting protein in breast cancer, we sought to manipulate SND1 expression and then perform transcriptomics analysis. SND1 was stably knocked down by two different shRNAs in the MDA-MB-231 sublines LM2 and SCP28 (Fig. 3, A and B). Microarray analyses were performed on RNA from control and SND1-KD1 SCP28 cell lines in duplicate. Widespread transcriptional changes were observed in SND1-KD cells with 132 unique genes showing a greater than 2-fold average change in expression (Fig. 3C).
FIGURE 3.
SND1 globally regulates metastatic and oncogenic signaling transcriptomic programs. A and B, SND1 was stably knocked down via shRNA in SCP28 and LM2 cells. The efficiency of SND1-KD was assessed at the RNA level via quantitative RT-PCR (A) and at the protein level via anti-SND1 immunoblotting (B). C, RNA harvested from vector or SND1-KD SCP28 cells was used for microarray analyses. Differentially expressed genes (n = 132) were used for hierarchical clustering and are displayed as a heat map. D–G, microarray data were used for GSEA. All gene sets from the C2 collection of the Molecular Signatures Database v3.0 passing the size threshold criteria (n = 2,484) were tested for enrichment in the list of genes ranked by expression change in SND1-expressing control SCP28 cells versus SND1-KD1 cells. GSEA plots of strongly enriched gene sets of relevance to metastasis and oncogenesis are displayed. The right panel in each figure shows the corresponding heat map of the differential expression of gene sets in the control and SND1-KD SCP28 cells. Ctrl, control; NES, normalized enrichment score. Data represent average ± S.E.
As the expression of a large number of genes was affected by SND1 silencing, we next investigated which functional classes of genes were overrepresented in the SND1-KD signature. Gene ontology category enrichment analyses indicated that the dominant functional classes of genes on the signature were those pertaining to secretion, extracellular matrix functionality, and growth (data not shown). To more robustly investigate potential signaling activities affected by SND1 knockdown, we performed GSEA to test groups of functionally related genes for their degrees of global up- or down-regulation following SND1 knockdown. Gene sets tested for enrichment included all sets from the C2 collection of the GSEA Molecular Signatures Database v3.0 as well as several gene sets added manually that were derived from published microarray signatures. The Molecular Signatures Database C2 collection includes a total of 3,272 gene sets, 2,484 of which passed size filtering criteria and were tested for enrichment. These gene sets are a combination of curated and canonical signaling pathways as well as microarray signatures obtained from experiments involving chemical and genetic perturbations. The list of gene sets globally enriched in SND1-expressing control cells versus SND1-KD cells was strikingly dominated by those involving genes up-regulated in some component of metastatic or oncogenic signaling. Specifically, of the 55 gene sets significantly enriched (i.e. down-regulated in SND1-KD) at p < 0.01 and a false discovery rate <0.25, almost one-third (n = 17) fit this category, including four of the top five most enriched gene sets overall (Table 1). Most notably, the most highly enriched gene set overall (normalized enrichment score, 2.25) was the 48-gene lung metastasis signature of genes most overexpressed in the process of breast-to-lung metastasis (29) (Fig. 3D). Conversely, genes that are underexpressed in lung metastasis were enriched in SND1-KD cells (Fig. 3E). Investigation of the “enrichment core” (Table 2) of the lung metastasis gene set revealed several known mediators of lung metastasis and/or chemoresistance that are part of the greater SND1-KD signature. For example, ANGPTL4, ID1, and EREG have been shown previously to promote breast cancer metastasis to the lungs specifically (40–42), and ALDH3A1 has been shown to be regulated by MTDH and functional in promoting MTDH-mediated chemoresistance (6). Altogether, transcriptomics analyses suggested that knockdown of SND1 leads to broad down-regulation of various transcriptional programs that may promote oncogenesis and metastasis in general and lung metastasis in particular.
TABLE 1.
SND1 global regulation of metastatic and oncogenic signaling programs
GSEA of control versus SND1-KD microarray analysis indicated 55 gene sets significantly down-regulated in the SND1-KD condition (p < 0.01 and false discovery rate (FDR) <0.25). Significantly down-regulated gene sets pertinent to metastasis and/or oncogenic signaling (n = 17) are shown. Gene sets tested for enrichment were obtained from the C2 collection provided in the Molecular Signatures Database v3.0 of the Broad Institute GSEA server. Nom, nominal; NES, normalized enrichment score; DMOG, dimethyloxalyl glycine.
| Gene set | Size | NES | Nom p value | FDR q value |
|---|---|---|---|---|
| Minn lung metastasis up-regulated genes | 43 | 2.25 | <0.001 | 0.006 |
| Amit EGF response 40 HeLa | 38 | 2.14 | <0.001 | 0.015 |
| Amit serum response 40 MCF10A | 28 | 2.13 | <0.001 | 0.013 |
| Bild HRas oncogenic signature | 220 | 1.97 | <0.001 | 0.076 |
| Amit EGF response 120 MCF10A | 39 | 1.83 | <0.001 | 0.117 |
| Nagashima EGF signature up | 52 | 1.82 | <0.001 | 0.116 |
| Elvidge hypoxia by DMOG up | 114 | 1.77 | <0.001 | 0.156 |
| Tomlins prostate cancer up | 34 | 1.77 | <0.001 | 0.151 |
| Mense hypoxia up | 80 | 1.77 | <0.001 | 0.149 |
| Elvidge hypoxia up | 147 | 1.72 | <0.001 | 0.162 |
| Farmer breast cancer cluster 7 | 19 | 1.72 | 0.005 | 0.160 |
| Wang Barrett esophagus cancer up | 25 | 1.70 | 0.005 | 0.179 |
| Winter hypoxia metagene | 215 | 1.67 | <0.001 | 0.191 |
| Croonquist NRas vs. stromal stimulation up | 33 | 1.64 | 0.006 | 0.211 |
| Rickman head and neck cancer | 78 | 1.64 | 0.003 | 0.213 |
| Winter hypoxia up | 81 | 1.61 | <0.001 | 0.235 |
| Wu cell migration | 166 | 1.59 | <0.001 | 0.244 |
TABLE 2.
Enrichment core of SND1-regulated lung metastasis signature genes
The lung metastasis signature gene set (29) is highly enriched in the control versus SND1-KD microarray GSEA-ranked list. Shown are the “core enrichment” genes for this GSEA plot (defined as those genes ranked at or higher than the gene at which the enrichment score peaks).
| Gene symbol | Rank in gene list | Rank metric score |
|---|---|---|
| C10orf116 | 13 | 2.79 |
| ANGPTL4 | 67 | 2.04 |
| ALDH3A1 | 165 | 1.60 |
| LTBP1 | 178 | 1.57 |
| KYNU | 204 | 1.53 |
| EREG | 207 | 1.53 |
| JAG1 | 256 | 1.46 |
| EMP1 | 286 | 1.41 |
| ETV1 | 495 | 1.18 |
| PTGS2 | 690 | 1.06 |
| LAPTM5 | 927 | 0.95 |
| ID1 | 1,236 | 0.86 |
| SLCO4A1 | 1,297 | 0.85 |
| SOX4 | 1,425 | 0.82 |
| QPCT | 1,997 | 0.70 |
| PTPRN2 | 2,864 | 0.58 |
| APOBEC3G | 3,372 | 0.53 |
| KRT81 | 3,836 | 0.49 |
| IL13RA2 | 4,477 | 0.44 |
| NR2F1 | 4,552 | 0.44 |
SND1 Promotes Experimental Lung Metastasis in Vivo
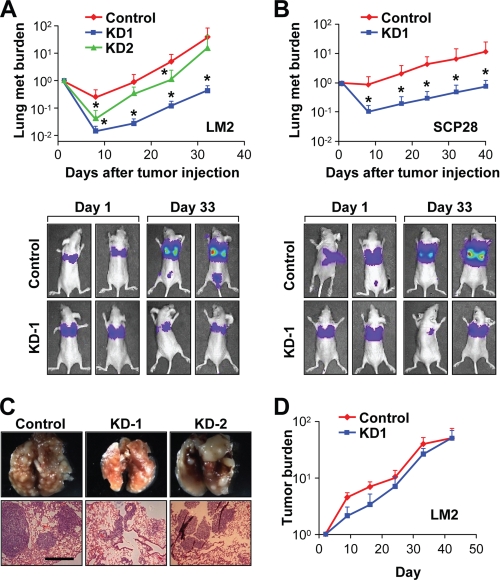
Given the bioinformatics prediction of the role of SND1 in organ-specific metastasis to lung, we thus decided to evaluate the role of SND1 in breast cancer lung metastasis. To this end, we performed experimental lung metastasis assays in nude mice using the LM2 cell line, a highly lung-metastatic subline of MDA-MB-231 (29). Two knockdown cell lines with different targeting shRNA sequences were used along with control cells. Following tail vein injections of tumor cells prelabeled with a luciferase-expressing construct, metastatic progression in the lungs was followed via quantitative bioluminescence imaging over the course of 5 weeks. SND1-KD cell lines had moderately to dramatically reduced metastatic burdens at 1 week postinjection, and these differences were maintained throughout the experiment (Fig. 4, A and C). LM2-SND1-KD-1 showed a dramatic reduction (>10-fold) in metastatic burden that was highly significant (p < 0.01, Mann-Whitney U test) at all time points. LM2-SND1-KD-2 also showed a decreased metastatic burden, although the reduction was intermediate between LM2-KD-1 and LM2-control with the difference being significant at two of four time points. Notably, quantitative RT-PCR and Western blot data (Fig. 3, A and B) indicated that SND1-targeting shRNA 1 was significantly more efficient than shRNA 2 in LM2 cells; thus, the magnitude of the loss of metastasis phenotype correlated with the efficiency of SND1 knockdown. To test whether SND1 knockdown could affect lung metastasis in a different cell line, experimental metastasis assays were repeated in SCP28 cells, which effectively colonize the lung albeit less aggressively than LM2 cells (43). Here again, SND1-KD led to a dramatic (>10-fold; p < 0.01, Mann-Whitney U test) reduction in metastatic burden at all time points over the course of 6 weeks (Fig. 4B).
FIGURE 4.
SND1 promotes lung metastasis in vivo. A and B, control and SND1-KD LM2 (A) and SCP28 (B) cells were xenografted into nude mice for experimental lung metastasis assays. Metastatic (met) burden was quantified by weekly whole body bioluminescence imaging. Representative images of mice from initial and late time points are displayed (lower panels). *, p < 0.05. C, whole lungs of mice used in LM2 experimental lung metastasis assays (top) and H&E staining of lung nodules (bottom). D, control and SND1-KD LM2 cells were used for mammary fat pad injections in nude mice, and primary tumor progression was measured over time via bioluminescence imaging and normalized to the signals at the start of the analysis. Scale bar in C, 500 μm. Data represent average ± S.E.
To test whether SND1-KD affected primary tumor growth, control and knockdown LM2 cell lines that were stably labeled with a luciferase gene were injected into the mammary fat pads of nude mice and subjected to bioluminescence imaging over the course of 6 weeks. Marginal reductions in primary tumor burden were observed in SND1-KD cells as compared with control cells; however, these differences did not reach statistical significance at any time points (Fig. 4D).
SND1 Functions to Oppose Apoptosis and Regulate Metastasis and Chemoresistance Genes
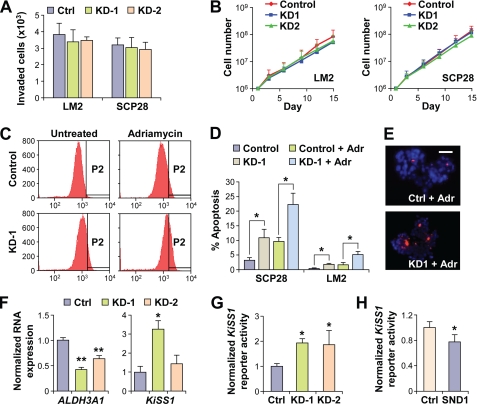
As a novel metastasis gene, SND1 could be promoting this phenotype in several possible ways. We first tested whether SND1 could affect proliferation or invasion by performing in vitro proliferation assays and modified Boyden chamber invasion assays. No significant differences were observed between control and SND1-KD cells in either assay in LM2 or SCP28 cells (Fig. 5, A and B). After ruling out proliferation and invasion as mechanisms that could explain the observed phenotypes, we next investigated the role of SND1 in apoptosis. MTDH has been shown in multiple studies to promote chemoresistance (6, 10–12) and apoptosis (13), and SND1 has been shown previously to be a substrate of caspase 3 with SND1 cleavage moderately sensitizing cells to apoptosis (27). To test whether SND1 could be playing a protective role in LM2 or SCP28 cells, we assessed basal and chemotherapeutic drug-induced rates of apoptosis via flow cytometry-mediated M30 CytoDEATH assays in control and SND1-KD1 SCP28 and LM2 cells. We found that silencing of SND1 led to significant sensitization of cells to apoptosis. In both cell lines, SND1 knockdown led to a robust 2–3-fold increase in apoptosis rates as compared with control both in basal and drug-induced states (Fig. 5, C–E).
FIGURE 5.
SND1 suppresses apoptosis and regulates expression of ALDH3A1 and KiSS1. A, modified Boyden chamber invasion assays using control or SND1-KD LM2 and SCP28 cells. B, in vitro growth curves of control or SND1-KD LM2 and SCP28 cells. C–E, control or SND1-KD LM2 and SCP28 cells were treated with either Adriamycin (Adr) or ethanol (control) and stained for apoptosis induction with M30 CytoDEATH. Flow cytometry (C) and immunofluorescence analyses (E) were used to monitor the extent of apoptosis induction. P2 indicates the apoptosis-positive populations. D, quantified data from apoptosis assays based on flow cytometry. F, expression of ALDH3A1 and KiSS1 in control or SND1-KD LM2 cells was determined via quantitative RT-PCR. G and H, KiSS1 promoter activity was assessed via a luciferase reporter assay of control versus SND1-KD LM2 cells (G) or control versus HA-SND1-transfected 293T cells (H). Scale bar in E, 50 μm. *, p < 0.05; **, p < 0.01. Ctrl, control. Data represent average ± S.E.
To identify potential mediators of SND1 phenotypes, we assessed the SND1-KD microarray profile data for the most relevant genes that could be under SND1 regulation. In the GSEA enrichment core of lung metastasis signature genes most strongly affected by SND1-KD, we observed one gene that has also been reported to be involved in promoting MTDH-mediated chemoresistance: ALDH3A1. To confirm SND1 regulation of ALDH3A1 expression, we performed quantitative RT-PCR for ALDH3A1 expression in SND1-KD lines. The ALDH3A1 expression level was decreased by roughly 2-fold in both SND1-KD lines as compared with control LM2 cells (Fig. 5F), highlighting its potential as a candidate mediator of the SND1 and MTDH prosurvival, prometastasis phenotypes. In addition to the global enrichment of the lung metastasis gene set in SND1-expressing cells, microarray data also revealed that the metastasis suppressor gene KiSS1 (44–47) was more than 6-fold overexpressed in SCP28-SND1-KD cells, making it the third most highly up-regulated gene in SND1-KD cells as compared with control cells. To confirm more rigorously the negative regulation of KiSS1 expression by SND1, we first validated microarray results via quantitative RT-PCR. KiSS1 expression was clearly shown to be up-regulated in both SND1-KD LM2 cell lines, albeit to differing extents depending on the efficiency of SND1-KD (Fig. 5F). To directly test regulation of the KiSS1 promoter by SND1, we performed KiSS1 promoter-luciferase reporter assays in both SND1-KD and overexpression conditions. SND1-KD led to a roughly 2-fold increase in KiSS1 promoter activity in LM2 cells (Fig. 5G), and transient transfection of SND1 into 293T cells led to a modest, but significant, decrease in KiSS1 reporter activity (Fig. 5H).
SND1 Expression Is Associated with Lung Metastasis and Reduced Metastasis-free Survival in Multiple Clinical Data Sets
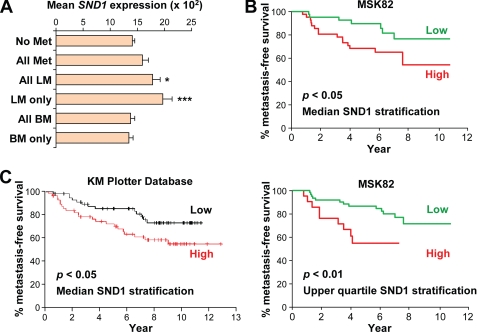
To investigate whether SND1 could be involved in promoting breast-to-lung metastasis in cases of clinical human breast cancer, we sought to investigate the expression patterns of SND1 in microarray data of primary tumors of breast cancer patients with known metastatic outcome. We first chose the previously published MSK-82 (29) data set for analysis as it is the only data set we were able to obtain that contains clinical, organ-specific metastasis data. In this data set, SND1 expression was found to be only slightly (and non-significantly) increased in expression when considering all patients who suffered metastatic relapse as compared with those who remained disease-free (Fig. 6A). However, mean SND1 expression was significantly higher (p < 0.01, Student's t test) in patients who developed metastasis specifically to the lungs (i.e. either lung metastasis only or lung and other organ metastasis). Furthermore, mean SND1 expression was higher still in patients who developed metastasis to the lungs only (without concomitant metastases at other sites) (p < 0.001, Student's t test) (Fig. 6A). In contrast, there was no increase in mean SND1 expression in patients who developed bone metastases as compared with those with no metastasis. To test whether SND1 was relevant to overall metastasis-free survival, we stratified patients by either the median or upper quartile level of SND1 expression and determined the correlation with metastasis-free survival by Kaplan-Meier curve analysis. Patients with greater than median expression of SND1 had significantly shorter overall metastasis-free survival (p < 0.05, log rank test) (Fig. 6B, upper panel), and patients with upper quartile SND1 expression had more dramatically and highly significant reduced metastasis-free survival as compared with all others (p < 0.01, log rank test) (Fig. 6B, lower panel). To investigate the correlation between SND1 expression and survival in an independent data set, we used the Kaplan-Meier Plotter database (37), which is a meta-analysis of many clinical data sets and includes microarray data from primary tumors of 1,908 breast cancer patients. In lymph node-positive patients, above median SND1 expression was significantly correlated with reduced distant metastasis-free survival (p = 0.042, log rank test) (Fig. 6C). Taken together, these data suggest that SND1 expression is relevant to breast cancer metastasis, particularly to the lungs, in both the clinical as well as the experimental setting.
FIGURE 6.
High SND1 expression correlates with increased risk of metastasis in breast cancer. A, expression of SND1 in primary tumors is shown in relevant subsets of breast cancer patients in the MSK-82 data set. Subsets include patients with either no metastasis (No Met) or metastasis to any organ (All Met), lung non-exclusively (All LM), lung exclusively (LM only), bone non-exclusively (BM), or bone exclusively (BM only). *, p < 0.05; ***, p < 0.001. B, Kaplan-Meier plot of metastasis-free survival of patients in the MSK-82 data set stratified by median (left) or upper quartile (right) SND1 expression. C, Kaplan-Meier plot of distant metastasis-free survival of node-positive patients stratified by median SND1 expression in the Kaplan-Meier Plotter breast cancer meta-analysis database. Data represent average ± S.E.
DISCUSSION
MTDH/AEG1 has generated high levels of interest in recent years as it has been shown to promote experimental lung metastasis as well as other oncogenic phenotypes of interest through incompletely understood mechanisms (19, 48). To date, investigations of how MTDH promotes such powerful phenotypes have generally focused on MTDH signaling in the context of classical oncogenic pathways, such as Ha-Ras (15), PI3K/AKT (13), ERK, and Wnt/β-catenin (8), and NF-κB (49). However, comparatively fewer protein-level data have been obtained regarding functional MTDH-interacting partners. Three MTDH-interacting proteins have been previously verified. MTDH was first shown to physically interact with the NF-κB subunit p65 (49) with the region of interaction later being mapped to MTDH amino acids 101–205 (16). Separate yeast two-hybrid-based approaches identified the transcriptional repressor PLZF (21) and the tumor suppressor BCCIPα (50) as additional MTDH-interacting proteins. The MTDH-BCCIPα interaction was mapped to MTDH amino acids 72–169, whereas PLZF was shown to interact with two regions of MTDH, amino acids 1–285 and 487–582. Additionally, MTDH interaction was shown to augment the transcriptional activity of NF-κB, oppose the transcriptional repression of PLZF, and target BCCIPα for proteosomal degradation. In a different approach, using an in vivo phage display method, Brown and Ruoslahti (3) identified an MTDH fragment, amino acids 378–440, as a mediator of attachment to the lung endothelium. This “lung-homing domain” was suggested to mediate interaction between surface-bound MTDH and a receptor protein expressed in the lung endothelium. However, the putative MTDH receptor has not yet been identified. Here we used unbiased screening and mass spectrometry-based approaches to identify SND1 as an MTDH-interacting protein. We have mapped the region of interaction to amino acids 364–470 of the MTDH coding sequence, which is very similar to the proposed MTDH lung-homing domain, highlighting this region as crucial for MTDH interactions and functionality. Through functional, in vivo analyses, we have further demonstrated that SND1 is in its own right a novel metastasis gene. Mechanistically, we have shown that SND1 functions to oppose apoptosis and also regulate the expression of genes associated with metastasis and chemoresistance.
SND1 was first identified as a transcriptional co-activator that could interact with EBNA-2 and help facilitate its transcriptional activity (51). Subsequent studies investigated the role of SND1 as a co-activator in other transcriptional programs, finding that it could enhance transcriptional activity of c-Myb (52), STAT5 (53), and STAT6. In contrast, several studies have also described SND1 as a mediator of multiple post-transcriptional processes via interactions with RNA and RNA-associated proteins. SND1 has been shown to associate with the U5 small nuclear ribonucleoprotein component of the spliceosome (39) and enhance spliceosome assembly and activity (26). A different approach identified SND1 as a component of the RNA-induced silencing complex and further determined SND1 to harbor broad scale (RNA and DNA) nuclease activity (25). Interestingly, SND1 was also shown to bind and degrade certain edited (adenosine to inosine) double-stranded RNA molecules after the editing-induced failure of Drosha recognition and processing (23, 24). Other molecular roles for cytoplasmic SND1 have been reported. Biochemical approaches demonstrated that SND1 is a cleavage substrate for caspase 3 and that inhibition of this cleavage reduces the overall efficiency of the programmed cell death process (27). Further implicating SND1 in cellular stress response programs, it was recently shown that SND1 interacts with the stress granule marker G3BP and that the two proteins strongly co-localize to stress granules upon induction of various cellular stresses (54).
Our current study provides several lines of evidence to link SND1 with lung metastasis and chemoresistance of breast cancer. First, GSEA analysis of the SND1-KD signature indicated very strong enrichment of lung metastasis signature genes, suggesting the possibility of global SND1 regulation of this transcriptional program. Specifically, several genes validated to promote different aspects of lung metastasis resided in the enrichment core of lung metastasis genes with the most significant elevation of expression in SND1-expressing control cells as compared with SND1-KD cells. For example, TGF-β-induced ANGPTL4 has been shown to enhance extravasation of breast cancer cells in lung metastasis (41), EREG has been shown to promote extravasation and lung colonization (42), and Id1 functions to promote the tumor initiation stage of lung metastasis colonization (40). KiSS1, although not part of the lung metastasis signature, has nonetheless been reported to repress metastasis (to the lung and other organs) in many tumor types. It has been suggested that KiSS1 antimetastasis functionality is rooted in suppression of motility and invasion (55). In addition to the potentially concerted regulation of such pro-lung metastasis genes, SND1 also promotes evasion of apoptosis. Apoptotic resistance is crucial for distant metastasis as breast cancer cells must survive for extended periods of time in the circulatory system to reach distant organs, such as the lung, a condition in which they are exposed to high risk of anoikis (56). A particularly interesting gene in the SND1-regulated lung metastasis enrichment core is ALDH3A1, which has been shown to promote resistance to various chemotherapeutic agents and other cell death-inducing toxins (57–59). Importantly, ALDH3A1 was also shown previously to promote MTDH-mediated resistance to chemotherapy-induced apoptosis (6). Thus, we envision the possibility that SND1 exerts its overall prometastatic phenotype via a combinatorial mechanism: antiapoptosis activity and gene regulation may allow survival in the circulation and resistance to chemotherapy, whereas regulation of multiple genes involved in both general and lung-specific seeding, extravasation, and colonization may allow directed metastasis to the lungs.
It is not known how SND1, in possible cooperation with MTDH, achieves this concerted regulation of prometastatic and antiapoptotic genes. However, likely possibilities are via transcriptional co-activation or post-transcriptional modulation of RNAs. Both SND1 and MTDH have been previously reported to possess a context-dependent transcriptional co-activation ability, and here we show via reporter assays that SND1 can activate the KiSS1 promoter. Thus, it is possible that SND1 and MTDH may be components of transcriptional complexes that regulate genes relevant to metastasis, apoptosis, and chemoresistance. Alternatively, SND1 has also been shown to be involved in RNA-induced silencing complex activity and degradation of edited double-stranded RNA molecules. Furthermore, MTDH was reported to post-transcriptionally regulate translation of the multidrug resistance protein 1 (MDR1) (11). Therefore, cytoplasmic interactions between MTDH and SND1 may regulate RNA silencing and/or editing machinery to modulate RNA stability, translation, or functionality. Future studies will be required to test these possibilities and further explore the molecular cooperation between SND1 and MTDH in promoting lung metastasis and chemoresistance.
Acknowledgments
We thank C. DeCoste for assistance with flow cytometry; M. Montminy for the gift of the KiSS1 reporter plasmid; M. Yuan, L. Wan, and R. Chakrabarti for technical assistance; and G. LeRoy, G. Hu, M. Korpal, N. Sethi, X. Lu, B. Tiede, and other members of our laboratory for insightful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA134519 (to Y. K.) and a National Research Service Award predoctoral fellowship (to M. A. B.). This work was also supported by grants from the United States Department of Defense and the Brewster Foundation.
- MTDH
- Metadherin
- SND1
- staphylococcal nuclease domain-containing 1
- SN
- staphylococcal nuclease
- eGFP
- enhanced GFP
- IP
- immunoprecipitation
- GSEA
- Gene Set Enrichment Analysis
- KD
- knockdown
- PLZF
- promyelocytic leukemia zinc finger
- BCCIPα
- BRCA2 and CDKN1A interacting protein alpha.
REFERENCES
- 1. Jemal A., Siegel R., Xu J., Ward E. (2010) CA Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Nguyen D. X., Bos P. D., Massagué J. (2009) Nat. Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 3. Brown D. M., Ruoslahti E. (2004) Cancer Cell 5, 365–374 [DOI] [PubMed] [Google Scholar]
- 4. Kang D. C., Su Z. Z., Sarkar D., Emdad L., Volsky D. J., Fisher P. B. (2005) Gene 353, 8–15 [DOI] [PubMed] [Google Scholar]
- 5. Britt D. E., Yang D. F., Yang D. Q., Flanagan D., Callanan H., Lim Y. P., Lin S. H., Hixson D. C. (2004) Exp. Cell Res. 300, 134–148 [DOI] [PubMed] [Google Scholar]
- 6. Hu G., Chong R. A., Yang Q., Wei Y., Blanco M. A., Li F., Reiss M., Au J. L., Haffty B. G., Kang Y. (2009) Cancer Cell 15, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emdad L., Lee S. G., Su Z. Z., Jeon H. Y., Boukerche H., Sarkar D., Fisher P. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21300–21305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoo B. K., Emdad L., Su Z. Z., Villanueva A., Chiang D. Y., Mukhopadhyay N. D., Mills A. S., Waxman S., Fisher R. A., Llovet J. M., Fisher P. B., Sarkar D. (2009) J. Clin. Investig. 119, 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu L., Wu J., Ying Z., Chen B., Han A., Liang Y., Song L., Yuan J., Li J., Li M. (2010) Cancer Res. 70, 3750–3759 [DOI] [PubMed] [Google Scholar]
- 10. Wei Y., Hu G., Kang Y. (2009) Cell Cycle 8, 2132–2133 [PubMed] [Google Scholar]
- 11. Yoo B. K., Chen D., Su Z. Z., Gredler R., Yoo J., Shah K., Fisher P. B., Sarkar D. (2010) Cancer Res. 70, 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoo B. K., Gredler R., Vozhilla N., Su Z. Z., Chen D., Forcier T., Shah K., Saxena U., Hansen U., Fisher P. B., Sarkar D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12938–12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S. G., Su Z. Z., Emdad L., Sarkar D., Franke T. F., Fisher P. B. (2008) Oncogene 27, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 14. Bhutia S. K., Kegelman T. P., Das S. K., Azab B., Su Z. Z., Lee S. G., Sarkar D., Fisher P. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 22243–22248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S. G., Su Z. Z., Emdad L., Sarkar D., Fisher P. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17390–17395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarkar D., Park E. S., Emdad L., Lee S. G., Su Z. Z., Fisher P. B. (2008) Cancer Res. 68, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 17. Li J., Yang L., Song L., Xiong H., Wang L., Yan X., Yuan J., Wu J., Li M. (2009) Oncogene 28, 3188–3196 [DOI] [PubMed] [Google Scholar]
- 18. Kikuno N., Shiina H., Urakami S., Kawamoto K., Hirata H., Tanaka Y., Place R. F., Pookot D., Majid S., Igawa M., Dahiya R. (2007) Oncogene 26, 7647–7655 [DOI] [PubMed] [Google Scholar]
- 19. Hu G., Wei Y., Kang Y. (2009) Clin. Cancer Res. 15, 5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sarkar D., Emdad L., Lee S. G., Yoo B. K., Su Z. Z., Fisher P. B. (2009) Cancer Res. 69, 8529–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thirkettle H. J., Mills I. G., Whitaker H. C., Neal D. E. (2009) Oncogene 28, 3663–3670 [DOI] [PubMed] [Google Scholar]
- 22. Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002) EMBO J. 21, 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scadden A. D. (2005) Nat. Struct. Mol. Biol. 12, 489–496 [DOI] [PubMed] [Google Scholar]
- 24. Yang W., Chendrimada T. P., Wang Q., Higuchi M., Seeburg P. H., Shiekhattar R., Nishikura K. (2006) Nat. Struct. Mol. Biol. 13, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caudy A. A., Ketting R. F., Hammond S. M., Denli A. M., Bathoorn A. M., Tops B. B., Silva J. M., Myers M. M., Hannon G. J., Plasterk R. H. (2003) Nature 425, 411–414 [DOI] [PubMed] [Google Scholar]
- 26. Yang J., Välineva T., Hong J., Bu T., Yao Z., Jensen O. N., Frilander M. J., Silvennoinen O. (2007) Nucleic Acids Res. 35, 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sundström J. F., Vaculova A., Smertenko A. P., Savenkov E. I., Golovko A., Minina E., Tiwari B. S., Rodriguez-Nieto S., Zamyatnin A. A., Jr., Välineva T., Saarikettu J., Frilander M. J., Suarez M. F., Zavialov A., Ståhl U., Hussey P. J., Silvennoinen O., Sundberg E., Zhivotovsky B., Bozhkov P. V. (2009) Nat. Cell Biol. 11, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 28. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 29. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 31. Luo Y., Li T., Yu F., Kramer T., Cristea I. M. (2010) J. Am. Soc. Mass Spectrom. 21, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greco T. M., Yu F., Guise A. J., Cristea I. M. (2011) Mol. Cell. Proteomics 10, M110.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 34. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nucleic. Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 37. Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z. (2010) Breast Cancer Res. Treat. 123, 725–731 [DOI] [PubMed] [Google Scholar]
- 38. Li C. L., Yang W. Z., Chen Y. P., Yuan H. S. (2008) Nucleic Acids Res. 36, 3579–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shaw N., Zhao M., Cheng C., Xu H., Saarikettu J., Li Y., Da Y., Yao Z., Silvennoinen O., Yang J., Liu Z. J., Wang B. C., Rao Z. (2007) Nat. Struct Mol. Biol. 14, 779–784 [DOI] [PubMed] [Google Scholar]
- 40. Gupta G. P., Perk J., Acharyya S., de Candia P., Mittal V., Todorova-Manova K., Gerald W. L., Brogi E., Benezra R., Massagué J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19506–19511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Padua D., Zhang X. H., Wang Q., Nadal C., Gerald W. L., Gomis R. R., Massagué J. (2008) Cell 133, 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gupta G. P., Nguyen D. X., Chiang A. C., Bos P. D., Kim J. Y., Nadal C., Gomis R. R., Manova-Todorova K., Massagué J. (2007) Nature 446, 765–770 [DOI] [PubMed] [Google Scholar]
- 43. Minn A. J., Kang Y., Serganova I., Gupta G. P., Giri D. D., Doubrovin M., Ponomarev V., Gerald W. L., Blasberg R., Massagué J. (2005) J. Clin. Investig. 115, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee J. H., Miele M. E., Hicks D. J., Phillips K. K., Trent J. M., Weissman B. E., Welch D. R. (1996) J. Natl. Cancer Inst. 88, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 45. Lee J. H., Welch D. R. (1997) Cancer Res. 57, 2384–2387 [PubMed] [Google Scholar]
- 46. Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., Ishibashi Y., Watanabe T., Asada M., Yamada T., Suenaga M., Kitada C., Usuki S., Kurokawa T., Onda H., Nishimura O., Fujino M. (2001) Nature 411, 613–617 [DOI] [PubMed] [Google Scholar]
- 47. Makri A., Pissimissis N., Lembessis P., Polychronakos C., Koutsilieris M. (2008) Cancer Treat Rev. 34, 682–692 [DOI] [PubMed] [Google Scholar]
- 48. Emdad L., Sarkar D., Su Z. Z., Lee S. G., Kang D. C., Bruce J. N., Volsky D. J., Fisher P. B. (2007) Pharmacol. Ther. 114, 155–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emdad L., Sarkar D., Su Z. Z., Randolph A., Boukerche H., Valerie K., Fisher P. B. (2006) Cancer Res. 66, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 50. Ash S. C., Yang D. Q., Britt D. E. (2008) Biochem. Biophys. Res. Commun. 371, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tong X., Drapkin R., Yalamanchili R., Mosialos G., Kieff E. (1995) Mol. Cell. Biol. 15, 4735–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leverson J. D., Koskinen P. J., Orrico F. C., Rainio E. M., Jalkanen K. J., Dash A. B., Eisenman R. N., Ness S. A. (1998) Mol. Cell 2, 417–425 [DOI] [PubMed] [Google Scholar]
- 53. Paukku K., Yang J., Silvennoinen O. (2003) Mol. Endocrinol. 17, 1805–1814 [DOI] [PubMed] [Google Scholar]
- 54. Gao X., Ge L., Shao J., Su C., Zhao H., Saarikettu J., Yao X., Yao Z., Silvennoinen O., Yang J. (2010) FEBS Lett. 584, 3525–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nash K. T., Welch D. R. (2006) Front. Biosci. 11, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpson C. D., Anyiwe K., Schimmer A. D. (2008) Cancer Lett. 272, 177–185 [DOI] [PubMed] [Google Scholar]
- 57. Moreb J. S., Mohuczy D., Ostmark B., Zucali J. R. (2007) Cancer Chemother. Pharmacol. 59, 127–136 [DOI] [PubMed] [Google Scholar]
- 58. Sládek N. E., Kollander R., Sreerama L., Kiang D. T. (2002) Cancer Chemother. Pharmacol. 49, 309–321 [DOI] [PubMed] [Google Scholar]
- 59. Townsend A. J., Leone-Kabler S., Haynes R. L., Wu Y., Szweda L., Bunting K. D. (2001) Chem. Biol. Interact. 130–132, 261–273 [DOI] [PubMed] [Google Scholar]