Abstract
Myotubularin is a 3-phosphoinositide phosphatase that is mutated in X-linked myotubular myopathy, a severe neonatal disorder in which skeletal muscle development and/or regeneration is impaired. In this report we provide evidence that siRNA-mediated silencing of myotubularin expression markedly inhibits growth factor-stimulated Akt phosphorylation, leading to activation of caspase-dependent pro-apoptotic signaling in HeLa cells and primary human skeletal muscle myotubes. Myotubularin silencing also inhibits Akt-dependent signaling through the mammalian target of rapamycin complex 1 as assessed by p70 S6-kinase and 4E-BP1 phosphorylation. Similarly, phosphorylation of FoxO transcription factors is also significantly reduced in myotubularin-deficient cells. Our data further suggest that inhibition of Akt activation and downstream survival signaling in myotubularin-deficient cells is caused by accumulation of the MTMR substrate lipid phosphatidylinositol 3-phosphate generated from the type II phosphatidylinositol 3-kinase PIK3C2B. Our findings are significant because they suggest that myotubularin regulates Akt activation via a cellular pool of phosphatidylinositol 3-phosphate that is distinct from that generated by the type III phosphatidylinositol 3-kinase hVps34. Because impaired Akt signaling has been tightly linked to skeletal muscle atrophy, we hypothesize that loss of Akt-dependent growth/survival cues due to impaired myotubularin function may be a critical factor underlying the severe skeletal muscle atrophy characteristic of muscle fibers in patients with X-linked myotubular myopathy.
Keywords: Akt PKB, Muscular Dystrophy, Phosphatidylinositol Phosphatase, Signal Transduction, Skeletal Muscle
Introduction
Myotubularin is the archetypical member of a family of phosphoinositide 3-phosphatases. Mutations in the myotubularin gene are causative for X-linked recessive myotubular myopathy (XLMTM),4 also known as “centronuclear myopathy.” XLMTM is a severe neonatal disorder in which the maturation and/or regeneration of skeletal muscle fibers is compromised (1, 2). Myofibers from affected individuals display abnormal centrally located nuclei and are severely atrophic. Mice in which the myotubularin gene has been deleted exhibit a skeletal muscle phenotype consistent with that of XLMTM disease. Like XLMTM patients, early stages of myogenesis in these mice appear to occur normally, and it has been suggested that skeletal muscle maintenance or regeneration is affected rather than early myogenic differentiation (3). Mutations in two other MTMR family proteins, MTMR2 and MTMR13, are causative for the demyelinating peripheral neuropathy type 4B Charcot-Marie-Tooth disease (4–6).
The myotubularin-related protein (MTMR) family consists of 14 proteins including 8 catalytically active lipid phosphatases and 6 forms that are inactive due to germ-line substitutions of essential catalytic residues (7). Active MTMRs specifically remove the 3-phosphate moiety from phosphatidylinositol 3-phosphate and phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), generating phosphatidylinositol (PI) and phosphatidylinositol 5-phosphate (PI(5)P), respectively (8–10). PI(3)P and PI(3,5)P2 play critical roles in membrane trafficking processes including trafficking through early/late endosomes and multivesicular bodies, lysosomal transport, trafficking to the trans-Golgi network, and autophagy (10–13). Recent work has linked myotubularin and/or MTMR2 to membrane remodeling and internalization and turnover of growth factor receptors (14–17). However, specific mechanisms of how impaired myotubularin function, aberrant levels of its substrate/product phosphoinositides, or altered trafficking events can lead to XLMTM disease have yet to be identified. Interestingly, recent studies have shown that MTMR6 can regulate KCa3.1 ion channels by a mechanism that is dependent on PI(3)P but not directly associated with alterations in early endosomal trafficking, suggesting that important functions exist for PI(3)P in addition to its roles in membrane trafficking (18, 19).
A growing body of evidence has linked MTMRs to cell survival and/or apoptotic processes. MTMR1, MTMR6, MTMR7, and MTMR8 active lipid phosphatases and MTMR12 (inactive) were identified as “survival” phosphatases using a gene silencing approach in HeLa cells that targeted all human kinases and phosphatases (20). In this study, depletion of MTMR7 or MTMR12 led to proteolytic cleavage and activation of the pro-apoptotic markers caspase-9 and poly(ADP-ribose) polymerase (PARP). In addition, MTMR2 knockdown has been shown to inhibit proliferation and to promote caspase-dependent apoptosis in cultured primary rat Schwann cells (21). Moreover, silencing of MTMR6 and MTMR9 in HeLa cells potentiated etoposide-induced apoptosis, whereas overexpression of these proteins blunted the effect of etoposide treatment (22). A mechanism to explain how MTMRs or their lipid substrates and/or products promote cell survival and oppose apoptosis has not been determined. However, these observations are part of an emerging picture that implicates important roles for MTMRs in regulating cell growth and survival.
In this work we have investigated the role of myotubularin as a potential regulator of cell survival and pro-apoptotic signaling. We find that myotubularin silencing in HeLa cells robustly activates caspase-dependent pro-apoptotic signaling, ultimately leading to nuclear DNA fragmentation and cell death. We show that impaired phosphorylation and activation of Akt is a major factor underlying these pro-apoptotic events. As a relevant model system for myotubularin function in skeletal muscle, we demonstrate that myotubularin depletion also inhibits Akt phosphorylation, activates caspase-dependent pro-apoptotic signaling, and promotes DNA fragmentation in primary human skeletal muscle myotubes. Myotubularin-deficient myotubes further exhibit aberrant signaling via Akt-dependent pathways including the mammalian target of rapamycin complex (mTORC1) and forkhead transcription factors (FoxOs). These are critical findings because mTORC1 and FoxOs are important regulators of skeletal muscle growth and atrophy and have been linked to muscle phenotypes similar to those observed in XLMTM disease in studies using genetically altered mice (23–25). Our findings represent the first evidence linking myotubularin to specific Akt-dependent signaling pathways and suggest potential mechanisms for the muscle pathology observed in XLMTM patients.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HeLa cells were maintained at 37 °C with 5% CO2 in DMEM supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. Human primary skeletal muscle myoblasts (HsMM) were maintained at 37 °C with 5% CO2 in SkBM-2 skeletal muscle basal medium containing 10% fetal bovine serum, gentamycin sulfate, amphotericin-B, epidermal growth factor, and dexamethasone as recommended by the supplier (Lonza). For differentiation into myotubes, HsMM primary skeletal muscle myoblasts were cultured in 35-mm dishes to ∼80% confluence then shifted to differentiation medium (DMEM/F-12 containing 2% horse serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin) for 96 h. At least 95% of the cells formed multinucleated myotubes under these conditions as assessed by phase contrast microscopy.
Transfections with siRNAs were carried out using Lipofectamine RNAi-MAX (Invitrogen) according to the manufacturer's protocol. Standard siRNA transfection conditions for HeLa cells were 45 pmol of siRNA and 6 μl of RNAi-MAX transfection reagent combined in 500 μl of Opti-MEM serum-free medium (Invitrogen). This transfection mixture was added to a 35-mm dish containing ∼6.0 × 104 cells in 2.5 ml of DMEM medium with fetal bovine serum and glutamine as above but without antibiotics. The final siRNA concentration in all HeLa samples was 15 nm. HeLa cells transfected with siRNAs were incubated 48 h before harvest and sample preparation. In HeLa cell experiments where myotubularin was depleted alone or in combination with lipid kinases, the total amount of siRNA used per sample was also 45 pmol, representing 22.5 pmol (each) of control and/or mRNA-specific siRNA pools (2 siRNAs per pool). The concentration of each of the 4 individual siRNAs in these samples was 3.75 nm, for a final siRNA concentration of 15 nm.
For transfection of HsMM primary human skeletal muscle myotubes, the cells were rinsed once in 2.5 ml of differentiation medium (DMEM/F-12 containing 2% horse serum and 2 mm glutamine) without antibiotics ∼16 h before siRNA transfection, then incubated overnight in 2.5 ml of differentiation medium without antibiotics. To achieve efficient myotubularin knockdown, myotube siRNA transfections were carried out using pools of three different siRNAs. The control pool included low GC, medium GC, and GFP negative control siRNA duplexes. The myotubularin-specific pool contained MTM1-1, MTM1-2, and MTM1-3 siRNA duplexes. For these transfections, 60 pmol (total) of control or myotubularin-specific siRNA pool (3 siRNAs per pool) and 9 μl of RNAi-MAX transfection reagent were used per 35-mm dish. The total siRNA concentration in these experiments was 20 nm (6.7 nm for each siRNA), and the cells were incubated 72 h before harvest and sample preparation for immunoblot analyses.
For sequential transfections of HeLa cells with expression plasmids and siRNA, plasmid transfections were carried out first followed by siRNA transfection 36 h later. Plasmid transfections were carried out using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. After ∼6 h, the cells were rinsed once, and the medium was replaced with fresh DMEM medium containing fetal bovine serum and glutamine as above but without antibiotics. Transfections with siRNAs were carried out the following day as described above.
Plasmids and siRNAs
The plasmids encoding FLAG-tagged wild-type human myotubularin and a severe myotubular myopathy (S376N) mutant were described previously (26). Plasmids encoding myr-HA-Akt1 (#9008, William Sellers) and myr-HA-Akt2 (#9016, William Sellers) were obtained through Addgene (27). The EGFP-2xFYVE construct containing tandem FYVE domains was generously provided by Dr. Harald Stenmark (28). All silencing experiments in these studies were carried out using control or protein-specific Stealth siRNAs (Invitrogen). The following siRNA duplexes were used (low and medium GC control sequences are not available): GFP negative control, 5′-GAGACCACAUGGUCCUUCUUGAGUU-3′ (sense) and 5′-AACUCAAGAAGGACCAUGUGGUCUC-3′ (antisense); MTM1-1, 5′-GCGACGAAUACAUAAAGCGGCUUGA-3′ (sense) and 5′-UCAAGCCGCUUUAUGUAUUCGUCGC-3′ (antisense); MTM1-2, 5′-GGCACCGCAUUAGAGCUCGAAAUAA-3′ (sense) and 5′-UUAUUUCGAGCUCUAAUGCGGUGCC-3′ (antisense); MTM1-3, 5′-GGACUGUUUCUUUAUGGUCACUGAU-3′ (sense) and 5′-AUCAGUGACCAUAAAGAAACAGUCC-3′ (antisense); PIK3C2A-1, 5′-GGAAUAUGGUGUGACAGGAUCCUUU-3′ (sense) and 5′-AAAGGAUCCUGUCACACCAUAUUCC-3′ (antisense); PIK3C2A-2, 5′-GGAUGUAGCAGAGUGUGAUCUUGUU-3′ (sense) and 5′-AACAAGAUCACACUCUGCUACAUCC-3′ (antisense); PIK3C2B-1, 5′-GAGAGAUGAGGAGGUUGCUGCAUUU-3′ (sense) and 5′-AAAUGCAGCAACCUCCUCAUCUCUC-3′ (antisense); PIK3C2B-2, 5′-GGUGACUGUGUUGUGUGACAGGCUU-3′ (sense) and 5′-AAGCCUGUCACACAACACAGUCACC-3′ (antisense); Vps34-1, 5′-CCCAUGAGAUGUACUUGAACGUAAU-3′ (sense) and 5′-AUUACGUUCAAGUACAUCUCAUGGG-3′ (antisense); Vps34-2, 5′-CCUCCACCUGCGAAGGUAUUCUAAU-3′ (sense) and 5′-AUUAGAAUACCUUCGCAGGUGGAGG-3′ (antisense); PIP5K3-1, 5′-GAGGCCAGGGAGAACAGCAGCCUUU-3′ (sense) and 5′-AAAGGCUGCUGUUCUCCCUGGCCUC-3′ (antisense); PIP5K3-2, 5′-GGAGACCUCCGAGCUUGCACAUAUU-3′ (sense) and 5′-AAUAUGUGCAAGCUCGGAGGUCUCC-3′ (antisense).
Protein Sample Preparation
After siRNA treatment and protein knockdown, cells were rinsed with ice-cold PBS and lysed in ∼150 μl (per 35-mm dish) of ice-cold lysis buffer (PBS containing 0.5% Triton X-100, 0.1% deoxycholate, protease inhibitor mixture (Complete + EDTA, Roche Applied Science), 1 mm PMSF, and 2 mm benzamidine) by scraping and vortexing. Soluble protein extracts were prepared from cell lysates by centrifugation (20 min, 18,000 × g, 4 °C). Protein determinations were carried out using Bradford reagent (Bio-Rad), and soluble protein extracts were normalized with lysis buffer to contain equal protein concentration. After the addition of 5× SDS-PAGE sample buffer, the samples were boiled for 10 min, frozen in liquid nitrogen, then stored at −80 °C. For analysis of phosphorylated proteins, cells were deprived of serum in DMEM for 2 h after siRNA treatment, then stimulated with 10 nm epidermal growth factor (EGF) for 10 min or 100 nm insulin for 5 min at 37 °C to promote Akt phosphorylation. Soluble protein extracts were prepared as described above except that the lysis buffer also contained phosphatase inhibitors (10 mm NaF, 2.5 mm sodium orthovanadate, 20 nm calyculin A (Cell Signaling)) to minimize protein dephosphorylation during sample processing. Except where listed, all chemicals and reagents were obtained from Sigma.
Immunoblotting
Approximately 5–8 μg of total protein per sample were resolved by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked in 5% milk in Tris-buffered saline containing 0.1% Tween 20, then immunoblotted using the procedures recommended by Cell Signaling.
Antibodies
Myotubularin polyclonal antiserum was produced in rabbits using recombinant human myotubularin purified as described (Cocalico) (29). Total IgG was enriched from the crude antiserum by ammonium sulfate precipitation followed by affinity purification using bacterial recombinant human myotubularin coupled to an NHS Hi-Trap column (Amersham Biosciences). Myotubularin antibodies were used at a concentration of 1 μg/ml. Primary antibodies against total Akt, phospho-Akt (Thr(P)-308, Thr(P)-450, Ser(P)-473)-cleaved caspase 9, cleaved PARP, 4E-BP1, phospho-4E-BP1, p70S6K, phospho-p70S6K, FoxO1, FoxO3a, FoxO4, phospho-FoxO1/phospho-FoxO3a/phospho-FoxO4, and Vps34 were obtained from Cell Signaling and used at 1:1000 dilution. Other primary antibodies used included tubulin (Sigma, 1:1000), FLAG-M2 (Sigma, 1 μg/ml), HA (Roche Applied Science, 1 μg/ml), PIK3C2A, PIK3C2B (BD Biosciences, 1:1000), EGFP (Clontech, 1:1000), and PIP5K3 (Santa Cruz, 1:200). Donkey anti-mouse IgG and sheep anti-rabbit IgG horseradish peroxidase conjugates (Amersham Biosciences, 1:5000) were used for secondary detection. Protein bands were visualized using LumiLight and LumiLight PLUS detection reagents (Roche Applied Science). Intensities of protein bands from developed films were quantified using a Bio-Rad GelDoc EQ system and Quantity One software.
TUNEL Assay
HeLa or HsMM cells were cultured in 2-well chamber slides and transfected with siRNAs as described above and processed using the In Situ Cell Death Detection kit, TMR Red (Roche Applied Science) according to the manufacturer's protocol. Cell nuclei were stained using Hoechst 33342 (Molecular Probes). Images were captured using a Leica DMI6000B fluorescence microscope and Leica Application Suite software.
PI(3)P Mass Assay
HeLa cells were seeded in 60-mm dishes at a density of ∼2.0 × 105 cells/dish and transfected with control or myotubularin siRNAs as described above. After serum-starvation and stimulation with EGF, the cells were harvested, and PI(3)P levels in lipid extracts from the cells were quantified using a PI(3)P Mass Assay kit (Echelon Biosciences) according to the manufacturer's protocol. Lipid extract samples were normalized for equal protein, and of the total lipid extract was spotted for each sample.
Graphing and Statistical Analyses
All graphs were generated using Kaleidagraph software for MacIntosh. For statistical probability determinations, data from at least three independent experiments were analyzed using a paired, two-tailed Student's t test (Excel). The immunoblot data shown in each figure represent one of the independent sample sets used for statistical analyses.
RESULTS
Myotubularin Depletion Activates Proapoptotic Signaling
To investigate the possible role(s) of myotubularin in endocytosis and vesicular trafficking, we used HeLa cells as a well characterized model system to study these events. We utilized siRNA gene silencing of myotubularin to mimic the loss of function effect caused by severe XLMTM mutations, many of which cause almost complete loss of myotubularin protein (30). The siRNAs employed in these studies correspond to the myotubularin mRNA coding region (MTM1-1, MTM1-3), or 3′-untranslated region (MTM1-2). Using these siRNAs, we routinely achieved >90% depletion of endogenous myotubularin after 48 h (Fig. 1A). No changes in the protein level of the closely related MTMR family member MTMR2 were observed, suggesting that these siRNAs were specific for myotubularin (Fig. 1A).
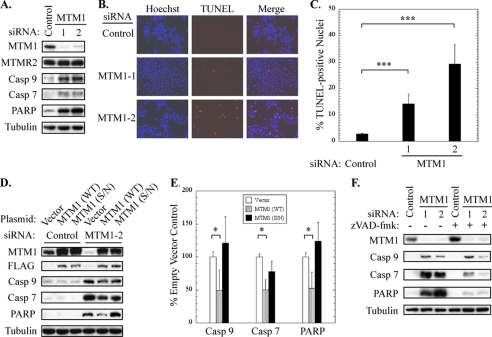
FIGURE 1.
Myotubularin gene silencing activates caspase-dependent pro-apoptotic signaling. A, soluble protein extracts from HeLa cells treated with control or myotubularin siRNAs (MTM1-1 and -2) were immunoblotted for myotubularin (MTM1), MTMR2, cleaved (activated) forms of caspase-9 and caspase-7, and cleaved (inactive) PARP. β-tubulin was used as loading control. B, control or myotubularin-deficient HeLa cells were analyzed using a red fluorescent TUNEL assay kit (center panels) for nuclear DNA strand breaks, indicative of DNA degradation in apoptotic cells. Nuclei were stained with Hoechst 33342 dye (left panels). Merged images are shown in the right panels. C, the percent of TUNEL-positive nuclei in control or myotubularin-deficient HeLa cells is represented graphically as the mean ± S.D. (n = 5, at least 100 cells were counted per replicate; ***, p < 0.005). D, myotubularin silencing was carried out in HeLa cells that had previously been transfected with expression plasmids encoding control (Vector), FLAG-tagged wild-type myotubularin (WT), or a catalytically inactive severe XLMTM S376N (S/N) myotubularin mutant. Soluble protein extracts were analyzed by immunoblotting for myotubularin, FLAG tag, cleaved caspase-9 and caspase-7, and cleaved PARP. E, cleaved caspase-9, caspase-7, and PARP bands from developed film were quantified by scanning densitometry. The data from 3 independent sample sets are shown graphically as the mean ± S.D. (n = 3; *, p < 0.05). F, the effect of the pan-caspase inhibitor benzyloxycarbonyl VAD- fluoromethyl ketone (zVAD-fmk; 40 μm final concentration, right three lanes) on pro-apoptotic signaling was determined in control or myotubularin-deficient HeLa cells. Soluble protein extracts were analyzed by immunoblotting for myotubularin, cleaved caspase-9, and caspase-7, and cleaved PARP.
Surprisingly, we observed many dead cells in myotubularin knockdown samples, suggestive of pro-apoptotic signaling. Therefore, we analyzed soluble protein extracts from these cells by immunoblotting with antibodies specific for cleaved (activated) caspase-9 and caspase-7 and cleaved (inactive) PARP, as markers of pro-apoptotic signaling. As shown in Fig. 1A, myotubularin depletion markedly activated caspase-9, caspase-7 and PARP cleavage as compared with cells treated with control siRNA. We also examined these cells for nuclear DNA fragmentation using a red fluorescent TUNEL assay. As shown in Fig. 1, B and C, myotubularin depletion significantly increased the number of TUNEL-positive nuclei, also indicative of pro-apoptotic signaling. Cell cycle analysis of myotubularin-deficient HeLa cells by Telford assay revealed no significant changes compared with control cells, suggesting that myotubularin depletion did not alter cell cycle progression (not shown).
To confirm that the pro-apoptotic effects we observed were not due to siRNA off-target effects, we knocked down endogenous myotubularin in cells that had been transfected 24 h before siRNA treatment with overexpression vectors encoding FLAG-tagged wild-type myotubularin or a myotubularin mutant (S376N). The S376N mutant was specifically chosen because it is associated with severe XLMTM and is catalytically inactive (26). Of six different severe XLMTM myotubularin point mutants that we tested, S376N was the only mutant that could be overexpressed at detectable levels in mammalian cells (not shown), which is consistent with the destabilizing effect of XLMTM disease mutations on the myotubularin protein previously reported (30). It should be noted that the mRNA from the myotubularin overexpression vectors is not sensitive to the MTM1-2 siRNA, which targets the 3′-UTR sequence of endogenous myotubularin mRNA. HeLa cells transfected with myotubularin wild-type and S376N expression vectors displayed ∼2.5-fold overexpression as compared with control cells transfected with empty vector (Fig. 1D). Overexpression of wild-type myotubularin before siRNA silencing suppressed the cleavage of caspase-9, caspase-7, and PARP caused by myotubularin depletion by ∼50%, as shown in Fig. 1, D and E. In contrast, overexpression of the inactive XLMTM S376N mutant did not significantly decrease cleavage of caspase-9, caspase-7, or PARP. It should be noted that using pEGFP vector and a fluorescently labeled siRNA, we determined that >98% of the HeLa cells were transfected with siRNAs under the transfection conditions used, whereas plasmid transfection efficiency was significantly lower at ∼75%. As a result, we estimate that the suppressive effect of the myotubularin expression plasmid was incomplete due to the differences in transfection efficiency. Importantly, because the inactive S376N mutant did not suppress cleavage of caspase-9, caspase-7, or PARP, these findings suggest that the pro-apoptotic signaling events we have observed are caused by loss of myotubularin function. The fact that levels of the closely related MTMR2 protein were not altered in these cells also underscores the idea that, although catalytically similar, active MTMRs do not necessarily exhibit functional redundancy (Fig. 1A).
We also tested the effect of the pan-caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone on pro-apoptotic signaling induced by myotubularin depletion. As shown in Fig. 1F, benzyloxycarbonyl-VAD-fluoromethyl ketone suppressed caspase-7 and PARP cleavage, whereas caspase-9 cleavage, which is carried out by Apaf-1 and therefore not affected by a general caspase inhibitor, remained unchanged. Together, our results indicate that myotubularin silencing activates a pro-apoptotic signaling pathway leading from caspase-9 through caspase-7 and PARP, ultimately resulting in nuclear DNA fragmentation and apoptosis. Activation of other caspases, including caspase-6, -8, and -12, was not detected in myotubularin-deficient cells (not shown).
Myotubularin Depletion Inhibits Growth Factor-stimulated Akt and Akt Effector Phosphorylation
Akt represents a nexus in signaling pathways that regulate cell survival and apoptosis in virtually all cell types (31, 32). In response to growth factor stimulation, Akt undergoes phosphoinositide-dependent translocation to the plasma membrane, where it becomes activated by phosphorylation on Thr-308 via PDK-1 (33). Phosphorylation of Akt at a second site within a C-terminal hydrophobic motif (Ser-473) leads to full activation (33). Constitutive Akt phosphorylation at a third site (Thr-450) has been shown to regulate its stability and turnover (34, 35). Akt phosphorylation at both Thr-450 and Ser-473 can be regulated by the mammalian target of rapamycin complex 2 (mTORC2) (34–36).
Due to its role as a critical regulator of cell survival and pro-apoptotic signaling, we investigated whether Akt phosphorylation might be affected by myotubularin depletion. Because Akt phosphorylation in HeLa cells is low under normal growth conditions (10% serum), we silenced myotubularin expression in HeLa cells, then serum-starved and stimulated the cells with EGF to promote Akt phosphorylation to detectable levels. Soluble protein extracts were analyzed by immunoblotting for phosphorylated forms of Akt. As shown in Fig. 2A, myotubularin depletion markedly decreased Akt phosphorylation at Thr-308 and Ser-473 by 40–60% as compared with the EGF-stimulated control siRNA sample (Fig. 2B). In contrast, myotubularin depletion had little effect on Akt Thr-450 phosphorylation (Fig. 2, A and B). Myotubularin depletion and decreased Akt phosphorylation at Thr-308 and Ser-473 could be detected as soon as 24 h after siRNA treatment. Consistent with these findings, myotubularin overexpression increased Akt Ser-473 phosphorylation in HeLa and HEK293 cells (data not shown).
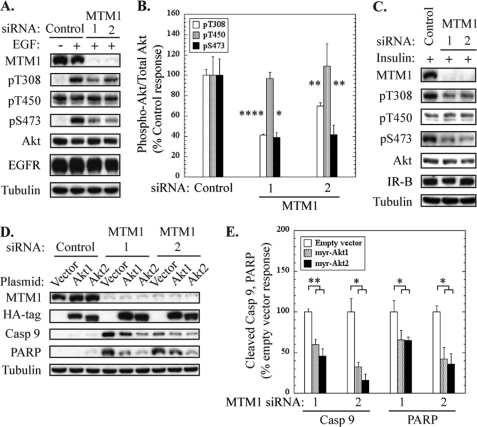
FIGURE 2.
Myotubularin silencing promotes pro-apoptotic signaling via inhibition of Akt activation. HeLa cells were transfected with control or myotubularin-specific siRNAs followed by serum starvation and stimulation with EGF to promote Akt phosphorylation. A, soluble protein extracts were analyzed by immunoblotting for myotubularin, phosphorylated Akt (Thr(P)-308 (pT308), Thr(P)-450 (pT450), Ser(P)-473 (pS473)), and total Akt. Tubulin immunoblotting was used as a loading control. B, bands on developed film corresponding to phosphorylated and total Akt were quantified by scanning densitometry. The data are represented graphically as phosphorylated Akt/total Akt in myotubularin-deficient samples as a percentage of stimulated control values (mean ± S.D., n = 3, *, p < 0.05; **, p < 0.01; ****, p < 0.001). Akt/pAkt values for Thr-308, Thr-450, and Ser-473, are shown as white, gray, and black bars, respectively. C, HeLa cells were transfected with control or myotubularin-specific siRNAs as described for panel A followed by serum starvation and stimulation with 100 nm insulin. Soluble protein extracts were analyzed by immunoblotting for myotubularin, phosphorylated Akt (Thr(P)-308 (pT308), Thr(P)-450 (pT450), Ser(P)-473 (pS473)), insulin receptor beta chain (IR-B), and total Akt. D, HeLa cells overexpressing myristoylated hemagglutinin-tagged (HA) constitutively active Akt1 and Akt2 were transfected with control siRNA (left three lanes) or myotubularin siRNAs 1 and 2 (center three lanes and right three lanes, respectively). Soluble protein extracts were analyzed by immunoblotting for myotubularin, HA (myr-HA-Akt1 or myr-HA-Akt2), cleaved caspase-9, and cleaved PARP. E, the bands on developed film corresponding to cleaved caspase-9 and PARP were quantified, and the values for myr-HA-Akt1 and myr-HA-Akt2 samples are represented graphically as a percentage of vector control values (mean ± S.D., n = 3; *, p < 0.05; **, p < 0.01).
We also wanted to determine whether myotubularin depletion might affect Akt phosphorylation after stimulation by a growth factor other than EGF. To address this issue, we analyzed Akt phosphorylation in control or myotubularin-deficient HeLa cells that were stimulated with insulin. As shown in Fig. 2C, myotubularin silencing also markedly decreased insulin-stimulated Akt phosphorylation at Thr-308 and Ser-473 as compared with the control siRNA sample. This result indicates that the inhibitory effect of myotubularin depletion on Akt activation is not limited to a specific growth factor receptor signaling pathway.
Previous reports have demonstrated that myotubularin and MTMR2 regulate internalization and turnover of growth factor receptors such as EGF receptor. Specifically, inhibition of myotubularin and/or MTMR2 function by gene silencing or overexpression of a catalytically inactive mutant resulted in delayed EGF receptor internalization and turnover (14). We observed a slight decrease in EGF receptor levels in myotubularin-deficient HeLa cells after EGF-stimulation, but this change was not statistically significant (Fig. 2A). Furthermore, we did not observe changes in insulin receptor levels in myotubularin-deficient cells that were stimulated with insulin (Fig. 2C). We also used FACS analysis to measure cell surface insulin receptor expression with an antibody specific to the extracellular domain of the insulin receptor in unstimulated and insulin-stimulated cells and did not detect marked changes (not shown). Therefore, in the short time points after growth factor stimulation (5–10 min), it appears that a mechanism other than altered growth factor receptor trafficking is responsible for the inhibition of Akt activation caused by myotubularin depletion.
Because inhibition of Akt activation is an established initiation event for caspase-dependent cell death, we next asked whether overexpression of constitutively active Akt proteins could suppress the pro-apoptotic effect of myotubularin depletion. We transfected HeLa cells with empty vector or vectors encoding Akt1 and Akt2 containing an N-terminal myristoylation sequence that promotes plasma membrane recruitment and constitutive activation (27). These cells were subsequently treated with control or myotubularin siRNAs, and soluble protein extracts were analyzed for cleaved caspase-9 and PARP as pro-apoptotic markers. As shown in Fig. 2, D and E, expression of constitutively active Akt1 and Akt2 decreased cleavage of caspase-9 by 40–70% in myotubularin-deficient cells and PARP cleavage by 40–60%. Although the pro-apoptotic response initiated by myotubularin depletion was markedly diminished by constitutively active Akt isoforms, it was not completely suppressed. It is likely that the lower transfection efficiency of Akt plasmids as compared with myotubularin siRNAs can account for part of the residual pro-apoptotic signaling in Akt-overexpressing cells as described above for incomplete rescue of the knockdown phenotype by myotubularin overexpression (Fig. 1, D and E). We also cannot rule out that an effect of myotubularin depletion in addition to impaired Akt activation contributes to increased pro-apoptotic signaling. Together, these data indicate that myotubularin plays an important role in regulating Akt phosphorylation and that impaired Akt activation is a major factor underlying the pro-apoptotic effect of myotubularin depletion.
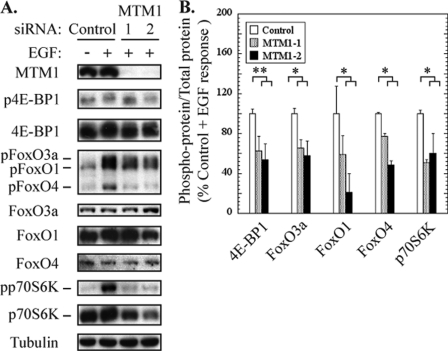
After activation of growth factor receptors, Akt signals are sent to numerous effector pathways that promote cell growth and survival and oppose pro-apoptotic signaling (31, 32). We next asked whether signaling through specific Akt downstream effector pathways might be impaired in myotubularin-deficient cells. We therefore analyzed the phosphorylation status of 4E-BP1, p70S6K, and FoxO transcription factors in myotubularin-deficient cells. Akt-dependent phosphorylation of 4E-BP1 and p70S6K is carried out by the mammalian target of rapamycin complex 1 (mTORC1), whereas FoxO transcription factors can be phosphorylated directly by Akt. As shown in Fig. 3A, myotubularin silencing caused a marked decrease in phosphorylation of 4E-BP1, FoxO1, FoxO3a, FoxO4, and p70S6K. When normalized for total protein levels, myotubularin-deficient HeLa cells displayed a 40–60% decrease in EGF-stimulated phosphorylation of these effectors. These findings suggest that loss of myotubularin function may differentially regulate distinct Akt effector pathways.
FIGURE 3.
Myotubularin silencing inhibits Akt signaling through downstream mTOR and FoxO effector pathways. HeLa cells were treated with control or myotubularin siRNAs, then serum-starved and stimulated with EGF. A, soluble protein extracts were analyzed by immunoblotting for the indicated proteins, and tubulin immunoblotting was used as a loading control. B, the bands on developed film corresponding to phosphorylated and total 4E-BP1, FoxO3a, FoxO1, FoxO4, and p70S6 kinase were quantified, and the values for phospho-protein/total protein in myotubularin-deficient samples are represented graphically as a percentage of EGF-stimulated control values (mean ± S.D., n = 3; *, p < 0.05; **, p < 0.01).
PI(3)P Mediates Akt Inhibition and Cell Death Caused by Myotubularin Depletion
A schematic diagram outlining the interconversion of PI(3)P, PI(3,5)P2, and PI(5)P by MTMR family phosphoinositide phosphatases and inositol lipid kinases in mammalian cells is illustrated in Fig. 4A. Although these pathways are not the only mechanisms by which these lipids can be generated or interconverted, they represent major routes for control of their levels in mammalian cells. Because myotubularin dephosphorylates PI(3)P and PI(3,5)P2 to generate PI and PI(5)P, respectively, overexpression of myotubularin decreases cellular levels of PI(3)P and PI(3,5)P2 and increases levels of the product PI(5)P (37). Conversely, myotubularin knockdown or overexpression of a catalytically inactive substrate-trapping myotubularin mutant causes approximately a 2-fold increase in total cellular PI(3)P (14, 26, 38). Based on these experimental data, it is expected that loss of myotubularin function in XLMTM disease would result in both increased levels of the substrate lipids PI(3)P and PI(3,5)P2 and in decreased levels of its product lipid PI(5)P. This is noteworthy because PI(5)P produced by pathogenic bacterial lipid phosphatases during infection has been shown to activate Akt (39). An important question is which of the myotubularin substrate/product lipids may be directly linked to inhibition of Akt and pro-apoptotic signaling induced by myotubularin depletion? We hypothesized that accumulation of the substrate PI(3)P in the absence of myotubularin causes impaired Akt phosphorylation.
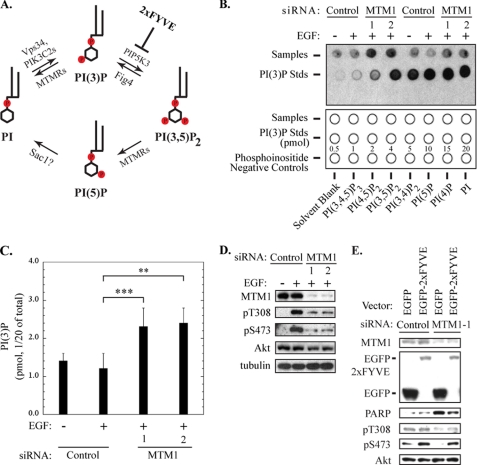
FIGURE 4.
Increased PI(3)P levels are linked to inhibition of Akt phosphorylation and pro-apoptotic signaling in myotubularin-deficient HeLa cells. A, routes for cyclic interconversion and regulation of the MTMR-specific phosphoinositides PI(3)P and PI(3,5)P2 are depicted. Phosphoinositide kinases associated with specific steps in this pathway (PIK3C2A, PIK3C2B, Vps34, PIP5K3) are listed above the arrows, and regulatory points for myotubularin family phosphatases (MTMRs) and other phosphatases that regulate these inositol lipids (Sac1) are shown below the arrows. The point at which the EGFP-2xFYVE PI(3)P-specific binding protein interferes with conversion of PI(3)P to PI(3,5)P2 is shown at the upper right. B, PI(3)P levels in control or myotubularin-deficient HeLa cells were quantified using a PI(3)P mass assay kit according to the manufacturer's protocol. An image of the developed film representing two independent sample sets and the PI(3)P standard curve is shown in the upper panel. The lower panel indicates the locations of HeLa samples (1/20 of total sample) (top row), PI(3)P standards (center row), and phosphoinositide negative control lipids (bottom row) that were spotted on the membrane for analysis. C, spots corresponding to PI(3)P derived from HeLa cell experimental samples and the PI(3)P standards on developed film were quantified by scanning densitometry. PI(3P levels from control and myotubularin-deficient HeLa samples were determined by comparison to PI(3)P standards and are represented graphically (mean ± S.D., n = 3; **, p < 0.01; ***, p < 0.005). D, soluble protein extracts from HeLa cells prepared in parallel with those used for PI(3)P quantification were analyzed by immunoblotting for myotubularin (MTM1), phosphorylated Akt (Thr(P)-308 (pT308), Ser(P)-473 (pS473)), and total Akt. Tubulin immunoblotting was used as a loading control. E, HeLa cells were transfected with EGFP or EGFP-2xFYVE expression vectors for 24 h, then transfected with control siRNA or myotubularin siRNA-1 for an additional 48 h. Before harvest, the cells were serum-deprived and stimulated with insulin. Soluble protein extracts were analyzed by immunoblotting for the indicated proteins.
To address this question, we first determined the effect of myotubularin depletion on cellular PI(3)P levels in HeLa cells using a mass assay for this lipid. As shown in Fig. 4B, myotubularin silencing caused a corresponding increase of PI(3)P relative to cells transfected with control siRNAs. Overall, the two myotubularin siRNAs increased relative levels of PI(3)P by 1.9- and 2.0-fold, respectively (Fig. 4C). This finding prompted us to ask whether impaired Akt activation and increased pro-apoptotic signaling in myotubularin-deficient cells could be attributed to increased levels of PI(3)P. Myotubularin silencing and the resultant inhibition of Akt phosphorylation in samples carried out in parallel are shown in Fig. 4D.
The highly conserved FYVE domain specifically binds PI(3)P (28). When overexpressed, EGFP fused to tandem FYVE domains (EGFP-2xFYVE) can function as a “scavenger” that can bind and sequester cellular PI(3)P, in effect decreasing the availability of PI(3)P within the cell. We reasoned that if excessive PI(3)P was a cause of impaired Akt activation and activated pro-apoptotic signaling in myotubularin-deficient cells, then sequestering excess PI(3)P using EGFP-2xFYVE should restore Akt activation and suppress apoptotic signaling. HeLa cells were transfected with vectors encoding EGFP or EGFP-2xFYVE. The cells were subsequently transfected with control or myotubularin siRNAs. After insulin stimulation, soluble protein extracts were analyzed by immunoblotting for Akt phosphorylation and PARP cleavage as an apoptotic index. As observed previously, depletion of myotubularin inhibited Akt phosphorylation at Thr-308 and Ser-473 and increased pro-apoptotic PARP cleavage (Fig. 4E). However, overexpression of EGFP-2xFYVE significantly increased Akt Thr-308 and Ser-473 phosphorylation and partially suppressed PARP cleavage in myotubularin-deficient cells compared with EGFP alone (Fig. 4E). Notably, EGFP-2xFYVE significantly increased insulin-stimulated Akt phosphorylation compared with EGFP alone even in the absence of myotubularin silencing (Fig. 4E, control siRNA lanes). These data suggest that suppressing PI(3)P accumulation reversed the effects of myotubularin depletion on Akt activation and apoptosis and implicate PI(3)P as the critical lipid species in myotubularin-dependent Akt signaling. These data also indicate that PI(5)P is unlikely to be the critical lipid component in myotubularin-dependent cell survival because it is not sequestered by the EGFP-2xFYVE scavenger.
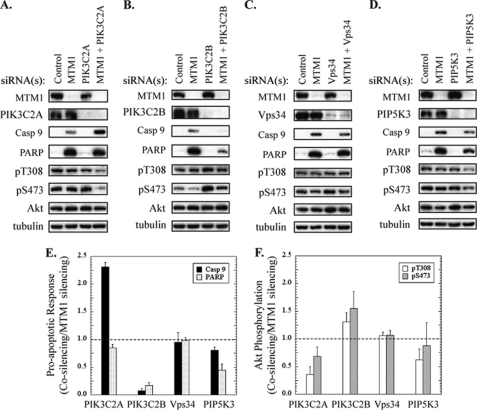
Next, we surmised that if accumulated PI(3)P was responsible for Akt inhibition and cell death, then knockdown of its source kinase should reduce PI(3)P accumulation in myotubularin-deficient cells and suppress pro-apoptotic signaling. We conducted co-silencing experiments in which three of the four PI 3-kinases that specifically generate PI(3)P from phosphatidylinositol in mammalian cells (PIK3C2A, PIK3C2B, and Vps34) were knocked down alone or in combination with myotubularin. PIK3C2G was not tested because it is not expressed at significant levels in HeLa cells. As shown in Fig. 5, A and C, co-silencing of PIK3C2A and Vps34 together with myotubularin failed to restore Akt phosphorylation or suppress PARP cleavage caused by myotubularin silencing alone. However, co-silencing of PIK3C2B together with myotubularin almost completely reversed the pro-apoptotic effect of myotubularin depletion alone (Fig. 5, B and E). At the same time, Akt phosphorylation at Thr-308 and Ser-473 in the myotubularin/PIK3C2B co-silencing samples was increased to levels significantly higher than observed in cells depleted of myotubularin alone (Fig. 5, B and F). Notably, silencing of PIK3C2B alone increased EGF-stimulated Akt phosphorylation to a level even above that of control siRNA-treated cells. These results further suggest that PI(3)P accumulation contributes to the pro-apoptotic signaling and cell death in myotubularin-deficient HeLa cells and support our interpretation that impaired Akt signaling is also a factor underlying these events.
FIGURE 5.
Co-silencing of PIK3C2B with myotubularin increases EGF-stimulated Akt phosphorylation and suppresses pro-apoptotic signaling. Myotubularin and phosphoinositide kinases involved in the MTMR phosphoinositide interconversion cycle (Fig. 4A) were knocked down separately or in combination in HeLa cells. A—D, HeLa cells were treated with control (medium GC negative control + GFP negative control) or myotubularin-specific (MTM1-1 and MTM1-2) siRNA pools in combination with siRNA pools (two siRNAs specific for each phosphoinositide kinase) against the phosphoinositide kinases PIK3C2A (panel A), PIK3C2B (panel B), Vps34 (panel C), and PIP5K3 (panel D). The control siRNA pool was used to equalize the total amount of siRNA in samples where myotubularin or a lipid kinase was silenced alone (second through fourth lanes, panels A–D). The cells were serum-starved and stimulated with EGF, and soluble protein extracts were analyzed by immunoblotting for the indicated proteins. Tubulin immunoblotting was used as a loading control. E, bands on developed film corresponding to cleaved caspase-9 and PARP were quantified by scanning densitometry. The data are represented graphically as the ratio of the myotubularin plus lipid kinase (co-silencing) response to myotubularin silencing (alone) response (n = 3, mean ± S.D.). The dashed line at 1.0 indicates the level of cleaved caspase-9 and PARP in samples where only myotubularin expression was silenced. F, bands on developed film corresponding to phosphorylated and total Akt were quantified by scanning densitometry, and values for phosphorylated Akt/total Akt were determined. The data are represented graphically as the ratio of phosphorylated Akt/total Akt values for myotubularin plus lipid kinase (co-silencing) to myotubularin silencing alone (n = 3, mean ± S.D.). The dashed line at 1.0 indicates the level of phosphorylated Akt/total Akt in samples where only myotubularin expression was silenced. pT308, Thr(P)-308; pS473, Ser(P)-473.
These experiments could not rule out the possibility that the myotubularin substrate PI(3,5)P2 affected Akt signaling and apoptosis. To address this question, we asked whether silencing of PIP5K3, the major source of PI(3,5)P2 in mammalian cells, could suppress the pro-apoptotic effect of myotubularin depletion. Therefore, we knocked down myotubularin alone or in combination with PIP5K3. As shown in Fig. 5, D–F, co-silencing of PIP5K3 with myotubularin did not significantly restore Akt phosphorylation or suppress PARP cleavage as compared with myotubularin silencing alone, suggesting that increased PI(3,5)P2 was unlikely to be the causative factor underlying Akt inhibition and pro-apoptotic signaling in myotubularin-deficient cells.
Although Akt phosphorylation at Ser-473, which is required for full Akt activation, was decreased in myotubularin-deficient cells by nearly 40% in these experiments as compared with control cells, it should be noted that this decrease in Akt phosphorylation was not as robust as previously shown in Fig. 2. The most likely explanation is that to keep total siRNA concentrations constant in this co-silencing experiment, the amount of myotubularin-specific siRNA was reduced by half. This likely reduced the efficiency of myotubularin knockdown and subsequent effects on Akt phosphorylation. Regardless, even small overall decreases in Akt phosphorylation over the time course of myotubularin knockdown were sufficient to induce apoptotic signaling through caspase 9 and PARP, and these effects were robustly suppressed through co-knockdown of PIK3C2B, but not PIK3C2A, Vps34, or PIK3C2B.
The cyclic interconversion of phosphoinositides and the presence of other phosphoinositide phosphatases and kinases make it challenging to unequivocally determine the critical lipid species responsible for our observations. Nevertheless, our results are consistent with PI(3)P as the likely candidate to mediate myotubularin-dependent Akt activation and survival signaling.
Myotubularin Depletion Promotes Caspase-dependent Pro-apoptotic Signaling in Primary Human Skeletal Muscle Myotubes
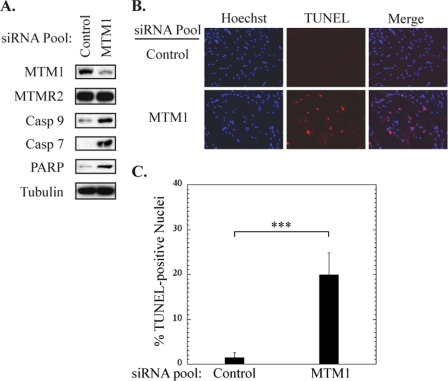
The findings we have presented thus far provide compelling evidence that myotubularin depletion inhibits Akt phosphorylation and triggers pro-apoptotic signaling in HeLa cells. We also wanted to employ a more relevant cell model system for understanding the role of myotubularin in skeletal muscle physiology. To do this we used HsMMs that were differentiated into multinucleated myotubes in culture. To achieve effective knockdown of myotubularin in these cells, we found it necessary to employ a myotubularin-specific siRNA pool that consisted of equimolar amounts of the three most effective myotubularin siRNAs (MTM1-1, MTM1-2, MTM1-3). Myotubularin silencing in HsMM myotubes, although not as efficient as in HeLa cells, consistently resulted in a >80% reduction in myotubularin protein levels after 72 h (Fig. 6A). Furthermore, myotubularin depletion markedly activated cleavage of caspases-9 and -7 and PARP (Fig. 6A). As in HeLa cells, myotubularin depletion had no effect on the level of the closely related MTMR2 protein, suggesting that increased caspase and PARP cleavage were specifically caused by myotubularin silencing. We also observed a significant increase in the number of TUNEL-positive nuclei in myotubularin-deficient myotubes as compared with those treated with the control siRNA pool (Fig. 6, B and C). Importantly, few myotubes could be detected that contained more than a single TUNEL-positive myonucleus. Together, these results demonstrate that myotubularin silencing also elicits a pro-apoptotic response in skeletal muscle myotubes.
FIGURE 6.
Myotubularin depletion activates pro-apoptotic signaling in skeletal muscle myotubes. Primary human skeletal muscle myotubes were treated with control or myotubularin-specific siRNA pools. A, soluble protein extracts were analyzed by immunoblotting for the indicated proteins and tubulin immunoblotting was used as a loading control. B, myotubes in two-well microscope slides were fixed and analyzed by fluorescence microscopy using a red fluorescent TUNEL assay (center panels). Nuclei were stained with Hoechst dye (left panels), and the merged images are shown at the right. C, the percent of TUNEL-positive nuclei in control and myotubularin-deficient myotubes are represented graphically as the mean ± S.D. (n = 5, at least 100 cells were counted per replicate; ***, p < 0.005).
Inhibition of Akt Phosphorylation in Myotubularin-deficient Skeletal Muscle Myotubes
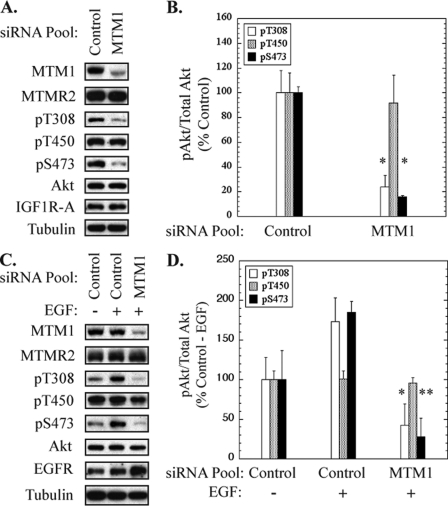
Because our studies indicated that impaired Akt activation was a critical factor underlying increased pro-apoptotic signaling in myotubularin-deficient HeLa cells, we wanted to assess Akt phosphorylation in skeletal muscle myotubes. We treated HsMM myotubes with control or myotubularin siRNA pools and analyzed Akt phosphorylation by immunoblotting soluble protein extracts from cells treated under two distinct conditions. First, we analyzed extracts from myotubes that were neither serum-starved nor stimulated with growth factor(s). In these samples, Akt phosphorylation is primarily dependent on autocrine signaling via the insulin-like growth factor receptor I induced by low serum differentiation conditions. Depletion of myotubularin inhibited basal levels of Akt phosphorylation at Thr-308 and Ser-473 by 80% (Fig. 7, A and B). Second, we analyzed phospho-Akt in extracts prepared from differentiated myotubes that were serum-deprived followed by stimulation with EGF. Here also, myotubularin depletion significantly decreased EGF-induced phosphorylation of Akt at Thr-308 and Ser-473 (Fig. 7, C and D). In both cases, phosphorylation at Thr-450 was unaffected. Furthermore, we did not detect changes in the level of either insulin-like growth factor receptor I or EGF receptor in myotubularin-deficient cells, suggesting that altered receptor turnover was not responsible for impaired Akt phosphorylation. These data further demonstrate that the effect was not unique to a single growth factor receptor, as EGF-dependent Akt phosphorylation was impaired in addition to the insulin-like growth factor receptor I-dependent Akt response. These results demonstrate that myotubularin silencing also elicits a marked decrease in growth factor-stimulated Akt phosphorylation in human skeletal muscle myotubes.
FIGURE 7.
Myotubularin silencing inhibits Akt activation in skeletal muscle myotubes. Primary human skeletal muscle myotubes were treated with control or myotubularin-specific siRNA pools. A, soluble protein extracts were prepared from cells that were not serum-starved or stimulated with growth factors and immunoblotted for the indicated proteins. Tubulin immunoblotting was used as a loading control. B, bands on developed film corresponding to phosphorylated and total Akt were quantified by scanning densitometry. The values for phosphorylated Akt/total Akt in myotubularin-deficient samples are represented graphically as a percentage of the control value (mean ± S.D., n = 3; *, p < 0.05). C, control and myotubularin-deficient myotubes were serum-starved and treated with EGF, and soluble protein extracts were immunoblotted for the indicated proteins. Tubulin immunoblotting was used as a loading control. D, bands on developed film corresponding to phosphorylated and total Akt were quantified by scanning densitometry. The values for phosphorylated Akt/total Akt in myotubularin-deficient samples are represented graphically as a percentage of the EGF-stimulated controls (mean ± S.D., n = 3; *, p < 0.05; ** p < 0.01). Values for phosphorylated Akt (Thr(P)-308 (pT308), Thr(P)-450 (pT450), Ser(P)-473 (pS473)) are represented as white, gray, and black bars, respectively.
Impaired Signaling through Akt Effectors in Myotubularin-deficient Skeletal Muscle Cells
We next analyzed the phosphorylation patterns of FoxO transcription factors that are regulated directly by Akt phosphorylation and proteins regulated indirectly via mTORC1 including 4E-BP1 and p70S6K. These proteins play critical roles in the growth and maintenance of many tissues including skeletal muscle and have been linked to myopathic phenotypes that are similar to those observed in XLMTM patients (23–25).
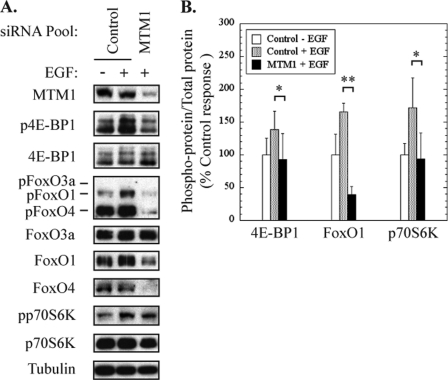
EGF stimulation in myotubes treated with control siRNA elicited a marked increase in phosphorylation of 4E-BP1, FoxO1, FoxO4, and p70S6K as compared with unstimulated control cells (Fig. 8A). The increase in phosphorylation (normalized for total phosphoprotein) of these proteins in response to EGF ranged from 30 to 75% greater than unstimulated control cells, as shown in Fig. 8B. In contrast, myotubularin-deficient myotubes exhibited significantly decreased levels of phosphorylation for 4E-BP1, FoxO1, and p70S6K (Fig. 8A). Compared with EGF-stimulated control cells, myotubularin-deficient myoblasts showed decreases of 33, 76, and 45% in 4E-BP1, FoxO1, and p70S6K phosphorylation, respectively (Fig. 8B). Even with long exposure times, we were unable to detect phosphorylation of FoxO3a in any of the samples, although total FoxO3a protein was readily detectable. In addition, myotubularin silencing caused a dramatic decrease in the total protein levels of FoxO1 and FoxO4 by an undetermined mechanism. Although we were able to take this decrease into account when determining the change in FoxO1 phosphorylation (Fig. 8B), we were unable to do so for FoxO4, which could not be detected by this method in myotubularin-deficient cells. Notably, FoxO1 is an important regulator of atrophy and apoptosis in skeletal muscle and can also function to repress transcription and expression of FoxO4 (40–43). Further studies will be necessary to determine how the function(s) of FoxO transcription factors is regulated by changes in their expression levels and phosphorylation status and how these changes may affect cell survival, growth, and maintenance in myotubularin-deficient myotubes.
FIGURE 8.
Myotubularin silencing inhibits signaling through Akt effector pathways in skeletal muscle myotubes. Control and myotubularin-deficient human skeletal muscle myotubes were serum-starved and treated with EGF as shown in Fig. 7C, and the phosphorylation status of Akt effector proteins was analyzed. A, soluble protein extracts were immunoblotted for the indicated proteins, and tubulin immunoblotting was used as a loading control. B, the bands on developed film corresponding to phosphorylated and total 4E-BP1, FoxO1, and p70S6 kinase as shown in panel A were quantified, and the values for phospho-protein/total protein in myotubularin-deficient samples are represented graphically as a percentage of EGF-stimulated control values (mean ± S.D., n = 3; *, p < 0.05; **, p < 0.01).
DISCUSSION
Loss-of-function myotubularin mutations are causative for X-linked recessive myotubular myopathy (1, 2). XLMTM is characterized by skeletal muscle fibers with numerous centrally located nuclei that are markedly atrophic/hypotrophic. Because early stages of myogenic differentiation and myoblast fusion into multinucleated myotubes appear to occur normally in XLMTM patients and myotubularin knock-out mice, it has been proposed that the XLMTM represents impaired skeletal muscle maturation and/or regeneration (3). Numerous studies have demonstrated that enzymatically active MTMR proteins specifically dephosphorylate the 3-phosphate of the inositol lipids PI(3)P and PI(3,5)P2 (7, 44). These phosphoinositides have well established roles in growth factor receptor internalization, endocytic vesicle trafficking, lysosomal transport, and initiation of autophagy (11, 12, 45). However, definitive evidence for how aberrant regulation of PI(3)P and/or PI(3,5)P2 might link these processes to the defects observed in XLMTM disease has remained elusive.
A growing body of evidence has emerged suggesting that MTMRs may regulate cell survival and pro-apoptotic signaling (20–22). In this study, we utilized siRNA-mediated gene silencing in HeLa and primary human skeletal muscle cells to mimic the cellular depletion of myotubularin protein observed in severe XLMTM patients (30). Our data show that myotubularin functions as a positive regulator of Akt-dependent cell growth and survival signaling in HeLa cells and primary human skeletal muscle myotubes. The pro-apoptotic effect of myotubularin silencing could be suppressed by expression of exogenous myotubularin but not by a catalytically inactive form of myotubularin associated with XLMTM, indicating that the effect is specifically due to loss of myotubularin function. We have further demonstrated that the pro-apoptotic effect of myotubularin depletion is due to impaired Akt activation. In myotubularin-deficient cells, growth factor-stimulated Akt phosphorylation at Thr-308 and Ser-473 was profoundly inhibited, resulting in impaired growth and survival signaling through mTORC1 and decreased phosphorylation of FoxO transcription factors. We have provided evidence that impaired Akt phosphorylation in myotubularin-deficient cells is caused by accumulation of the MTMR substrate PI(3)P. Moreover, our findings have identified the type II PI 3-kinase PIK3C2B as a likely source of accumulated PI(3)P in myotubularin-deficient cells and a potential negative regulator of Akt phosphorylation. Our work has provided the first evidence linking impaired Akt activation to myotubularin deficiency and aberrant regulation of its inositol lipid substrate, PI(3)P.
Our studies highlight a potential role for impaired Akt activation and cell growth and survival signaling in the pathology of XLMTM. The remarkable specificity of active MTMR family lipid phosphatases toward PI(3)P and PI(3,5)P2 substrates suggests that aberrant activation of Akt may also represent an important link between other MTMR family proteins and altered cell survival and apoptosis. In support of this hypothesis, a recent study by Berger et al. (46) has provided biochemical and genetic evidence that MTMR2 and MTMR13 also regulate Akt activity. We detected no change in MTMR2 protein levels in myotubularin-deficient cells, suggesting that the effect of myotubularin silencing on Akt and cell survival in both HeLa cells and skeletal muscle myotubes was independent of an effect of MTMR2 on these processes. Collectively, these findings support the idea that, although they are highly similar enzymatically, active MTMRs may possess non-redundant functions, possibly by regulating distinct subcellular pools of their substrate phosphoinositides.
Although Vps34 has long been considered the major source of endosomal PI(3)P, numerous recent studies have demonstrated that class II PI 3-kinases also produce this lipid. For example, a recent study by Velichkova et al. (47) demonstrated that the sole Drosophila ortholog of the mammalian MTM1/MTMR1/MTMR2 subfamily (mtm) regulates an endosomal pool of PI(3)P that plays distinct roles in endocytic trafficking and cortical remodeling. It is probable that mammalian MTMRs have evolved non-redundant functions to spatially and temporally regulate such distinct events. Notably, Velichkova et al. (47) also found that Pi3K68D, the Drosophila ortholog of the mammalian type-II PI 3-kinase PIK3C2B, was directly linked to a PI(3)P pool regulated by Drosophila mtm. While this manuscript was under revision, the same group also showed that depletion of Pi3K68D could suppress integrin trafficking defects associated with loss of mtm function (48). In that study, Ribeiro et al. (48) found that altered regulation of integrin trafficking in mtm-deficient flies was linked to muscle attachment and maintenance defects that are remarkably similar to those observed in XLMTM disease. This observation is especially intriguing in light of our finding that myotubularin regulates Akt signaling because disruption of integrin trafficking has previously been linked to impaired Akt signaling, pro-apoptotic signaling, and skeletal muscle atrophy (49, 50). In addition, PIK3C2B has also been shown to regulate integrin trafficking and inhibit Akt phosphorylation in mammalian cells (51). Although it does not rule out a role for myotubularin as a regulator of Vps34-dependent signals, our finding that co-silencing of PIK3C2B, but not Vps34, together with myotubularin can suppress Akt inhibition and pro-apoptotic signaling induced by silencing of myotubularin alone is consistent with the idea that these signals result from accumulated PI(3)P generated by PIK3C2B.
An important question that stems from these findings is that of the mechanism(s) by which Akt activation is impaired in myotubularin-deficient cells. Akt signaling has been directly linked to endocytic vesicle trafficking by several independent studies. For example, Akt can be recruited to a subset of Rab5-positive endosomes by an adaptor protein containing a PH domain, PTB domain, and leucine zipper motif (Appl1), thereby controlling its specificity toward distinct effectors (52). Furthermore, WDFY2, a protein containing WD repeats and a PI(3)P binding FYVE domain has also been established as an Akt interactor (53). PI(3)P on endocytic vesicles can serve as a molecular switch to coordinately regulate the endosomal localization of Appl1 and WDFY2. After growth factor stimulation, Appl1 is predominantly localized to pre-early endosomes that are deficient in PI(3)P (54). PI(3)P accrues on the surface of these structures as they mature into early endosomes, correlating temporally with the acquisition of EEA1 and WDFY2 and release of Appl1. Together, these findings have established a strong correlation between PI(3)P-dependent endosomal trafficking events and Akt activation and substrate specificity. The inhibition of Akt activation and up-regulation of pro-apoptotic signaling caused by myotubularin depletion are consistent with the idea that aberrant accumulation of PI(3)P may shift the balance from pre-early endosomes to favor WDFY2- and EEA1-containing early endosomes, thereby displacing Appl1 and inhibiting Akt activation.
Our findings also raise the question of how impaired Akt signaling might underlie the skeletal muscle defects associated with XLMTM. Recent studies have revealed how Akt signaling can promote maintenance of skeletal muscle cell size and prevent muscle atrophy through the actions of FoxO transcription factors and mTORC1 as downstream Akt effectors (23–25). Under normal growth conditions, phosphorylation of FoxO transcription factors by Akt inhibits their nuclear translocation and prevents transcriptional activation of target genes that are essential for autophagy. Under conditions where Akt and mTORC1 activities are low, FoxOs can translocate to the nucleus to stimulate target gene transcription. FoxO1 and FoxO3a are both known to play critical roles in the induction of autophagy and protein turnover and have been associated with atrophy in skeletal muscle (23, 24, 55, 56). Myotubularin depletion led to decreased Akt activation and subsequent downstream phosphorylation of FoxO transcription factors in HeLa cells and skeletal muscle myotubes. Abnormal autophagic flux has been associated with skeletal muscle phenotypes similar to those observed in XLMTM skeletal muscle (23, 24, 57). In addition, other MTMR family proteins have been linked to altered regulation of autophagy (58). Taken together with our results, these findings suggest that altered autophagic flux may play a role in skeletal muscle atrophy in XLMTM disease.
A second Akt-dependent effector pathway relevant to skeletal muscle maintenance and XLMTM disease is signaling through mTORC1. The mTOR complex 1 regulates initiation of protein translation and can oppose autophagy. Notably, impaired signaling through mTORC1 has also been linked to phenotypes that are remarkably similar to those of XLMTM disease. For example, skeletal muscle fibers from the raptor-deficient (mTORC1-deficient) mice were severely atrophic and contained many centrally located nuclei, phenotypes that are hallmarks of XLMTM (25). Our finding that myotubularin depletion inhibits Akt-dependent activation of the mTORC1 effectors p70 S6-kinase and 4E-BP1 is consistent with the concept that impaired mTORC1 signaling can promote the atrophy and centrally located nuclei that are important clinical markers for XLMTM.
The signaling events that initiate programmed cell death are highly conserved among most cell types. However, skeletal muscle is unique among mammalian tissues in that it is composed of multinucleated myofibers. Unlike mononucleated cells, activation of pro-apoptotic signaling in skeletal muscle results in selective destruction of myonuclei and decreased myofiber size (atrophy) rather than overall death of the myofiber (59). This distinct outcome of pro-apoptotic signaling in skeletal muscle myofibers is termed “nuclear apoptosis” and is consistent with the concept of the myonuclear domain, defined as the volume of cytoplasm that can be supported by a single muscle fiber nucleus (60). Pathological factors such as age, diet, injury, or disease states including myopathies and dystrophinopathies, activate pro-apoptotic signaling and cause decreased numbers of myonuclei, overall reduction in cytoplasmic volume, and corresponding atrophy of muscle fibers. Our finding that myotubularin depletion leads to increased pro-apoptotic signaling suggests that decreased myofiber size in XLMTM patients may be linked to selective nuclear apoptosis.
In summary, our data collectively implicate myotubularin as a positive regulator of Akt survival and growth signaling and identify aberrant mTORC1 and FoxO signaling as potential mechanisms underlying the pathology of XLMTM disease. These findings provide new insight into how Akt-dependent signaling events may be linked to human neuromuscular diseases such as XLMTM and Charcot-Marie-Tooth 4B, altered phosphoinositide metabolism, endocytic vesicle trafficking, and myotubularin family lipid phosphatases. Our ongoing efforts are aimed at further clarifying the roles of MTMR proteins in the regulation of Akt growth and survival signaling pathways and how they affect neuromuscular tissues.
Acknowledgments
We express our sincere appreciation to Jack Dixon, Richard MacDonald, Parmender Mehta, Fred Robinson, and Carolyn Worby for critical evaluation of this manuscript and helpful suggestions regarding its preparation. We also thank Margaret Wheelock and the Nebraska Center for Cellular Signaling. Very special thanks go to Harold and Pauline Taylor for their contributions to this work.
This work was supported, in whole or in part, by a National Institutes of Health CoBRe grant (National Center for Research Resources; to the Nebraska Center for Cellular Signaling (to G. S. T.).
- XLMTM
- X-linked myotubular myopathy
- MTMR
- myotubularin-related protein
- PI(3)P
- phosphatidylinositol (PI) 3-phosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PI(5)P
- phosphatidylinositol 5-phosphate
- PARP
- poly(ADP-ribose) polymerase
- mTORC1
- mammalian target of rapamycin complex 1
- FoxO
- forkhead family transcription factor
- HsMM
- primary human skeletal muscle myoblasts
- mtm
- Drosophila ortholog of mammalian MTM1/MTMR1/MTMR2 subfamily proteins.
REFERENCES
- 1. Spiro A. J., Shy G. M., Gonatas N. K. (1966) Arch. Neurol. 14, 1–14 [DOI] [PubMed] [Google Scholar]
- 2. van Wijngaarden G. K., Fleury P., Bethlem J., Meijer A. E. (1969) Neurology 19, 901–908 [DOI] [PubMed] [Google Scholar]
- 3. Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J. F., Mandel J. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15060–15065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolino A., Muglia M., Conforti F. L., LeGuern E., Salih M. A., Georgiou D. M., Christodoulou K., Hausmanowa-Petrusewicz I., Mandich P., Schenone A., Gambardella A., Bono F., Quattrone A., Devoto M., Monaco A. P. (2000) Nat. Genet. 25, 17–19 [DOI] [PubMed] [Google Scholar]
- 5. Azzedine H., Bolino A., Taïeb T., Birouk N., Di Duca M., Bouhouche A., Benamou S., Mrabet A., Hammadouche T., Chkili T., Gouider R., Ravazzolo R., Brice A., Laporte J., LeGuern E. (2003) Am. J. Hum. Genet. 72, 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Senderek J., Bergmann C., Weber S., Ketelsen U. P., Schorle H., Rudnik-Schöneborn S., Büttner R., Buchheim E., Zerres K. (2003) Hum. Mol. Genet. 12, 349–356 [DOI] [PubMed] [Google Scholar]
- 7. Robinson F. L., Dixon J. E. (2006) Trends Cell Biol. 16, 403–412 [DOI] [PubMed] [Google Scholar]
- 8. Previtali S. C., Quattrini A., Bolino A. (2007) Expert Rev. Mol. Med. 9, 1–16 [DOI] [PubMed] [Google Scholar]
- 9. Suter U. (2007) Cell. Mol. Life Sci. 64, 3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nicot A. S., Laporte J. (2008) Traffic 9, 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deretic V., Singh S., Master S., Kyei G., Davis A., Naylor J., de Haro S., Harris J., Delgado M., Roberts E., Vergne I. (2007) Biochem. Soc. Symp. 74, 141–148 [DOI] [PubMed] [Google Scholar]
- 12. Shisheva A. (2008) Cell Biol. Int. 32, 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Botelho R. J. (2009) Bioessays 31, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 14. Cao C., Backer J. M., Laporte J., Bedrick E. J., Wandinger-Ness A. (2008) Mol. Biol. Cell 19, 3334–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolis A., Coviello S., Visigalli I., Taveggia C., Bachi A., Chishti A. H., Hanada T., Quattrini A., Previtali S. C., Biffi A., Bolino A. (2009) J. Neurosci. 29, 8858–8870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dowling J. J., Vreede A. P., Low S. E., Gibbs E. M., Kuwada J. Y., Bonnemann C. G., Feldman E. L. (2009) PLoS Genet. 5, e1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A. H., Allard B., Mandel J. L., Laporte J., Jacquemond V., Buj-Bello A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18763–18768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Srivastava S., Li Z., Lin L., Liu G., Ko K., Coetzee W. A., Skolnik E. Y. (2005) Mol. Cell. Biol. 25, 3630–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srivastava S., Choudhury P., Li Z., Liu G., Nadkarni V., Ko K., Coetzee W. A., Skolnik E. Y. (2006) Mol. Biol. Cell 17, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacKeigan J. P., Murphy L. O., Blenis J. (2005) Nat. Cell Biol. 7, 591–600 [DOI] [PubMed] [Google Scholar]
- 21. Chojnowski A., Ravisé N., Bachelin C., Depienne C., Ruberg M., Brugg B., Laporte J., Baron-Van Evercooren A., LeGuern E. (2007) Neurobiol. Dis. 26, 323–331 [DOI] [PubMed] [Google Scholar]
- 22. Zou J., Chang S. C., Marjanovic J., Majerus P. W. (2009) J. Biol. Chem. 284, 2064–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 24. Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 25. Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 26. Taylor G. S., Maehama T., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S. A., Taylor G. S., Torgersen K. M., Dixon J. E. (2002) J. Biol. Chem. 277, 4526–4531 [DOI] [PubMed] [Google Scholar]
- 30. Laporte J., Kress W., Mandel J. L. (2001) Ann. Neurol. 50, 42–46 [DOI] [PubMed] [Google Scholar]
- 31. Bhaskar P. T., Hay N. (2007) Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 32. Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 34. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 37. Tronchère H., Laporte J., Pendaries C., Chaussade C., Liaubet L., Pirola L., Mandel J. L., Payrastre B. (2004) J. Biol. Chem. 279, 7304–7312 [DOI] [PubMed] [Google Scholar]
- 38. Blondeau F., Laporte J., Bodin S., Superti-Furga G., Payrastre B., Mandel J. L. (2000) Hum. Mol. Genet. 9, 2223–2229 [DOI] [PubMed] [Google Scholar]
- 39. Pendaries C., Tronchère H., Arbibe L., Mounier J., Gozani O., Cantley L., Fry M. J., Gaits-Iacovoni F., Sansonetti P. J., Payrastre B. (2006) EMBO J. 25, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Southgate R. J., Neill B., Prelovsek O., El-Osta A., Kamei Y., Miura S., Ezaki O., McLoughlin T. J., Zhang W., Unterman T. G., Febbraio M. A. (2007) J. Biol. Chem. 282, 21176–21186 [DOI] [PubMed] [Google Scholar]
- 41. Waddell D. S., Baehr L. M., van den Brandt J., Johnsen S. A., Reichardt H. M., Furlow J. D., Bodine S. C. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E785–E797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLoughlin T. J., Smith S. M., DeLong A. D., Wang H., Unterman T. G., Esser K. A. (2009) Am. J. Physiol. Cell Physiol. 297, C548–C555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi X., Bowlin K. M., Garry D. J. (2010) Stem Cells 28, 462–469 [DOI] [PubMed] [Google Scholar]
- 44. Clague M. J., Lorenzo O. (2005) Traffic 6, 1063–1069 [DOI] [PubMed] [Google Scholar]
- 45. Dove S. K., Johnson Z. E. (2007) Biochem. Soc. Symp. 74, 129–139 [DOI] [PubMed] [Google Scholar]
- 46. Berger P., Tersar K., Ballmer-Hofer K., Suter U. (2009) J. Cell Mol. Med. 15, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Velichkova M., Juan J., Kadandale P., Jean S., Ribeiro I., Raman V., Stefan C., Kiger A. A. (2010) J. Cell Biol. 190, 407–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ribeiro I., Yuan L., Tanentzapf G., Dowling J. J., Kiger A. (2011) PLoS Genet. 7, e1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marino J. S., Tausch B. J., Dearth C. L., Manacci M. V., McLoughlin T. J., Rakyta S. J., Linsenmayer M. P., Pizza F. X. (2008) Am. J. Physiol. Cell Physiol. 295, C1026–C1036 [DOI] [PubMed] [Google Scholar]
- 50. Gurpur P. B., Liu J., Burkin D. J., Kaufman S. J. (2009) Am. J. Pathol. 174, 999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Domin J., Harper L., Aubyn D., Wheeler M., Florey O., Haskard D., Yuan M., Zicha D. (2005) J. Cell. Physiol. 205, 452–462 [DOI] [PubMed] [Google Scholar]
- 52. Schenck A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., Zerial M. (2008) Cell 133, 486–497 [DOI] [PubMed] [Google Scholar]
- 53. Fritzius T., Burkard G., Haas E., Heinrich J., Schweneker M., Bosse M., Zimmermann S., Frey A. D., Caelers A., Bachmann A. S., Moelling K. (2006) Biochem. J. 399, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zoncu R., Perera R. M., Balkin D. M., Pirruccello M., Toomre D., De Camilli P. (2009) Cell 136, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., Zhu W. G. (2010) Nat. Cell Biol. 12, 665–675 [DOI] [PubMed] [Google Scholar]
- 57. Masiero E., Sandri M. (2010) Autophagy 6, 307–309 [DOI] [PubMed] [Google Scholar]
- 58. Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T., Laporte J., Deretic V. (2009) EMBO J. 28, 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dupont-Versteegden E. E., Strotman B. A., Gurley C. M., Gaddy D., Knox M., Fluckey J. D., Peterson C. A. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1730–R1740 [DOI] [PubMed] [Google Scholar]
- 60. Cheek D. B. (1985) Early Hum. Dev. 12, 211–239 [DOI] [PubMed] [Google Scholar]