Abstract
As in all other eukaryotic organisms, endoplasmic reticulum (ER) stress triggers the evolutionarily conserved unfolded protein response in soybean, but it also communicates with other adaptive signaling responses, such as osmotic stress-induced and ER stress-induced programmed cell death. These two signaling pathways converge at the level of gene transcription to activate an integrated cascade that is mediated by N-rich proteins (NRPs). Here, we describe a novel transcription factor, GmERD15 (Glycine max Early Responsive to Dehydration 15), which is induced by ER stress and osmotic stress to activate the expression of NRP genes. GmERD15 was isolated because of its capacity to stably associate with the NRP-B promoter in yeast. It specifically binds to a 187-bp fragment of the NRP-B promoter in vitro and activates the transcription of a reporter gene in yeast. Furthermore, GmERD15 was found in both the cytoplasm and the nucleus, and a ChIP assay revealed that it binds to the NRP-B promoter in vivo. Expression of GmERD15 in soybean protoplasts activated the NRP-B promoter and induced expression of the NRP-B gene. Collectively, these results support the interpretation that GmERD15 functions as an upstream component of stress-induced NRP-B-mediated signaling to connect stress in the ER to an osmotic stress-induced cell death signal.
Keywords: Cell Death, ER Stress, Oxidative Stress, Plant, Transcription Factors, Abiotic Stresses, ER Stress, Integration of Signaling Pathways, Plant Biology, Transcription Factors
Introduction
The endoplasmic reticulum (ER)6 is a key organelle that serves as the gateway for newly synthesized proteins into the secretory pathway. Following synthesis, secretory proteins are exported from the ER to various cellular compartments, where they fulfill their inherent biological roles. Under normal conditions, the processing capacity of the ER is dynamically balanced with the protein synthesis rate. Disruption of the equilibrium between the secretory activity of the cell and the processing and folding capacities of the ER promotes a condition that is known as ER stress. In general, perturbation of ER homeostasis by ER stress leads to the accumulation of unfolded proteins in the lumen of the organelle, triggering a cytoprotective signaling pathway designated as the unfolded protein response (UPR) (1, 2). In mammals, ER stress is sensed, and the UPR is transduced by three distinct classes of ER transmembrane proteins; inositol-requiring enzyme 1, protein kinase activated by dsRNA (PKR)-like ER kinase, and activating transcription factor 6 (1). Components of the plant UPR, functioning as proximal sensors, include Ire1p homologs and the activating transcription factor 6-related proteins AtbZIP28 and AtbZIP60 (2, 3). Upon activation, these receptors act in concert to transiently attenuate protein synthesis, up-regulate ER folding capacity, and degrade misfolded proteins. However, if the stress persists and the UPR is unable to restore ER homeostasis, a cell death signal is activated as an ultimate attempt for survival.
In mammalian cells, the UPR has been shown to be connected to other stress response pathways through shared components. For instance, the UPR-transducer PKR-like ER kinase, which phosphorylates eIF2α to down-regulate translation, also mediates phosphorylation of the transcription factor NRF2, which in turn translocates into the nucleus to up-regulate genes involved in redox maintenance (4). The ASK1/JNK stress-activated kinase pathway can be induced by both ER stress and oxidative stress to promote apoptosis (5). In addition, Ca2+ released from mitochondria causes UPR induction in ER (6). In plants, the potential of the ER stress response to accommodate adaptive pathways and its connection with other environmentally induced responses have been the subjects of studies in recent years (3, 7). One plant-specific, ER stress shared response is the ER stress- and osmotic stress-integrating signaling that converges on N-rich proteins (NRPs) to transduce a cell death signal (8). This integrated pathway was first identified by genome-wide expression profiling studies, which revealed the existence of a modest overlap between the ER stress- and osmotic stress-induced transcriptomes in soybean seedlings treated with PEG (an inducer of osmotic stress) or tunicamycin and azetidine-2-carboxylic acid, potent inducers of ER stress (9). The co-regulated genes, displaying similar induction kinetics and a synergistic response to the combination of osmotic and ER stress-inducing treatments, represent a shared response that integrates the two signaling pathways. Among them, a small family of NRPs (NRP-A, NRP-B, and NRP-C) shows the strongest synergistic effect of induction by multiple stresses. NRPs harbor a conserved cell death and developmental domain (10), are able to induce apoptosis in plants, and integrate the ER stress and osmotic stress responses (8). As an integrated pathway, the NRP-mediated cell death signaling is activated by either ER stress and osmotic stress but requires both signals for full activation.
Although NRP-mediated cell death signaling may allow versatile adaptation of cells to different stress conditions through the integration of stress responses, knowledge about this pathway is limited. In this investigation, we used a yeast one-hybrid system to identify transcriptional factors that are upstream components of the NRP-mediated cell death response. We isolated a soybean ERD15 (Early Responsive to Dehydration 15) homolog by its capacity to interact with the NRP-B promoter. ERD15 was first described in Arabidopsis as a dehydration-induced gene (11) that encodes a small acidic and hydrophilic protein belonging to the PAM2 domain-containing protein family (11–13). The PAM2 conserved domain enables interactions of ERD15 with the C terminus of poly(A)-binding proteins (PABP) (14, 15). More recently, ERD15 has been shown to function as a negative regulator of the abcisic acid (ABA)-mediated response (13). Overexpression of ERD15 reduces the ABA sensitivity of Arabidopsis, whereas silencing of ERD15 by RNAi promotes hypersensitivity to the hormone. The negative effect of ERD15 on ABA signaling enhances salicylic acid-dependent defense because overexpression of ERD15 was associated with increased resistance to the bacterial necrotroph Erwinia carotovora, and the enhanced induction of marker genes for systemic acquired resistance. These results are consistent with the observed antagonistic effect of ABA on salicylic acid-mediated defense and may implicate ERD15 as a shared component of these responses.
Based on the presence of the PAM2 domain, a possible function for ERD15 in some aspect of RNA metabolism has been proposed to underlie a biochemical role for the protein in the ABA signaling response (13). In this model, ERD15 would act at the post-transcriptional level to regulate ABA signal transduction. In contrast, we show here that the soybean ERD15 homolog defines a novel class of plant-specific transcriptional factors that bind to the NRP-B promoter in vitro and in vivo to activate the NRP-B-mediated cell death response.
EXPERIMENTAL PROCEDURES
cDNA Library Construction
A cDNA library was prepared from mRNA isolated from tunicamycin- and PEG-treated soybean leaves (Glycine max, variety Conquista). The plants were initially germinated in soil, acclimatized in a greenhouse, and then transferred to a solution containing 10 μg/ml tunicamycin (Sigma) and 10% (w/v) PEG (molecular weight, 8000) one week after germination. Plant material was collected after 2 h of treatment, immediately frozen in liquid nitrogen, and stored at −80 °C until use. Polyadenylated mRNAs were purified with the Fast Track mRNA isolation kit from Invitrogen. cDNAs were synthesized and cloned into yeast expression plasmids with a SuperScript plasmid system and gateway technology for cDNA synthesis and cloning (Invitrogen), using 5 μg of poly(A) mRNA. Then the cDNAs were cloned as SalI-NotI inserts into the pEXP-AD502 vector (Invitrogen) and transformed into Escherichia coli DH5α cells by electroporation to yield a cDNA expression library fused downstream of a GAL4 activation domain. The average size of the inserts was ∼1 kb. After plating of the E. coli transformants, plasmid DNA was purified with the Qiaprep kit (Qiagen) to a concentration of 250 ng/μl.
NRP-B Promoter Reporter Constructs
A 1000-bp promoter fragment of the GmNRP-B gene was amplified from soybean DNA with the appropriate primers (supplemental Table S1) and cloned into the TOPO-pCR4 vector (Invitrogen) to yield pUFV1457. Likewise, 700- and 350-bp fragments upstream of the translational initiation codon of the GmNRP-B promoter were amplified from pUFV1457 with specific primers (supplemental Table S1) and cloned into the same vector to yield pUFV1458 and pUFV1459, respectively. The 1000-bp promoter fragment from pUFV1457 was cloned directionally (5′ → 3′) into the EcoRI-SacI sites of the pHis2.1 vector, a Trp-marked plasmid, in front of a HIS3 reporter gene under the control of the minimal, inactive promoter GAL1 to generate pUFV1466. For the second reporter gene construct, the 1000-, 700-, and 350-bp GmNRP-B promoter fragments were excised from TOPO-pCR4 with EcoRI-SalI digestion and cloned into the pLacZi integrative vector, a URA-marked plasmid, in front of the lacZ reporter gene under the control of a minimal, inactive CYC1 (cytochrome C1) promoter, resulting in pUFV1463, pUFV1464, and pUFV1465, respectively. A 350-bp fragment of the GmBiPD promoter was obtained from the clone pUFV228 (16) by digestion with EcoRI and SalI restriction enzymes and cloned into the pLacZi vector to be used as a negative control. The resulting clone, pUFV1226, contains the lacZ reporter gene under the control of an active soyBiPD promoter (16).
To construct a NRP-B promoter-Gus fusion, a 1-kb promoter sequence was released from pUFV1457 with EcoRI/SalI and cloned into the same sites of pCAMBIA1381Z to yield −1000pNRP-B::Gus. An internal deletion in the NRP-B promoter was obtained by releasing the 263-bp XbaI fragment of −1000pNRP-B::Gus (XbaI, positions −666 and −403) and circularizing it in vitro to give Δ(−666/−403)pNRP-B::Gus.
GmERD15 Cloning and Constructs
To clone the complete GmERD15 cDNA sequence into different expression vectors, the full-length cDNA was amplified using the primers Fw-GmERD15 and GmERD15st-Rv (supplemental Table S1) and inserted by recombination into the entry vector pDONR201 (Invitrogen) to yield pDONR201-GmERD15. Then GmERD15 cDNA was transferred from pDONR201-GmERD15 (pUFV1358.1) to different expression vectors (pDEST17, pDEST22, pDEST32, pK7WG2, and pCAMBIA1300-YFP-pnos), by recombination, using the enzyme LR clonase (Invitrogen). The resulting clones include the following: pBD-GmERD15 (pUFV1379) and pAD-GmERD15 (pUFV1425), for expression in yeast; pHis-GmERD15 (pUFV1380), for expression in bacteria; and p35S-GmERD15 (pUFV1469) and pYFP-GmERD15 (pUFV1470), for expression in plants.
Yeast One-hybrid Cloning and Detection System
The reporter yeast strain was constructed by co-transforming strain W303 (MATa/MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+]) with both pUFV1466 and pUFV1463 constructs (1 μg each), using a lithium acetate protocol (18), resulting in the modified yeast strain W303 PNRPB-HIS3/LACZ (pUFV1325). The reporter yeast was transformed with the cDNA library. Double screening was performed sequentially, as described by Kim et al. (18). Putative positive yeast clones were grown for 2 days and assayed for β-galactosidase activity colorimetrically, according to Amberg et al. (19). Yeast plasmid DNA was prepared and transferred to E. coli DH5α cells by electroporation. After the plating of E. coli transformants, plasmid DNA was prepared and purified with the Qiaprep kit (Qiagen). DNA sequencing was performed by Macrogen (Seoul, Korea), and database searches were performed using the Basic Local Alignment Search Tool (BLAST) algorithm.
To verify the influence of deletions in the GmNRP-B promoter in the yeast one-hybrid system, reporter yeasts strains were constructed by transforming strain W303 with pUFV1466, pUFV1467 and pUFV1468. The resulting modified strains W303 pNRPB-1000bp/HIS3/LACZ (pUFV1325), W303 pNRPB-700bp/LACZ (pUFV1597), and W303 pNRPB-350bp/LACZ (pUFV1598), respectively, were transformed with pAD-GmERD15 (pUFV1425). The modified yeast strains were grown for 2 days and assayed for β-galactosidase activity, as previously described (19).
Electrophoretic Mobility Shift Assay
A 187-bp fragment of 5′-flanking sequences of GmNRP-B was amplified from genomic DNA using specific oligonucleotides (gaattcTTATCTAGCATCAGTGCTTG and gtcgacCCAAAACGCGTACACGTGAA) and then biotinylated using the 3′ OH biotin labeling kit from Pierce. Electrophoretic mobility shift assays were performed using a LightShift Chemiluminescent EMSA kit (Pierce), according to the manufacturer's protocols. Each 20-μl binding reaction contained 20 mm HEPES buffer at pH 7.9, 100 mm KCl, 5% (v/v) glycerol, 1 mm MgCl2, 1 mm DTT, 0.1% (v/v) Tween 20, 25 ng/ml poly(dI-dC), 20 fmol biotin-labeled probe, and 2 μg of His-GmERD15. Unlabeled DNA was used as a competitor, and 1 μl of anti-His-GmERD15 antibody was added after the incubation for identification of the protein-DNA complex. The binding reactions were incubated at room temperature for 20 min. A 6% polyacrylamide minigel (37.5:1 acrylamide: bisacrylamide in 0.5 mm Tris borate-EDTA containing 3% glycerol) was prerun for 1 h at 100 V with 0.5 mm Tris borate-EDTA (TB buffer), and 5 μl of loading buffer was added to the binding reaction before 20 μl of the reaction was separated by gel electrophoresis at 80 V for 90 min. The gel was then transferred to a charged nylon membrane (Amersham Biosciences Bioscience) for 1 h at 320 mA, UV cross-linked to the membrane at 120 mJ/cm2 for 1 min, and visualized by chemiluminescence.
Transactivation Assays in Yeast Cells
The presence of a functional transactivation domain in GmERD15 was monitored through a yeast transactivation assay, as described previously (20). The GmERD15 coding region was fused to the GAL4-binding domain (pUFV1379), and the transactivation of reporter genes was assayed in Saccharomyces cerevisiae strain AH109 (MATa, Trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, LYS2::GAL1UAS-GAL1TATAHIS3, MEL1 GAL2UAS-GALTATA::MELUAS-MEL1TATA-lacZ). AH109 competent cells were transformed with pBD-GmERD15 (pUFV1379) using the lithium acetate method (17). Transformants were plated on synthetic dropout medium lacking leucine and histidine, but supplemented with 10 mm 3-aminotriazole, and cultured for 3 days at 28 °C. β-Galactosidase activity was monitored colorimetrically using cultures grown for 2 days, according to Amberg et al. (19).
Soybean Suspension Cells and Stress Treatments
Soybean suspension cells were cultured in MS-soybean medium (21) and subcultured weekly by diluting 5 ml of suspension cells into 50 ml of fresh medium. Treatments were performed on 5-day-old subcultures. The cells were treated with 5 μg/ml of tunicamycin or 10% (w/v) polyethylene glycol for 30, 60, 90, and 120 min. After treatment, the cells were filtrated under vacuum, washed with distilled water, and immediately frozen in liquid N2.
cDNA Synthesis and Real Time PCR Analysis
Total RNA was extracted from tissues and cells with TRIzol (Invitrogen) and treated with 3 units of RNase-free DNase (Promega). Purified RNA was quantified spectrophotometrically (Thermo Fisher Scientific EVO 60) and analyzed in denaturing 1.5% (w/v) agarose gel. The cDNA synthesis was performed with 4 μg of total RNA, oligo(dT), and reverse transcriptase Moloney murine leukemia virus (Invitrogen) according to the protocol of the manufacturer.
All of the procedures for real time PCR, including tests, validations, and experiments, were conducted according to the recommendations of Applied Biosystems. Real time PCRs were performed on an ABI7500 instrument (Applied Biosystems), using specific primers (supplemental Table S2), cDNAs from the treatments, and SYBR® Green PCR Master Mix (Applied Biosystems). The amplification reactions were performed under the following conditions: 10 min at 95 °C, 40 cycles of 94 °C for 15 s, and 60 °C for 1 min. Gene expression was quantified using the comparative methods Ct: 2 −ΔCt and 2−ΔΔCt. For quantitation of gene expression in soybean cells and seedlings, we used RNA helicase (9) as the endogenous control gene for data normalization in real time PCR analysis.
Transient Expression in Soybean Protoplasts
Protoplasts were prepared from soybean suspension cells essentially as described by Costa et al. (8). Transient expression assays were performed by electroporation (250 V, 250 microfarads) of 10 μg of expression cassette DNA and 30 μg of sheared salmon sperm DNA into 2 × 105–5 × 106 protoplasts in a final volume of 0.8 ml. Protoplasts were diluted into 8 ml of MS medium supplemented with 0.2 mg/ml 2,4-dichlorophenoxyacetic acid and 0.6 m mannitol at pH 5.5. After 36 h of incubation in the dark, the protoplasts were washed with 0.6 m mannitol with 20 mm MES at pH 5.5 and frozen in liquid N2 until use.
Transient Overexpression in Nicotiana tabacum by Agrobacterium Infiltration and Subcellular Localization of YFP-GmERD1
Three- to four-week-old tobacco leaves were infiltrated with Agrobacterium strain GV3101 carrying pYFP-GmERD15 (pUFV 1470), as described (22). Three days after inoculation, transformed leaves were observed with an inverted Zeiss LSM 510 META laser scanning microscope (Jena, Germany), using a 40× oil immersion lens and an argon laser. For imaging of YFP, the excitation line was 514 nm, and the emission data were collected at 535–590 nm. The pinhole was usually set to give a 1.5–2.0-μm optical slice. Post-acquisition image processing was done using the LSM Image Browser 4 (Zeiss) and Adobe Photoshop 7.0.1 software. To obtain a GFP-NSP fusion to be used as a nuclear marker, the NSP cDNA was transferred from the entry vector pDON-NSP (23) to the binary vector pK7FWG2, generating pGFP-NSP, which contains the GFP gene fused in-frame after the last codon of the NSP cDNA.
His6-tagged Protein Expression and Purification
To express GmERD15 in bacteria, the E. coli BL21-pLys host strain was transformed with the clone pHis-GmERD15 (pUFV1380). After induction with 2 mm isopropyl β-d-thiogalactopyranoside, each culture was incubated for an additional 6 h. Total proteins were extracted under both nondenaturing and denaturing conditions, and His-GmERD15 was purified by nickel-nitrilotriacetic acid agarose affinity chromatography and eluted under native conditions according to the QIAexpressionist protocol (Qiagen). Purified His-GmERD15 was used as antigen for antibody production in rabbits, which were immunized through subcutaneous injections during 2-week intervals three times (24).
ChIP Assay
The ChIP assay was performed using a chromatin immunoprecipitation assay kit (Imprint ChIP kit; Sigma) following the manufacturer's instructions. Briefly, protoplasts derived from soybeans suspension cells were treated with tunicamycin for 30 min, the proteins bound to DNA were cross-linked with 1% formaldehyde, and cells were subsequently resuspended in the lysis buffer, followed by sonication. The protein extracts were incubated with antibody against GmERD15, human RNA polymerase II, or normal mouse IgG. Immunocomplexes were extensively washed, and the DNA was recovered by using a chromatography column and eluted in 50 μl of 10 mm Tris-HCl, pH 8.5. The DNA was used as a template for PCR amplification with sets of primers corresponding to the 187-bp GmNRP-B promoter fragment identified in the EMSA assay (5′-GAATTCTTATCTAGCATCAGTGCTTG-3′ and 5′-GtcgacCCAAAACGCGTACACGTGAA-3′). As a negative control, PCR was carried out with a set of primers for a 1000-bp GmNAC6 promoter fragment.
Sequence Analysis
The ERD15 protein sequences were obtained from the NCBI website. Multiple sequence alignments were performed using ClustalW.
RESULTS
A Tunicamycin- and PEG-induced Soybean ERD15 Homolog Associates Stably with the NRP-B Promoter in Yeast
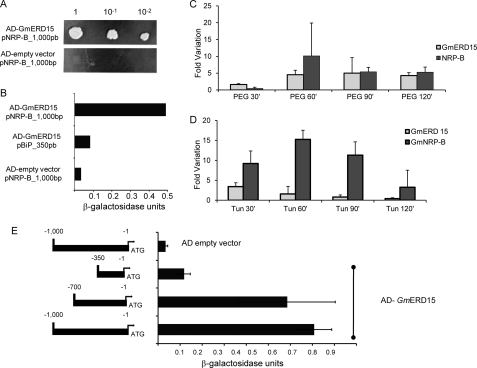
Because NRP-B (8) and NRP-A (accession number AJ875407) are strongly induced by tunicamycin (a potent inducer of ER stress) and PEG (an inducer of osmotic stress), we rationalized that the transcriptional factors controlling NRP expression would accumulate at higher levels under these induced conditions (8, 9). Therefore, we prepared a GAL4-fused cDNA library derived from soybean leaves treated with tunicamycin and PEG, and we used a 1-kb upstream sequence of the NRP-B gene as bait to isolate transcriptional factors mediating NRP-B expression through the yeast one-hybrid system. From ∼4,000 yeast clones screened, one candidate displaying histidine prototrophy was selected for further testing, followed by retransformation of the yeast modified strain, W303 pNRPB-HIS3/LACZ, with the isolated plasmid. The resulting transformants were able to activate histidine/adenine auxotrophy (Fig. 1A) and high levels of β-galactosidase expression (Fig. 1B). The isolated cDNA-encoded product associated specifically with NRP-B promoter sequences, because the expression of the GAL4-fused cDNA did not activate the lacZ reporter gene under the control of a soybean BiP promoter (Fig. 1B).
FIGURE 1.
A tunicamycin- and PEG-induced ERD15 soybean homolog associates stably with the NRP-B promoter in yeast. A, isolation of GmERD15 by one-hybrid screening in yeast. An S. cerevisiae strain carrying the pNRPB-HIS3 reporter was transformed with the GAL4 activation domain-encoded vector or GAL4AD vector containing the cDNA of GmERD15. After growth for 12 h, the yeast culture was diluted, as indicated in the figure, and plated on medium lacking histidine but supplemented with 100 mm 3-aminotriazole and incubated for 3 days. B, LacZ reporter expression. β-Galactosidase activity was determined from total protein extracts of yeast strains carrying the indicated combinations of the empty expression vector and AD-GmERD15. As a control, a pBip_350bp-LacZ integrated strain was transformed with AD-GmERD15. Thin bars indicate the standard deviation of three independent experiments. C, kinetics of GmERD15 and GmNRP-B induction by PEG-induced osmotic stress in soybean-cultured cells. Levels of RNA were examined at various times after PEG treatment by quantitative RT-PCR. Expression values were calculated using the 2 − ΔCt method with helicase used as an endogenous control. cDNA was prepared from three biological replicates. Thin bars indicate the standard deviation. D, induction of GmERD15 and NRP-B by tunicamycin. Soybean-cultured cells were treated with tunicamycin for the indicated times. Levels of RNA were examined by quantitative RT-PCR, as described in C. E, GmERD15 associates to a discrete region of NRP-B promoter in yeast. The schematic representation of the promoter constructs indicates the 5′-flanking sequences of NRP-B fused to LacZ that were integrated into the W303 strain and transformed with AD-GmERD15 or the empty vector. The activity of the β-galactosidase was expressed in units as described under “Experimental Procedures.” Thin bars indicate the standard deviation of three independent experiments.
The isolated clone harbors a full-length cDNA that encodes a protein that is 100% identical to an ERD15-related soybean protein (accession Glyma 02g42860.1) of unknown function. The deduced protein from the isolated cDNA, here designated GmERD15 (G. max ERD15), contains 125 amino acid residues (calculated Mr 14,696 and pI 5.00), harbors a PABP-interacting motif, PAM2, at the N terminus (positions 6–27), and shares sequence conservation with ERD15-like proteins from other plant species (ranging from 36 to 58% identity; supplemental Fig. S1). The N terminus, encompassing the PAM2 domain, is highly conserved among plant proteins (average identity, 80–90%) and less conserved in GmERD15 (average identity, 57%). Like the ERD15 gene from Arabidopsis (13), GmERD15 is rapidly induced by the osmotic stress inducer PEG (Fig. 1C). It is also up-regulated by the ER stress inducer tunicamycin (Fig. 1D), mimicking the pattern of NRP expression, although with slightly different kinetics (8, 9).
PEG-induced GmERD15 transcript accumulation began as early as 30 min after treatment and increased rapidly until saturation occurred after 1 h (Fig. 1C). Induction of NRP-B by PEG occurred with slightly delayed response kinetics, because increased levels of NRP-B transcripts in PEG-treated seedlings were first detected after 1 h and then declined. In tunicamycin-treated seedlings, GmERD15 RNA levels were rapidly increased with maximal accumulation 30 min post-treatment (Fig. 1D). NRP-B RNA induction was initially detected by 30 min post-treatment, reached full induction after 1 h, and declined shortly after that. Thus, for both tunicamycin and PEG treatments, an increased expression of GmERD15 and transcript accumulation preceded induced expression of the NRP-B gene. ER stress and osmotic stress marker genes were included in the assay to ensure the efficiency of the tunicamycin and PEG treatments (data not shown).
We have found that GmERD15 stably associates with the NRP-B promoter in yeast. Nevertheless, ERD15-like proteins have not been implicated as DNA-binding proteins; hence, conserved cis-regulatory elements for ERD15 binding on promoters of plant genes are unknown. To map the localization of ERD15 binding sites on the NRP-B promoter, the lacZ reporter gene was placed under the control of different extensions of the 5′-flanking sequences of NRP-B, up to positions −1000, −700, and −350 relative to the translational initiation codon. The yeast strain W303 was transformed with the resulting DNA constructs, and reporter gene transcriptional activation was monitored upon expression of a GmERD15-GAL4 activation domain fusion (Fig. 1E). Transactivation of the NRP-B 1-kb 5′-flanking sequences by expression of GmERD15-GAL4 fusion promoted high levels of β-galactosidase activity (0.7074 ± 0.08). Deletion of promoter sequences up to position −700 caused only a small reduction in ERD15-GAL4-mediated transactivation of the NRP-B promoter, and β-galactosidase activity was kept at high levels (0.5864 ± 0.217). In contrast, deletion of NRP-B upstream sequences up to position −350 drastically affected the capacity of the GmERD15-GAL4 fusion to transactivate the NRP-B promoter, and β-galactosidase activity was reduced to 0.1171 ± 0.03. These results suggest the presence of ERD15 binding sites within position −700 to −350 and further demonstrate that GmERD15 associates specifically with NRP-B promoter sequences.
GmERD15 Is a DNA-binding Protein That Exhibits Transactivation Activity in Yeast
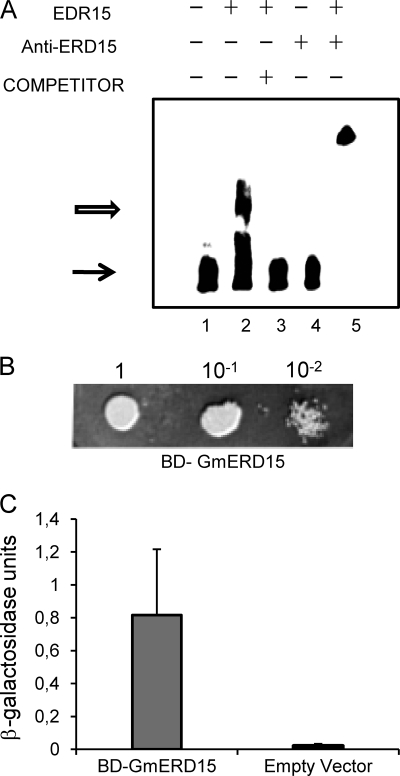
The finding that GmERD15 binds to the NRP-B promoter and induces the expression of reporter genes in yeast prompted us to investigate the DNA binding and transactivation properties of this protein that does not contain any of the typical motifs found in most known DNA-binding proteins. To confirm that GmERD15 binds specifically to NRP-B sequences limited by positions −700 to −350, we performed an EMSA using a 187-bp fragment of the NRP-B promoter (from position −340 to −517). Incubation of the labeled 187-bp promoter fragment with a purified, His-tagged GmERD15 (supplemental Fig. S2) resulted in DNA-protein complex formation, as judged by the shift in the electrophoretic mobility of the labeled DNA (Fig. 2A, lane 2). The DNA-protein interactions were specific as introduction of a 40-fold excess of unlabeled probe competed for binding (lane 3). Furthermore, the DNA-protein complex was supershifted to a higher position by an anti-GmERD15 serum, indicating that the protein contained in the complex was indeed GmERD15 and not an E. coli contaminant (lane 5). This interpretation was substantiated by the observation that the anti-GmERD15 antibodies did not bind directly to the labeled DNA (lane 4). The specificity of GmERD15 binding to the NRP-B promoter was further confirmed by the incapacity of GmERD15 to bind to a soybean BiP promoter, used as a negative control (supplemental Fig. S3A). We have also shown that the unrelated GST protein did not bind to the 187-bp NRP-B promoter fragment (supplemental Fig. S3B). Collectively, these results establish that GmERD15 binds specifically to the NRP-B promoter in vitro.
FIGURE 2.
GmERD15 is a DNA-binding protein that exhibits transactivation activity in yeast. A, gel shift assays. A 187-bp biotin-labeled DNA fragment of the GmNRP-B 5′-flanking sequences (positions −330 to −517; lane 1) was incubated with purified, His-tagged GmERD15 in the absence (lane 2) and presence of 4 pmol of unlabeled DNA probe (lane 3) for 20 min at RT. The products were separated by electrophoresis in a 5% polyacrylamide gel in TB buffer. Lane 4 indicates probe plus anti-GmERD15 antisera, and lane 5 indicates probe plus anti-GmERD15 antisera plus recombinant GmERD15 protein. The solid arrow indicates the free DNA, and the open arrow designates the DNA-protein complexes. B, transactivation activity of the GmERD15 protein in yeast. pBD-GmERD15 and the empty vector (pDEST32) were introduced separately into yeast strain AH109. Overnight growth cultures were diluted as indicated in the figure and plated on medium lacking histidine but supplemented with 100 mm 3-aminotriazole and incubated for 3 days. C, GmERD15 transactivates expression of a lacZ reporter gene. β-Galactosidase activity was determined from total protein extracts of overnight growth yeast strains carrying the indicated DNA construct. Thin bars indicate the standard deviation of the three independent experiments.
A close inspection of the primary structure of GmERD15 using the BLOCKS program revealed the presence of a conserved sequence of 13 amino acids at positions 71–83 (71DEDEKERKEGKEV83), similar to that found on ssDNA-binding transcriptional regulators (25). To examine whether GmERD15 harbors a functional transactivator domain, the coding region of GmERD15 was fused to the binding domain of GAL4 (BD-GAL4) and expressed as a fusion protein in a yeast strain harboring the reporter genes HIS3 and lacZ under the control of the GAL1 promoter. Expression of BD-GAL4 fused to GmERD15 promoted growth of yeast in the absence of histidine and presence of 3-aminotriazole (Fig. 2B) and induction of lacZ expression as measured by β-galactosidase activity (0.817 ± 0.013; Fig. 2C). These results demonstrate that GmERD15 exhibits transactivation activity in yeast.
GmERD15 Localizes in the Nucleus of Tobacco Leaf Cells and Binds to the NRP-B Promoter in Vivo
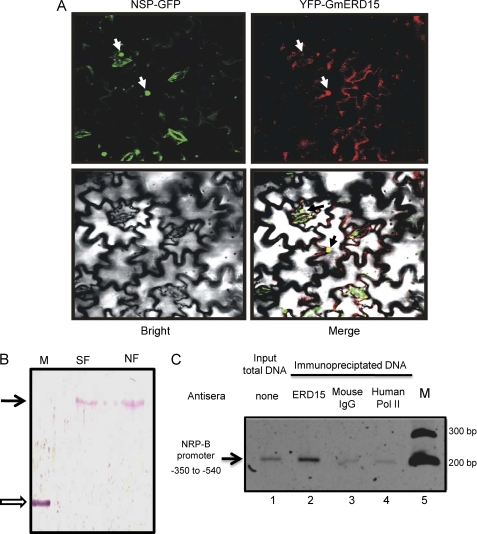
To determine the subcellular localization of GmERD15, we utilized an Agrobacterium-mediated transient expression assay in epidermal cells from tobacco leaves and analyzed by confocal microscopy the localization of a chimeric GmERD15 protein fused to YFP (Fig. 3A). Although relatively strong GFP signals accumulated in the cytoplasm, the YFP-GmERD15 fusion was also detected in the nucleus of transfected cells. The bottom right panel of Fig. 3A is an overlay of the fluorescent images of YFP-GmERD15 and NSP-GFP with the light microscopy image; the figure shows the nuclear YPF-GmERD15 fluorescent signal co-localized with a geminivirus NSP that accumulates in the nucleus of transfected tobacco leaf cells (26) and served here as a nuclear marker. The nucleocytoplasmic distribution of YFP-GmERD15 was further confirmed by immunoblotting nuclear extracts and soluble fractions from transfected tobacco leaves with an anti-ERD15 serum (Fig. 3B). An intact, native GmERD15 was also detected in nuclear extracts prepared from tunicamycin-treated soybean cells (data not shown). The presence of the protein both in the cytoplasm and nucleus would be consistent with dual roles for this protein in distinct subcellular compartments.
FIGURE 3.
GmERD15 localizes to the nucleus of tobacco leaf cells and binds to the NRP-B promoter in vivo. A, confocal fluorescence image of transiently expressed GmERD15-YFP in epidermal cells of tobacco leaves. Tobacco leaves were infiltrated with Agrobacterium tumefaciens carrying 35S::GFP-NSP or 35S::GmERD15-YFP DNA constructs and observed by confocal microscopy 3 days after agroinoculation. The white arrows show the nuclear positions of the nuclear viral protein NSPs (left panel) and GmERD15 (right panel). The black arrows in the bottom right panel show the co-localization signal of GmERD15 with the nuclear marker NSP. B, immunoblotting of nuclear fractions of tobacco leaves expressing YFP-GmERD15. Soluble (SF) and nuclear fractions (NF) of tobacco leaves expressing YFP-GmERD15 were immunoblotted with anti-GmERD15 antibodies. The open arrow indicates the position of purified His-GmERD15, and the black arrow indicates the position of YFP-GmERD15. M is molecular mass. C, ChIP with tunicamycin-treated soybean suspension cells using anti-GmERD15 antibodies. The 187-bp DNA fragment of NRP-B 5′-flanking sequences was detected by PCR amplification when immunoprecipitation was mediated by anti-GmERD15 antibodies (lane 2) but not by unrelated anti-polymerase (Pol) II antibodies (lane 4) or IgG (lane 3). Lane 1 represents the amplification from input total DNA, and M represents the molecular mass standards.
ChIP experiments were performed to examine whether GmERD15 binds to the NRP-B target promoter in vivo using tunicamycin-induced soybean cells and anti-GmERD15 antibodies (Fig. 3C). Although no significant amount of NRP-B promoter was immunoprecipitated from control soybean protoplasts (date not shown), an NRP-B promoter fragment was pulled down from the tunicamycin-induced soybean protoplasts (second lane). This result was consistent with the up-regulation of both GmERD15 and NRP-B expression by tunicamycin. The immunoprecipitation of NRP-B promoter DNA was mediated by the specific binding of GmERD15, as evidenced by the fact that the anti-GmERD15 antibody did not immunoprecipitate an NAC6 promoter, used as a negative control (data not shown), and neither anti-human RNA polymerase II antibodies nor an IgG antibody mediated immunoprecipitation of the NRP-B promoter (third and fourth lanes). These results further substantiate the argument that GmERD15 functions as a DNA-binding protein that specifically associates with the NRP-B promoter in vivo.
GmERD15 Activates the NRP-B Promoter and Induces Expression of the NRP-B Gene in Soybean Protoplasts
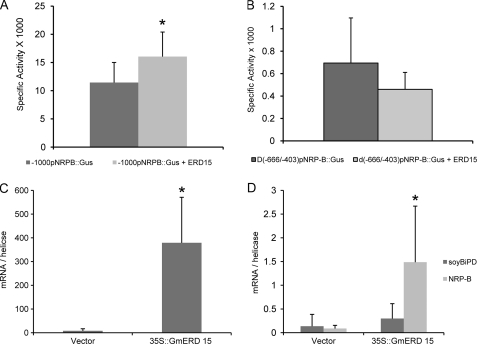
Because GmERD15 binds to the NRP-B promoter both in vitro and in vivo and exhibits transcriptional activity in yeast, we tested whether GmERD15 would activate transcription in a transient expression assay in soybean protoplasts with an NRP-B promoter::β-glucuronidase reporter construct. Expression of GmERD15 resulted in increased reporter gene expression (Fig. 4A), indicating that GmERD15 activates the NRP-B promoter in vivo. Because NRP-B expression has been shown to be induced by wounding (supplemental Fig. S4), the relatively high expression of the NRP-B promoter in the absence of ectopic expression of GmERD15 in soybean protoplasts is most likely due to a wounding signal that is undoubtedly generated during protoplast preparation. In addition, NRPs have also been shown to be induced by infection with bacterial pathogens (27), and treatment of plant cells with plant cell wall-degrading enzymes during the protoplasting procedure has been shown to mimic bacterial pathogen attack and induce a defense response (28). Thus, it is not surprising that ectopic expression of GmERD15 caused only a small, but statistically significant, enhancement in promoter activity under these conditions. This interpretation is further supported by the fact that the activity of the NRP-B promoter was monitored by measuring the enzymatic activity of a highly stable reporter protein, and the low transfection efficiency of soybean protoplasms was not normalized to an internal standard. In contrast, a much higher fold induction of NRP-B expression by GmERD15 expression (Fig. 4C) can be detected by directly measuring NRP-B transcript accumulation (Fig. 4D). This is consistent with the fact that NRP-B transcripts are rapidly and transiently induced by wounding and cell wall-degrading enzyme treatment (supplemental Fig. S4); hence, at 48 h after transfection, these stress signals are not expected to mask GmERD15-mediated NRP-B transcript accumulation. We also demonstrated that deletion of an internal fragment (positions −660 to −403) in the NRP-B promoter abolished completely transactivation of the NRP-B promoter by GmERD15 (Fig. 4B). This deleted fragment overlaps partially with sequences of the 187-bp fragment that interacted with GmERD15 in vitro (Fig. 2A). Taken together, these results indicate that GmERD15 directly binds to and regulates expression of the NRP-B gene in vivo.
FIGURE 4.
GmERD15 activates the NRP-B promoter and induces NRP-B expression. A, transient expression of GmERD15 in soybean protoplast activates a NRP-B promoter::β-glucuronidase gene. Soybean protoplasts were co-electroporated with plasmids carrying −1000pNRP-B::β-glucuronidase (Gus) gene and either 35S::GmERD15 DNA constructs (light gray bars) or empty vector (dark gray bars). After 48 h, β-glucuronidase activity (nmol/min/mg protein) was measured from the total protein extracts of transfected soybean cells. The thin bars indicate the standard deviation of 15 biological replicates. The asterisks indicate the mean values statistically different from the control treatment according to Student's t test with the two-tailed p value 0.000846. B, an internal deletion in the promoter sequence of NRP-B abolishes ERD15 transactivation of NRP-B promoter in soybean cells. Soybean protoplasts were co-electroporated with plasmids carrying Δ(−666/−403)pNRP-B::β-glucuronidase gene and either 35S::GmERD15 DNA constructs (light gray bars) or empty vector (dark gray bars). The mean values do not differ from each other according to Student's t test with the two-tailed p value 0.083458. C, transient expression of GmERD15 in soybean protoplasts. Plasmid containing a GmERD15 expression cassette was electroporated into soybean protoplasts, and gene expression was monitored by quantitative RT-PCR. Expression values were calculated using the 2−ΔCt method and helicase as an endogenous control. The thin bars indicate the standard deviation of five biological replicates. The asterisks denote mean values statistically different from the control treatment according to Student's t test with the two-tailed p value 0.0000527. D, transient expression of GmERD15 in soybean protoplasts induces GmNRP-B expression but not soyBiPD expression. GmNRP-B (light gray bars) and soyBiPD (dark gray bars) transcript levels were determined by quantitative RT-PCR in protoplasts transformed with empty vector pK7WG2 (PK7) or a GmERD15 expression cassette (PK7 GmERD-15). Expression values were calculated using the 2−ΔCt method and helicase as an endogenous control. The thin bars indicate the standard deviations of five biological replicates. The asterisks denote the mean values statistically different from the control treatment according to Student's t test with the two-tailed p value 0.00563.
ERD15 Binds Specifically to a 12-bp Palindromic Sequence on NRP-B Promoter to Activate Transcription
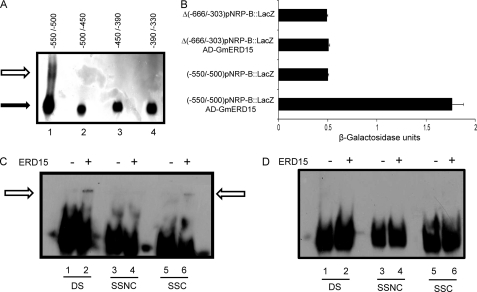
To map precisely the binding sites for GmERD15 on the NRP-B promoter, we first performed gel shift assay using 50- and 60-bp sequences (positions −550/−500; −500/−450; −450/−390; and −390/−330) covering the 187-bp fragment that interacted with GmERD15, as probes (Fig. 5A). GmERD15 bound stably with the −550/−500 fragment of the NRP-B promoter and failed to interact with the other three fragments covering positions −500 to −330 on the NRP-B promoter. We next asked whether the 50-bp (−550/−500) fragment would be functional and sufficient to activate expression of a minimal promoter in the presence of GmERD15 in yeast. This fragment was linked to a yeast minimal promoter of CYC1, and we analyzed its capacity to activate reporter gene expression when GmERD15 was provided in trans (Fig. 5B). We found that the 50-bp binding site of GmERD15 was sufficient to support GmERD15-mediated activation of a minimal promoter in yeast. Consistent with these data, a promoter fragment lacking the 50-bp (−550/−500) fragment by deleting the −666/−403 region of NRP-B failed to be transactivated by GmERD15 (Fig. 5B).
FIGURE 5.
ERD15 binds specifically to an inverted repeat sequence to activate NRP-B promoter. A, GmERD15 binds to a 50-bp sequence limited by positions −550 to −500. Sequential 50- and 60-bp biotin-labeled DNA fragments of the NRP-B promoter spanning from positions −550 to −330 (as indicated in the figure) were incubated with purified, His-tagged GmERD15 for 20 min at room temperature. The products were separated by electrophoresis in a 5% polyacrylamide gel in TB buffer. The solid arrow indicates the free DNA, and the open arrow designates the DNA-protein complexes. B, the 50-bp fragment (−550/−500) is sufficient to promote GmERD15-mediated transactivation of the minimal promoter of the yeast CYC1 gene. The 50-bp fragment of NRP-B promoter, positions −550 to −500, was cloned into pLACZi upstream to the minimal promoter of CYC1 and to the LacZ gene. β-Galactosidase activity was determined from total protein extracts of overnight growth yeast strains carrying the indicated DNA construct. C and D, GmERD15 binds to the −12-bp −511AGCAnnnnTGCT−500 palindromic sequence but not to a −25-bp palindromic sequence (−424ACGATTCTAnnnnnnnnTAGAACGT−399) of the NRP-B promoter. The 12-bp (C) and the 25-bp (D) palindromic sequences were biotin-labeled and incubated with purified, His-tagged GmERD15 in the absence (lanes 1, 3, and 5) and presence of 4 pmol of unlabeled DNA probe as double-stranded form (lanes 2), noncoding strand (lanes 4), and coding strand (lanes 6) for 20 min at room temperature. The products were separated by electrophoresis in a 5% polyacrylamide gel in TB buffer. The open arrows designate the DNA-protein complexes. DS, double-stranded DNA; SSC, single-stranded coding sequence; SSNC, single-stranded noncoding sequence.
Inspection of the 50-bp binding site (position −550/−500) revealed the presence of the 12-bp palindromic sequence −511AGCAnnnnTGCT−500 (supplemental Fig. S5). Given that inverted repeat sequences have been demonstrated to function as binding sites for ssDNA-binding proteins (33), we examined whether GmERD15 would bind to the double-stranded form, noncoding strand, or coding strand of the palindromic sequence by gel shift (Fig. 5C). Protein-DNA complexes were formed by incubation of GmERD15 with −511AGCAnnnnTGCT−500 as double-stranded DNA (lane 2, DS) and single-stranded coding sequence (lane 6, SSC) but not with the single-stranded noncoding sequence (lane 4, SSNC). The predicted potential of the 12-bp palindromic sequence to form a hairpin structure (ΔG = −2.3; supplemental Fig. S5B) is not the only determinant of GmERD15 binding because GmERD15 does not bind to a second 25-bp palindromic sequence (−424ACGATTCTAnnnnnnnnTAGAACGT−399) of the NRP-B promoter (Fig. 5, A and D) that also is predicted to fold into a stable hairpin structure (supplemental Fig. S5B). Thus, other intrinsic and inherent features of the 12-bp palindromic sequence also contribute to specific binding of GmERD15.
DISCUSSION
NRPs have been characterized as targets of a novel adaptive pathway that integrates ER and osmotic stress signals to potentiate a cell death response (8). Despite the potential of this integrative signaling pathway to protect plant cells against different stress conditions, the mechanisms by which NRPs integrate both signals are unknown. Here, we used one-hybrid screening to isolate upstream components of the ER stress- and osmotic stress-induced NRP-mediated cell death response. We isolated an ER stress- and osmotic stress-induced soybean ERD15 homolog and described GmERD15 (G. max ERD15) as a novel transcription factor that activates expression of NRP genes. We showed that GmERD15 binds directly to the NRP-B promoter in vivo and in vitro. Furthermore, transient expression of GmERD15 in soybean protoplasts activates the NRP-B promoter and enhances NRP expression. Our data support the interpretation that ER and osmotic stress induce GmERD15 to activate NRP-B expression and trigger a cell death response.
In addition to binding to and activating expression of the NRP-B promoter, the induction kinetics of GmERD15 gene expression in tunicamycin- and PEG-treated soybean cells implicates GmERD15 as an upstream component of the NRP-mediated signaling pathway that integrates ER and osmotic stress cell death signals. In fact, we have shown that induction of GmERD15 gene expression by PEG and tunicamycin treatment preceded the increase in mRNA accumulation for the NRP-B gene (Fig. 1, C and D), as would be expected for a transcription factor acting upstream of NRP-B in the stress-induced cell death signaling pathway. These results further support the involvement of GmERD15 in NRP-B expression.
Confocal microscopy and subcellular fractionation studies revealed that GmERD15 is localized to both the cytoplasm and nucleus at relatively high levels (Fig. 3). This nucleocytoplasmic distribution of the protein may imply that GmERD15 plays dual roles in different subcellular compartments. GmERD15 harbors a conserved PAM2 domain in its N terminus that is one of the two poly(A)-binding protein-interacting motifs that mediates interactions with the C terminus of PABP. PABP has been implicated in several aspects of post-transcriptional control of gene expression and RNA metabolism, including biosynthesis, turnover, and export of mRNA (29, 30). Cytoplasmic PABP is particularly involved in regulation of translation and stabilization of mRNA transcripts (31). It will be relevant to confirm whether GmERD15 binds to PABP through a functional PAM2 domain, an activity that would be consistent with a cytoplasmic location. Although GmERD15 lacks a nuclear localization signal, GmERD15-YFP fusions were also found in the nuclei of transfected tobacco leaves (Fig. 3), and GmERD15 was detected in nuclear extracts of tunicamycin-treated soybean cells (data not shown). The nuclear import of GmERD15 that is required for its transcriptional-activation function may be mediated by a nuclear protein partner through protein-protein interaction.
We discovered that GmERD15 is a transcription factor that binds to the NRP-B promoter both in vivo and in vitro. Nevertheless, GmEDR15 does not harbor any typical motif found in most known DNA-binding proteins. The only exception is the presence of a conserved sequence, consisting of 13 amino acids, at positions 71–83 (DEDEKERKEgKEv), representing a putative motif of ssDNA-binding transcriptional regulators. This motif is part of a tripartite motif domain that has been derived from several ssDNA-binding transcriptional regulators from distinct kingdoms: RNA polymerase II transcriptional co-activators KELP and KIWI from Arabidopsis (32); the DNA-binding protein p24 from Solanum tuberosum (33); the human, mouse, and chicken nuclear factor 1 C-type; the human poly(C)-binding protein 4 protein and its SUB1 homolog from S. cerevisiae (34); the hnRBP protein K (35); and many others. These proteins have been shown to function as ssDNA-binding proteins to activate or suppress gene activation (36–39). One class of ssDNA-binding proteins, the poly(C)-binding proteins, are generally known as RNA- and DNA-binding proteins that interact in a sequence-specific fashion with single-stranded poly(C) and CT-rich sequences, the CT element (34). Another class of ssDNA-binding protein interacts specifically with palindromic sequences, such as the mammalian nuclear factor 1 C-type and plant PR-10a binding factor 2, that bind the palindromic sequences 5′-TTGGCnnnnnGCCAA-3′ and 5- TGACAnnnnTGTCA-3′, respectively (30). We showed that GmRD15 belongs to the latter class of ssDNA-binding proteins because it recognizes specifically the 12-bp palindromic sequence −511AGCAnnnnTGCT−500 in both single-stranded (coding sequence) and double-stranded configurations.
Despite the presence of a putative motif of ssDNA-binding transcriptional regulators in the GmERD15 sequence and a ssDNA-binding protein site on the target promoter fragment, GmERD15 binds stably and specifically to the 187-bp dsDNA fragment in vitro (Figs. 1E and 2A). Based on the thermodynamic estimation for stability of hairpin formation, one thought is that the 12-bp palindromic sequence folds into a hairpin structure (ΔG = −2.3; supplemental Fig. S5B), providing a single-stranded DNA loop for preferential GmERD15 binding. Alternatively, GmERD15 would bind to the double-stranded form of the palindromic sequence, followed by strand separation facilitated by binding to single-stranded DNA. This second model for sequence-specific interactions of ssDNA-binding proteins with target promoters has been previously invoked to understand the mechanism by which poly(C)-binding proteins binding to single- and double-stranded DNA mediates gene expression (40). This alternative is consistent with the high affinity of GmERD15 binding to the double-stranded form of the 12-pb palindromic sequence that was mapped as the GmERD15 DNA binding site (Fig. 5C).
In general, ssDNA-binding proteins are capable of activating or suppressing transcription through coordinated action with other transcriptional factors assembled, or not, on target promoters (34, 36, 38, 41). For instance, poly(C)-binding proteins act as transcriptional co-activators by binding to specific elements in gene promoters to facilitate assembly of the TFDII complex (42). In contrast, the Arabidopsis protein LEUNIG, which harbors a ssDNA-binding domain, functions as a transcriptional co-repressor and associates with SEUSS to suppress transcription of target promoters (38). Likewise, the regulated association of the ssDNA-binding protein DIP1 (DBF1 interactor protein) with the maize dehydration-responsive element-binding factor, DBF1, controls the levels of target gene expression during osmotic stress conditions (43). DEF1 is a member of the AP2 (Apetala 2)/ethylene response factor transcription factor family, which has been shown to regulate the ABA-responsive gene rab17 in response to osmotic stress. In this scenario, GmERD15 could potentially function in concert with transcription factors that recognize dsDNA cis-regulatory elements on the NRP-B promoter to activate expression in response to stresses. In silico analysis of 1-kb 5′flanking sequences of GmNRP-B revealed the presence of putative cis-regulatory elements responsive to ER stress, osmotic stress, and drought that could potentially be binding sites for soybean transcription factor homologs (supplemental Fig. S5A). These include a putative unfolded protein response element, associated with ER stress (UPRE: CCNNNNNNNNNNNNCCACG, yellow box), and MYC recognition sites (pink boxes) found in the promoters of the dehydration-responsive gene RD22/DREB1 (CANNTG), in addition to four putative binding sites for RAV1 (in red). The RAV family comprises transcriptional factors that harbor both a B3-like DNA-binding motif and an AP2 motif. AP2 domain-containing proteins, like maize DEF1, belong to the classes of ethylene-responsive (EREBs) and dehydration-responsive transcription factors (DREBs) (44). These putative cis-regulatory elements on the ER stress and osmotic stress-induced NRP-B promoter illustrate potential sites for assembly of transcription factors, which might constitute targets for GmERD15 action.
ERD5 from Arabidopsis has been described as a negative regulator of ABA signaling (13). Conservation between GmERD15 and its counterpart from Arabidopsis is limited to sequences at the N terminus of the proteins overlapping the PAM2 domain (supplemental Fig. S1). The C-terminal regions of the proteins are highly divergent, and ERD15 from Arabidopsis lacks the putative motif of ssDNA-binding transcriptional regulators found in GmERD15. Consistent with these data, ERD15 from Arabidopsis does not associate with the NRP-B promoter in yeast and, more importantly, does not transactivate a GAL4-dependent promoter when fused to the GAL-4 binding domain (data not shown). These results suggest that unlike GmERD15, ERD15 from Arabidopsis does not exhibit a transcriptional activation function and may not be involved in control of gene expression at the transcriptional level. Whether GmERD15 also functions as a regulator of ABA signaling in soybean remains to be determined.
In summary, our data implicate GmERD15 as a novel transcription factor that binds and activates expression of the ER stress- and osmotic stress-induced NRP-B promoter to transduce a stress-induced cell death signal. Although GmERD15 harbors a conserved motif of ssDNA-binding transcriptional regulators, it lacks two additional conserved motifs that compose their ssDNA-binding domain and is not predicted to fold into a ssDNA-binding transcriptional regulator-like structure (25, 45). GmERD15, therefore, defines a new class of plant-specific transcription factors that may encompass other members of the early responsive to dehydration protein family. We propose that GmERD15 is an upstream component of the ER stress-induced NRP-mediated signaling and hence connects stress in the ER with an osmotic stress-induced cell death signal. Both GmERD15 and its target gene NRP-B are coordinately induced by several biotic and abiotic stimuli. Thus, NRP-mediated cell death signaling may represent a general plant response to multiple stresses.
Supplementary Material
Acknowledgments
We thank the Universidade Federal de Viçosa microscopy core facility for the use of the laser scanning confocal microscope (LSCM) and Prof. Chris Hawes for the 35S-YFP-casseteA-Nos-pCAMBIA1300 binary vector.
This work was supported by Brazilian Government Agency Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Grants 559602/2009-0, 573600/2008-2, and 470878/2006-1 (to E. P. B. F.), as well as by Fundac̦ão de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) Grant CBB-APQ-00070-09 and Financiadora de Estudos e Projetos (FINEP) Grant 01.09.0625.00 (to E. P. B. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- ER
- endoplasmic reticulum
- NRP
- N-rich protein
- UPR
- unfolded protein response
- PABP
- poly(A)-binding proteins
- ABA
- abcisic acid
- MES
- 4-morpholineethanesulfonic acid
- NSP
- nuclear shuttle protein.
REFERENCES
- 1. Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Urade R. (2009) BioFactors 35, 326–331 [DOI] [PubMed] [Google Scholar]
- 3. Liu J. X., Howell S. H. (2010) Plant Cell 22, 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cullinan S. B., Diehl J. A. (2006) Int. J. Biochem. Cell Biol. 38, 317–332 [DOI] [PubMed] [Google Scholar]
- 5. Matsukawa J., Matsuzawa A., Takeda K., Ichijo H. (2004) J. Biochem. 136, 261–265 [DOI] [PubMed] [Google Scholar]
- 6. Xu W., Liu L., Charles I. G., Moncada S. (2004) Nat. Cell Biol. 6, 112(–1134 [DOI] [PubMed] [Google Scholar]
- 7. Valente M. A., Faria J. A., Soares-Ramos J. R., Reis P. A., Pinheiro G. L., Piovesan N. D., Morais A. T., Menezes C. C., Cano M. A., Fietto L. G., Loureiro M. E., Aragão F. J., Fontes E. P. (2009) J. Exp. Bot. 60, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costa M. D., Reis P. A., Valente M. A., Irsigler A. S., Carvalho C. M., Loureiro M. E., Aragão F. J., Boston R. S., Fietto L. G., Fontes E. P. (2008) J. Biol. Chem. 283, 20209–20219 [DOI] [PubMed] [Google Scholar]
- 9. Irsigler A. S., Costa M. D., Zhang P., Reis P. A., Dewey R. E., Boston R. S., Fontes E. P. (2007) BMC Genomics 8, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tenhaken R., Doerks T., Bork P. (2005) BMC Bioinformatics 6, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. (1994) Plant Physiol. 106, 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albrecht M., Lengauer T. (2004) Biochem. Biophys. Res. Commun. 316, 129–138 [DOI] [PubMed] [Google Scholar]
- 13. Kariola T., Brader G., Helenius E., Li J., Heino P., Palva E. T. (2006) Plant Physiol. 142, 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozlov G., De Crescenzo G., Lim N. S., Siddiqui N., Fantus D., Kahvejian A., Trempe J. F., Elias D., Ekiel I., Sonenberg N., O'Connor-McCourt M., Gehring K. (2004) EMBO J. 23, 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., Grumet R. (2004) Plant Mol. Biol. 54, 85–98 [DOI] [PubMed] [Google Scholar]
- 16. Buzeli R. A., Cascardo J. C., Rodrigues L. A., Andrade M. O., Almeida R. S., Loureiro M. E., Otoni W. C., Fontes E. P. (2002) Plant Mol. Biol. 50, 757–771 [DOI] [PubMed] [Google Scholar]
- 17. Geitz R. D., Woods R. A. (1993) in Molecular Genetics of Yeast: A Practical Approach (Johnston J. R. ed) pp. 121–134, IRL Press, Oxford, UK [Google Scholar]
- 18. Kim S. Y., Chung H. J., Thomas T. L. (1997) Plant J. 11, 1237–1251 [DOI] [PubMed] [Google Scholar]
- 19. Amberg D. C., Burke D. J., Strathern J. N. (2005) Methods in Yeast Genetics, pp. 135–137, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20. Pinheiro G. L., Marques C. S., Costa M. D., Reis P. A., Alves M. S., Carvalho C. M., Fietto L. G., Fontes E. P. (2009) Gene 444, 10–23 [DOI] [PubMed] [Google Scholar]
- 21. Cascardo J. C., Almeida R. S., Buzeli R. A., Carolino S. M., Otoni W. C., Fontes E. P. (2000) J. Biol. Chem. 275, 14494–14500 [DOI] [PubMed] [Google Scholar]
- 22. Carvalho C. M., Santos A. A., Pires S. R., Rocha C. S., Saraiva D. I., Machado J. P., Mattos E. C., Fietto L. G., Fontes E. P. (2008) PLoS Pathog. 4, e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontes E. P., Santos A. A., Luz D. F., Waclawovsky A. J., Chory J. (2004) Genes Dev. 18, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delú-Filho N., Pirovani C. P., Pedra J. H. F., Matrangolo F. S. V., Macêdo J. N. A., Otoni W. C., Fontes E. P. B. (2000) Plant Physiol. Biochem. 38, 353–361 [Google Scholar]
- 25. Desveaux D., Allard J., Brisson N., Sygusch J. (2002) Nat. Struct. Biol. 9, 512–517 [DOI] [PubMed] [Google Scholar]
- 26. Carvalho C. M., Fontenelle M. R., Florentino L. H., Santos A. A., Zerbini F. M., Fontes E. P. (2008) Plant J. 55, 869–880 [DOI] [PubMed] [Google Scholar]
- 27. Ludwig A. A., Tenhaken R. (2001) Eur. J. Plant Pathol. 107, 323–336 [Google Scholar]
- 28. Vidal S., Eriksson A. R., Montesano M., Denecke J., Palva E. T. (1998) Mol. Plant-Microbe Interact. 11, 23–32 [Google Scholar]
- 29. Chekanova J. A., Belostotsky D. A. (2003) RNA 9, 1476–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mangus D. A., Evans M. C., Jacobson A. (2003) Genome Biol. 4, 223.1–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z., Kiledjian M. (2000) Mol. Cell. Biol. 20, 6334–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cormack R. S., Hahlbrock K., Somssich I. E. (1998) Plant J. 14, 685–692 [DOI] [PubMed] [Google Scholar]
- 33. Desveaux D., Després C., Joyeux A., Subramaniam R., Brisson N. (2000) Plant Cell 12, 1477–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi H. S., Hwang C. K., Song K. Y., Law P. Y., Wei L. N., Loh H. H. (2009) Biochem. Biophys. Res. Commun. 380, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takimoto M., Tomonaga T., Matunis M., Avigan M., Krutzsch H., Dreyfuss G., Levens D. (1993) J. Biol. Chem. 268, 18249–18258 [PubMed] [Google Scholar]
- 36. Duncan R., Bazar L., Michelotti G., Tomonaga T., Krutzsch H., Avigan M., Levens D. (1994) Genes Dev. 8, 465–480 [DOI] [PubMed] [Google Scholar]
- 37. Kamada S., Miwa T. (1992) Gene 119, 229–236 [DOI] [PubMed] [Google Scholar]
- 38. Sridhar V. V., Surendrarao A., Gonzalez D., Conlan R. S., Liu Z. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michelotti E. F., Tomonaga T., Krutzsch H., Levens D. (1995) J. Biol. Chem. 270, 9494–9499 [DOI] [PubMed] [Google Scholar]
- 40. Ritchie S. A., Pasha M. K., Batten D. J., Sharma R. K., Olson D. J., Ross A. R., Bonham K. (2003) Nucleic Acids Res. 31, 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smidt M. P., Russchen B., Snippe L., Wijnholds J., Ab G. (1995) Nucleic Acids Res. 23, 2389–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. (1996) Mol. Cell. Biol. 16, 2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saleh A., Lumbreras V., Lopez C., Dominguez-Puigjaner E., Kizis D., Pagès M. (2006) Plant J. 46, 747–757 [DOI] [PubMed] [Google Scholar]
- 44. Shinozaki K., Yamaguchi-Shinozaki K. (2007) J. Exp. Bot. 58, 221–227 [DOI] [PubMed] [Google Scholar]
- 45. Brandsen J., Werten S., van der Vliet P. C., Meisterernst M., Kroon J., Gros P. (1997) Nat. Struct. Biol. 4, 900–903 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.