Abstract
Alternative splicing is typically controlled by complexes of regulatory proteins that bind to sequences within or flanking variable exons. The identification of regulatory sequence motifs and the characterization of sequence motifs bound by splicing regulatory proteins have been essential to predicting splicing regulation. The activation-responsive sequence (ARS) motif has previously been identified in several exons that undergo changes in splicing upon T cell activation. hnRNP L binds to this ARS motif and regulates ARS-containing exons; however, hnRNP L does not function alone. Interestingly, the proteins that bind together with hnRNP L differ for different exons that contain the ARS core motif. Here we undertake a systematic mutational analysis of the best characterized context of the ARS motif, namely the ESS1 sequence from CD45 exon 4, to understand the determinants of binding specificity among the components of the ESS1 regulatory complex and the relationship between protein binding and function. We demonstrate that different mutations within the ARS motif affect specific aspects of regulatory function and disrupt the binding of distinct proteins. Most notably, we demonstrate that the C77G polymorphism, which correlates with autoimmune disease susceptibility in humans, disrupts exon silencing by preventing the redundant activity of hnRNPs K and E2 to compensate for the weakened function of hnRNP L. Therefore, these studies provide an important example of the functional relevance of combinatorial function in splicing regulation and suggest that additional polymorphisms may similarly disrupt function of the ESS1 silencer.
Keywords: Protein Phosphatase, RNA, RNA-binding Protein, RNA-Protein Interaction, RNA Splicing, Alternative Splicing, CD45, hnRNP, Splicing Regulation
Introduction
Proper control of protein expression is essential for human health and development. A critical step in determining protein expression is that of pre-mRNA splicing in which non-coding intronic sequences are removed and exonic sequences are joined together through the action of the spliceosome (1). Importantly, the pattern of pre-mRNA splicing for any given gene is not a static choice; rather the inclusion or exclusion of an exon or group of exons can be highly variable (2, 3). Such alternative splicing of an exon is dictated by splicing enhancer or silencer sequences located within the exon or the flanking introns. These regulatory sequences typically bind to trans-acting protein factors that, in turn, interact with components of the spliceosome in such a way that promotes or inhibits the activity of the spliceosome on the exon substrate (2, 3).
Alternative splicing alters the protein coding potential of the vast majority of human genes, often in a cell type-specific manner or in response to environmental cues (4, 5). Alternative splicing is especially prevalent in genes expressed in the nervous and immune systems, in which functional diversity and cellular responsiveness are particularly critical (6). Disruption of normal alternative splicing in such cell types has been linked to an increasing number of human diseases, underscoring the physiologic significance of this mode of gene regulation (7, 8).
An excellent model system to illustrate the mechanisms and consequences of regulated alternative splicing is the human CD45 gene. CD45 encodes a transmembrane protein-tyrosine phosphatase that is expressed on the surface of T cells and other lymphocytes (9). In T cells, CD45 functions to maintain T cell receptor signaling by removing inhibitory phosphates on T cell receptor-proximal signaling proteins such as Lck (9, 10). CD45 has three variable exons (exons 4, 5, and 6), which are skipped with some frequency from the final mRNA in naive or resting T cells and skipped from the majority of the mRNAs in activated cells (9, 11) (see Fig. 1A). Increased skipping of the variable exons leads to increased homodimerization of CD45, which in turn results in an intermolecular inhibition of the CD45 phosphatase activity (10, 12, 13). T cells that express a form of CD45 engineered to prevent dimerization-induced inhibition are hyper-reactive to antigen (14), and mice expressing this constitutively active form of CD45 are prone to develop autoimmune disease and lymphoma (12), demonstrating a requirement for activation-induced dimerization (driven by alternative splicing) in maintaining T cell homeostasis.
FIGURE 1.
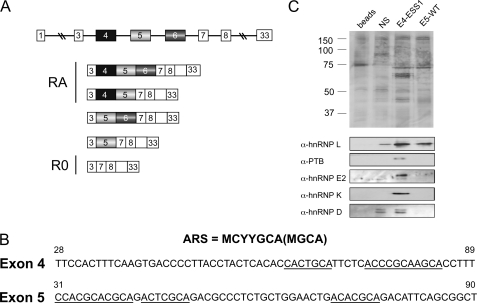
CD45 exons 4 and 5 share sequence similarity but recruit distinct proteins. A, schematic of CD45 alternative splicing, showing variable exons 4, 5, and 6 in the genomic context and the five isoforms that have been identified in human T cells. Based on antibody reactivity, isoforms encoded by mRNAs containing exon 4 are also termed RA, whereas the smallest isoform is RO. Boxes correspond to exons, and lines designate introns. B, sequence of the silencer regions of CD45 exons 4 and 5, as defined previously (Rothrock et al. (15) and Motta-Mena et al. (19)), with the ARS core element underlined and the ARS motif consensus shown above. Numbers indicate nucleotide position with respect to full exon. C, silver stain (top) and Western blot (bottom) of proteins isolated by RNA affinity using the exons 4 and 5 silencer elements (as shown in panel B), unrelated silencer sequence (NS; Melton et al. (21)), or beads alone control.
We have previously identified a conserved sequence motif common to all three CD45 variable exons that drives repression of these exons in both resting and activated T cells (15). This motif, termed the activation-responsive sequence (ARS),3 consists of imperfect tandem repeats of the sequence MCYYGCA (M = C/A, Y = C/T). A human polymorphism at nucleotide 77 in CD45 exon 4 (C77G) falls within one of the pyrimidine (Y) residues of the ARS motif. Strikingly, this C77G change in exon 4, which is silent with regard to protein sequence, results in aberrantly high inclusion of exon 4 (16). Consistent with the aforementioned model in which CD45 exon skipping leads to attenuation of T cell activity, the presence of this polymorphism contributes to a hyperactive immune system. Specifically, studies have correlated the presence of the C77G polymorphism with susceptibility to autoimmune disease and HIV infection, at least in some genetic backgrounds (17, 18). Despite the impact of this C77G polymorphism on human health, the mechanism by which it abrogates exon silencing has yet to be determined.
Interestingly, the ARS core motif is imbedded in distinct sequence contexts in each of the CD45 variable exons, with the context of exon 4 (termed exonic splicing silencer 1 or ESS1) being the most complex (15, 19) (see Fig. 1B). Understanding the detailed sequence requirements of the ARS and surrounding sequence is essential to interpret the effect of mutations such as C77G. Here we carried out a systematic mutational analysis of sequences within the ESS1. We demonstrate that mutations within the ESS1 element can be grouped into distinct functional classes, which to at least some extent can be explained by disruption of binding of distinct proteins. Most notably, we demonstrate that the C77G polymorphism weakly alters binding of the primary CD45 regulatory protein hnRNP L but greatly abrogates binding of hnRNPs E2 and K. Although neither hnRNP K nor hnRNP E2 play a prominent role in CD45 splicing under wild-type conditions, both proteins have a compensatory role when the activity of hnRNP L is compromised. Thus, the loss of redundant control by the hnRNPs K and E2 provides a molecular mechanism for the effect of the C77G polymorphism. Together our data demonstrate the importance of combinatorial control within a splicing regulatory complex and suggest that other polymorphisms could alter CD45 splicing as does C77G.
EXPERIMENTAL PROCEDURES
Minigenes and RNAs
Splicing minigenes CD4, CD4ΔESS1, and CD5 were previously described (19) and contain CD45 variable exon 4, exon 4 with ESS1 replaced, and exon 5, respectively. The Glo and GloESS constructs were previously described (15). Mutants were generated in the GloESS background using PCR-based mutagenesis. The constructs used for in vitro splicing also utilized PCR mutagenesis to introduce mutations into the CD4 vector backbone. Templates for RNAs for gel shift assays were made by cloning just the ESS1 sequence, or mutants thereof, immediately downstream of a T7 promoter.
Nuclear Extract and Recombinant Proteins
Nuclear extract was purified from JSL1 cells using a standard protocol described previously (19). Recombinant hnRNPs L, E2, and PTB were expressed and purified as described previously (20). His-hnRNP K was purified from Escherichia coli using standard methods with a nickel-nitrilotriacetic acid resin.
Cell Culture
JSL1 cells and the generation of sublines that stably express minigenes have been described previously (15). For knockdown experiments, 10 × 106 JSL1 cells were transfected by electroporation with up to 5 nmol of a morpholino oligonucleotide (Gene Tools) blocking the translation start site of the respective target. Cells were grown for 48 h after transfection and then harvested, and total RNA was extracted using RNABee (Tel-Test). Morpholino sequences were: hnRNP L, 5′-CGCCCGCCGCCGCCATCTTCACCAT-3′; hnRNP E2, 5′-CCGGTGTCCATGTCGAGCAGTGTTC-3′; hnRNP K, 5′-TTTCTTCTGGCTGTTCAGTTTCCAT-3′; hnRNP D, 5′-GCCGAACTGCTCCTCCGACATAGTG-3′; PTB, 5′-CTATATCTGGGACAATGCCGTCCAT-3′.
RT-PCR Assay
RT-PCR and analysis were performed and analyzed using vector- or gene-specific primers as described previously (15). In brief, the PCR step was performed under conditions of limiting cycle number with one primer that contained a 5′-end 32P radiolabel. RT-PCR products were resolved on a denaturing polyacrylamide gel, which was then exposed to a PhosphorImager plate and quantified using a Typhoon (GE Healthcare) and associated imaging software ImageQuant to obtain ratios of alternate isoform expression after correcting for background signal.
RNA Mobility Shift Assays
Standard binding reactions were done as described previously (20) with the indicated recombinant proteins and 32P-labeled RNA and then resolved on a 4.5% native gel (acrylamide/bis 29:1, Bio-Rad).
In Vitro Splicing
Approximately 1 fmol of unlabeled RNA substrate was incubated with 30% JSL1 nuclear extract plus the indicated recombinant proteins in a total volume of 12.5 μl as described previously (20). Reactions were incubated for 2 h at 30 °C, and then the RNA was recovered and analyzed by RT-PCR as described above.
RNA Affinity Purification
RNA affinity purification was done as described in detail previously (19) using 50 pmol of 5′-biotinylated RNA (Dharmacon) incubated with 100 μg of JSL1 nuclear extract in a 500-μl binding reaction.
Western Blotting
Western blotting was done as described previously (11) using the following antibodies: anti-hnRNP L (4D11, Abcam), anti-hnRNP E2 (rabbit polyclonal, kind gift of R. Andino), anti-PTB (rabbit polyclonal, kind gift of D. Black), anti-hnRNP K (Bethyl Laboratories, A300-676), anti-hnRNP D (ab61193, Abcam), and anti-U1A (rabbit polyclonal, kind gift of I. Mattaj).
RESULTS
The ESS1 Co-associated Protein hnRNP K Contributes to the Silencing of CD45 Exon 4
Previously, we have shown that hnRNP L is the predominant protein that binds to the ARS repeat sequences under resting conditions in both exon 4 and exon 5 (19, 20). This binding is specific as only a background amount of hnRNP L appears to bind to an unrelated control sequence (Fig. 1C, NS) despite the high abundance of hnRNP L in the nuclear extract (see also Refs. 19–21). However, mass spectrometry, silver stain, and Western blot of RNA affinity-purified complexes reveal that additional proteins bind along with hnRNP L specifically to the ARS-containing silencer element of exon 4 (21) (Fig. 1C). The additional proteins that are reproducibly observed to associate with the exon 4 ESS1 RNA are the hnRNPs D, E2 (aCP or PCBP2), K, and I (PTB). With the exception of hnRNP D, these proteins show significant specificity for the exon 4 ESS1 element over a control sequence from CD45 exon 14 (Fig. 1C). However, the association of these proteins is presumably dependent on the sequence context surrounding the ARS motifs in exon 4 as they are not observed to associate with the ARS-containing sequence from exon 5 (Fig. 1C).
Although the ARS motif has been implicated in both basal and signal-induced exon repression, we chose to focus this study on the proteins that bind to the ARS under resting conditions. We have previously identified additional activation-specific proteins PSF and hnRNP LL and have largely characterized their binding determinants (19). By contrast, the sequence determinants for the proteins bound under resting conditions are less well characterized. Moreover, the spectrum of proteins bound to distinct ARS-containing exons is most dissimilar under resting conditions. Therefore, analysis of the proteins that associate with the ARS in resting cells is an optimal system to investigate the impact of sequence context on differential binding and function.
To determine the functional relevance of the proteins that associate with the exon 4 ARS-containing silencer element (ESS1) in resting cells, we used morpholino oligonucleotides to deplete each of these individually from the JSL1 T cell line. As in our previous studies (21), we used a minigene that consists of exon 4 flanked by constitutive CD45 exons 3 and 7. Consistent with our previous results, even partial knockdown of hnRNP L results in a dramatic decrease in the skipping of exon 4 and exon 5 (Fig. 2, A–C). This activity of hnRNP L is largely dependent on the ESS1 as substitution of this 60-nucleotide element with a non-functional control sequence significantly reduces the responsiveness of the minigene to hnRNP L depletion (Fig. 2D).
FIGURE 2.
hnRNPs L and K have the majority of the silencing activity among the ESS1-associated proteins. A, the percentage of skipping of wild-type exon 4 in JSL1 cells upon individual knockdown of hnRNPs L, K, E, D, and PTB by transfection of the indicated amount of morpholino oligonucleotide. The average of multiple independent experiments is graphed. Corresponding S.D. and p values are shown in supplemental Table 1. Double asterisks indicate p value of less than 0.01. B, Western blots demonstrating knockdown from representative experiment used for graph in A. MO, morpholino oligonucleotide. C, the percentage of skipping of exon 5 in JSL1 cells. D, the percentage of skipping of exon 4 derivative lacking ESS1 done as for panel A. E, the percentage of skipping of exon 4 done as for panel A with 0.5 nmol of morpholino against each of the specified proteins alone or in combination. F, quantification of splicing of the endogenous CD45 gene in cells treated as in panel E. Statistics are shown in supplemental Table 1. The corresponding Western blot for panels E and F and a representative gel for panel F are shown in supplemental Fig. 1.
Interestingly, we observe that knockdown of hnRNP K has a modest but statistically significant effect on exon 4 inclusion that is dependent on the presence of the ESS1 (Fig. 2, A–D). Depletion of hnRNP E2 also results in a modest decrease in exon 4 skipping (Fig. 2, A and B), although this is neither statistically significant (supplemental Table 1) nor specific for the ESS1 element in exon 4 (Fig. 2D). Importantly, depletion of hnRNP K or hnRNP E2 has no notable effect on the skipping of exon 5 in JSL1 cells (Fig. 2C), confirming the functional relevance of the distinct protein binding profile that we observed between exons 4 and 5 (Fig. 1C).
Notably, however, not all proteins that bind to the ESS1 contribute to repression. We observe no evidence for a functional role of either PTB or hnRNP D in exon 4 repression, despite reducing expression of both of these proteins to nearly undetectable levels (Fig. 2, A and B). The lack of function of PTB is consistent with our previous in vitro and in vivo data (20) and suggests that binding of this protein to the ESS1 sequence is not relevant for regulation. Similarly, the lack of function of hnRNP D, together with the lack of specificity observed in Fig. 1, suggests that the presence of this protein in the RNA affinity experiments is likely an artifact of the assay, and we have not pursued further studies with this protein.
hnRNPs K and E2 are both poly(C)-binding proteins and share significant similarity in their domain structure (22). Therefore, we wondered whether these two proteins might be playing a redundant function with respect to ESS1 silencing activity. Knockdown of both proteins together resulted in no further loss of silencing than was observed for either protein on its own (Fig. 2E and supplemental Fig. 1A). Moreover, knockdown of either hnRNP K or hnRNP E2 alone in the background of the hnRNP L morpholino resulted in no more decrease in exon skipping as compared with knockdown of hnRNP L alone. Strikingly, however, depletion of both hnRNP K and hnRNP E2 did show a cooperative effect with depletion of hnRNP L to result in another ∼2-fold drop in exon skipping (Fig. 2E).
As discussed in Fig. 1, the hnRNPs L, K, and E2 were initially identified as interacting with ESS1 in resting cells, and no change in binding was observed upon stimulation (21). However, to determine whether these proteins had a different functional role in stimulated cells, we also quantified the effect of depletion of these proteins on the responsiveness of exon 4 to stimulation. Consistent with these proteins playing predominantly a role in setting the basal level of repression, we find no change in the -fold increase in repression upon stimulation (-fold repression) upon knockdown of hnRNPs L, K, or E2 alone or in combination (supplemental Fig. 1B). In other words, the level of repression conferred by these proteins is constant between resting and activated conditions, and they are not specifically involved in the “activation response.”
Finally, to confirm the physiologic significance of hnRNPs L, K, and E2 in the regulation of CD45, we assayed splicing of the endogenous CD45 gene in our JSL1 cells. Specifically, we quantified the ratio between the “RO” isoforms to the “RA” isoforms (Fig. 1A), which represent skipping or inclusion of exon 4, as these are the most biologically significant and are the isoforms that are altered in patients with the C77G polymorphism (16, 17). As shown in Fig. 2F, the effect of the morpholino oligonucleotides on the expression of exon 4 in the endogenous CD45 (as measured by RO/RA) is strikingly similar to that which we observe in the minigene. Consistent with previous studies from our group (24), depletion of hnRNP L results in a partial decrease in exon 4 skipping (reduced RO/RA). By contrast, hnRNPs K, E, D, or PTB have little effect on the RO/RA ratios. Importantly, however, the hnRNP L-dependent decrease in RO/RA is significantly augmented by a simultaneous reduction in hnRNPs K and E2. These results underscore the physiologic significance of hnRNPs K and E in the combinatorial regulation of CD45 expression.
We also assayed splicing of three additional endogenous genes that have alternative exons known to be regulated by distinct RNA-binding proteins. None of these genes (CELF2, LEF1, Bcl-X) displayed any effect from the knockdown of hnRNPs L, K, or E2 alone or in combination (supplemental Fig. 1D), demonstrating the specificity of the regulation of CD45 by these proteins. Taken together, these data confirm that hnRNP L is the primary silencer protein functioning through ESS1 but suggest that hnRNPs K and E2 serve a redundant “backup” role to silence exon 4 under conditions in which hnRNP L function is compromised.
Mutations within the ESS1 Element Differentially Alter Basal and Activation-induced Repression
The ability of exon 4 to recruit more functionally relevant proteins than exon 5 suggests that the sequences outside the ARS core motifs influence protein association and silencer function. To examine this, we engineered mutations to disrupt sequence elements that had the hallmarks of characterized binding sites for hnRNPs or were conserved in the ARS repeats (Fig. 3A). The ESS1 and mutants thereof were then inserted into a heterologous β-globin minigene so as to fully separate the functional contribution of the ESS1 from any native flanking sequence and stably transfected into JSL1 cells to analyze splicing.
FIGURE 3.
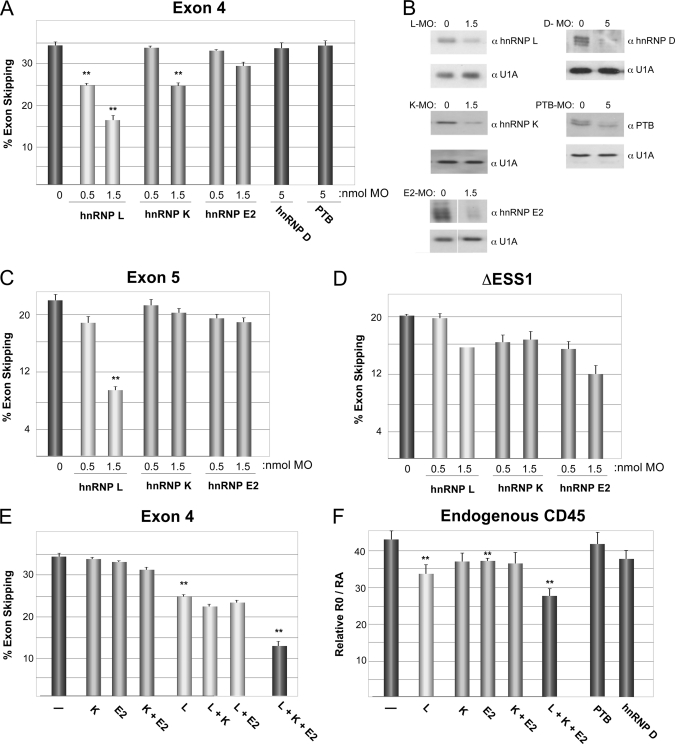
Mutation of sequences in ESS1 uncouples basal and activation-induced silencing. A, schematic of mutations used, grouped according to activity. Names of mutations are described under “Results”; nr = no response. The percentage of inclusion of mutant-containing exon in resting cells (% inc) and -fold repression difference between resting and stimulated cells (FR) is shown as an average of multiple experiments, as shown in panel B. FR is calculated as (%Exon exclusion/%Exon inclusion)activated/(%Exon exclusion/%Exon inclusion)resting. Mutations are categorized based on whether they primarily alter basal silencing (basal), the -fold repression in response to activation (act resp), or both. B, representative RT-PCR from JSL1 cells expressing the indicated minigenes grown under resting (phorbol 12-myristate 13-acetate-negative (−PMA)) or stimulated (phorbol 12-myristate 13-acetate-positive (+PMA)) conditions for 70 h. Quantification of splicing is shown in panel A. C, RT-PCR analysis of indicated minigenes spliced in vitro in nuclear extract (NE) prepared from cells grown under resting (R) or stimulated (S) conditions. The percentage of inclusion of exon 4 derivative in resting nuclear extract and -fold repression between resting and stimulated nuclear extract is shown. Statistics are shown in supplemental Table 2. In most cases, the values that deviate from ESS have a p value < 0.02.
As we have observed previously, the ESS1 causes almost complete exon silencing in the β-globin background (15) (Fig. 3B). Mutations in the CU-rich regions at either end of the ESS1 do not significantly alter either the basal or the activation-induced repressive activity of this element (Fig. 3A, nr1 and nr2). These CU sequences match the preferred binding site for PTB (23). Thus, the lack of functional effect of these mutations is consistent with the conclusion that PTB is not functionally relevant for ESS1 repression (Fig. 2A). We also see no loss of exon silencing when we mutate the G residues that are part of the highly conserved GCA triplet in the ARS consensus element (Figs. 1 and 3A, nr3). By contrast, mutating the neighboring C residues of the ARS core abolishes all silencing activity of ESS1 (Fig. 3, A and B, mESS). Strikingly, we observe a third phenotype upon mutation of the conserved A residues of the GCA triplets. An exon containing the mAx3 mutation (Fig. 3A) is repressed as efficiently as wild-type exon 4 (ESS) in resting cells; however, there is no increase in the exon skipping of this mutant construct between resting and stimulated cells (Fig. 3B).
Interestingly, each of the GCAs appear to contribute differently to the overall function of ESS1 (Fig. 3, A and B). Mutation of the first C (M1) primarily weakens basal silencing as inclusion of exon 4 is increased ∼10-fold by this mutation (3–38%), with only modest loss of activation responsiveness (-fold repression 3.2–2.8). On the other hand, mutation of the last C (M3) almost entirely abolishes activation responsiveness, with little effect on basal silencing (3–6%), whereas mutation of the middle C (M2) has an effect on both silencing activities.
We also find functional consequences from mutation of sequences outside the ARS repeats. Mutation of the pyrimidine stretch that separates the ARS repeats has a splicing phenotype essentially indistinguishable from that of mAx3 or M3 in eliminating signal-responsive silencing with only a minimal effect on the level of exon skipping in resting cells (Fig. 3, A and B; mPy). The activation-specific effect of the mAx3, mPy, and M3 mutations is also recapitulated in vitro. As shown in Fig. 3C, splicing of these minigenes in vitro is identical to wild-type exon 4 when incubated in nuclear extract from resting cells; however, inclusion of the mutant exon 4 is much greater than wild type in extract from stimulated cells.
By contrast to the activation-specific effect of mAx3, M3, and mPy, mutation of the poly(C) stretch upstream of the ARS repeats (m4C) dramatically increases exon inclusion in resting cells without altering signal responsiveness. Most notably, the disease-associated human polymorphism C77G, which falls in the ARS consensus but at a less conserved position, causes a 20-fold increase in exon 4 inclusion under resting conditions (Fig. 3, A and B). However, minigenes harboring this mutation still show a 3.5-fold reduction in exon inclusion upon cellular activation. The basal-specific effect of the C77G and m4C mutations is also observed in extracts (Fig. 3C), further demonstrating that the in vitro system faithfully recapitulates the mechanism by which exon 4 is regulated in vivo.
Multiple Mutations Selectively Affect Binding and Function of hnRNPs L, K, and E2
The fact that we have found multiple mutations that each disrupt either basal or activation-responsive repression independently demonstrates that these activities are at least partially separable. The most straightforward explanation for these results is that distinct proteins mediate the basal versus activation-responsive exon silencing and that certain mutations disrupt association of only one class of protein. Previously, we have shown that the activation-specific mutations mAx3 and mPy abolish binding of the stimulation-specific silencing factors hnRNP LL and PSF, respectively (21, 24). Furthermore, we have shown that the mESS mutation affects the binding and function of hnRNPs L, LL, and PSF (20, 21, 24). However, there has been no investigation as to how mutations that alter ESS1 basal repression influence the binding of the basal-associated and functional proteins hnRNPs K, E2, and L. We therefore assayed hnRNPs L, K, and E2 individually in in vitro binding and functional assays with mutant RNAs from each class designated in Fig. 3 that displayed the greatest functional effects.
Consistent with previous results, purified recombinant hnRNP L binds efficiently to both the wild-type ESS1 sequence, as well as the exon 5 regulatory sequence (E5), and represses inclusion of both wild-type exons 4 and 5 when supplemented into resting nuclear extract in in vitro splicing assays (Fig. 4, A–C) (19). In contrast, both binding and activity of hnRNP L are significantly abrogated by the mESS mutation (Fig. 4, A–C) (20). Surprisingly, the mAx3 mutation abolishes virtually all of the binding of purified hnRNP L on exon 4, although some weak repressive activity is retained in splicing assays (Fig. 4, A–C). In previous studies, we have shown that hnRNP L retains some binding to the mAx3 mutant in the context of nuclear extract, as measured by UV cross-linking (24). Therefore, we conclude that although the mAx3 mutation greatly impairs the inherent binding activity of hnRNP L, additional proteins present in nuclear extract must facilitate binding of hnRNP L in such a way as to partially compensate for this loss of affinity.
FIGURE 4.
Specificity of hnRNP L binding and activity. A, quantification of gel shift assays performed with purified recombinant hnRNP L and purified RNAs as indicated. B, representative gel shifts used for graph in panel A. For all panels, titration of protein is at 1, 3, 10, and 30 ng of hnRNP L per reaction. C, quantification of in vitro splicing assays in which purified recombinant hnRNP L was added to nuclear extract derived from resting cells and then incubated with minigene RNAs in which the middle exon contained wild-type exon 4, wild-type exon 5, or mutants of exon 4 as in Fig. 3. S.D. and p values are shown in supplemental Table 3.
We note that although both the activation-specific mutation mPy and the basal-specific mutations C77G and 4mC only modestly reduce the overall affinity of hnRNP L for ESS1 (Fig. 4A), the pattern of binding is different (Fig. 4B and supplemental Fig. 2). Binding of hnRNP L to wild-type ESS1 resolves on a native gel as two main species, perhaps indicative of distinct binding conformations (Fig. 4B, labeled c1 and c2). By contrast, hnRNP L bound to C77G resolves as a single band with a migration similar to the smallest species observed on wild-type ESS1 (Fig. 4B, c1 and supplemental Fig. 2). Conversely, hnRNP L bound to mPy predominantly migrates similarly to the larger species (Fig. 4B, c2 and supplemental Fig. 2). These data suggest that the C77G and mPy mutations constrain the way in which hnRNP L associates with ESS1, perhaps altering which or how many of the four RNA recognition motifs of hnRNP L are contacting the RNA. This altered binding likely causes the modestly reduced exon silencing activity of hnRNP L observed in the functional assay (Fig. 4C). Together these data demonstrate that the GCAs are the primary determinants of hnRNP L binding, with additional sequences surrounding the GCA repeats functioning to fine tune the interactions of this protein with the RNA.
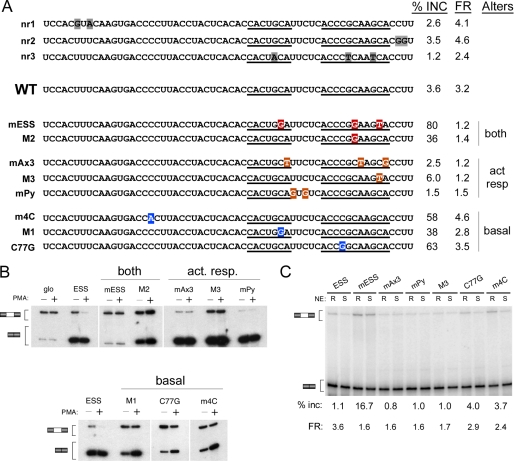
Mutation of the GCA motifs, at either the C or the A positions, also dramatically reduces the binding and function of hnRNP K, relative to wild-type ESS1 (Fig. 5, mESS and mAx3). However, unlike hnRNP L, the binding and function of hnRNP K is highly sensitive to the 4mC and C77G mutations (Fig. 5). The loss of hnRNP K binding in the 4mC and C77G mutants is consistent with the fact that both of these mutations disrupt a stretch of 3–4 cytosine residues, identified as the optimal binding site for hnRNP K (22). Unexpectedly, we find that hnRNP K does bind to the exon 5 regulatory sequence with only about a 3-fold loss in affinity over ESS1; however, hnRNP K only marginally represses exon 5 in the nuclear extract-based in vitro splicing system (Fig. 5, A and C). We also observe a similar phenotype with the mPy mutant of ESS1. These results, together with the lack of hnRNP K observed in the E5 affinity purifications from nuclear extract (Fig. 1), suggest that although purified hnRNP K is able to bind E5 or mPy, the affinity of hnRNP L for these sequences is greater than that of hnRNP K, such that when both proteins are present, hnRNP L prevails over hnRNP K. Based on the data above, we conclude that the binding and function of hnRNP K require the precise intact ARS consensus sequence in the specific context of exon 4, whereas hnRNP L is more permissive to sequence variations.
FIGURE 5.
Specificity of hnRNP K binding and activity. A, quantification of gel shift assays performed with purified recombinant hnRNP K and purified RNAs as indicated. B, representative gel shifts used for graph in panel A. For all panels, titration of protein is at 1, 3, 10, 30, 100, and 300 ng of hnRNP K per reaction. C, quantification of in vitro splicing assays in which purified recombinant hnRNP K was added to nuclear extract derived from resting cells and then incubated with minigene RNAs in which the middle exon contained wild-type exon 4, wild-type exon 5, or mutants of exon 4 as in Fig. 3. S.D. and p values are shown in supplemental Table 3.
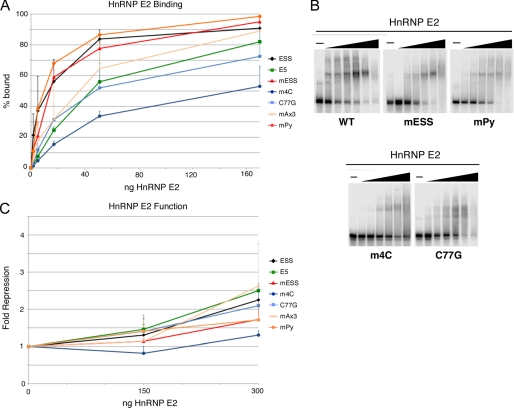
In striking contrast to hnRNP K, hnRNP E2 shows very little binding specificity to ESS1 (Fig. 6, A and B). Binding assays with recombinant hnRNP E2 protein revealed that neither mutations in the ARS core nor the intervening sequence have an effect on the affinity of hnRNP E2 (Fig. 6, A and B). Further, consistent with the in vivo functional studies in Fig. 2, hnRNP E2 exhibits only weak repressive activity on CD45 exon 4 in vitro (Fig. 6C), and this is not highly sequence-dependent as most mutations did not significantly diminish the weak repressive activity (Fig. 6C). The only mutations that had any significant, albeit mild, effect on hnRNP E2 binding and function were 4mC and C77G. Similar to hnRNP K, hnRNP E2 has been shown to bind preferentially to multiple short runs of cytosine residues (22). Therefore, the four-cytosine stretch disrupted by 4mC is likely the highest affinity site for hnRNP E2 among additional redundant lower affinity sites such as the three cytosines at positions 75–77, thereby allowing hnRNP E2 to associate with ESS1 with overall little sequence discrimination.
FIGURE 6.
Specificity of hnRNP E2 binding and activity. A, quantification of gel shift assays performed with purified recombinant hnRNP E2 and purified RNAs as indicated. B, representative gel shifts used for graph in panel A. For all panels, titration of protein is at 0.56, 1.7, 5.6, 17, 56, and 170 ng of hnRNP E2 per reaction. C, quantification of in vitro splicing assays in which purified recombinant hnRNP E2 was added to nuclear extract derived from resting cells and then incubated with minigene RNAs in which the middle exon contained wild-type exon 4, wild-type exon 5, or mutants of exon 4 as in Fig. 3. S.D. and p values are shown in supplemental Table 3.
C77G Mutation Disrupts Combinatorial Silencing by hnRNPs
The finding that the binding and activity of both hnRNP K and hnRNP E2 on ESS1 are reduced by mutation of poly-cytosine stretches in ESS1 is particularly interesting with respect to the disease-causing C77G polymorphism. Although previous studies have demonstrated that the C77G SNP causes a profound shift in CD45 isoform expression in patient samples and in tissue culture (11, 16), the molecular mechanism by which exon silencing is disrupted has remained a mystery. The observation that the C77G polymorphism only marginally reduces the binding and activity of the primary exon 4 silencer protein hnRNP L seems initially incongruous with the significant loss of exon skipping conferred by this mutation. However, we reasoned that perhaps the reduction of binding of hnRNPs K and E2 caused by C77G is sufficient to explain its dramatic functional effect.
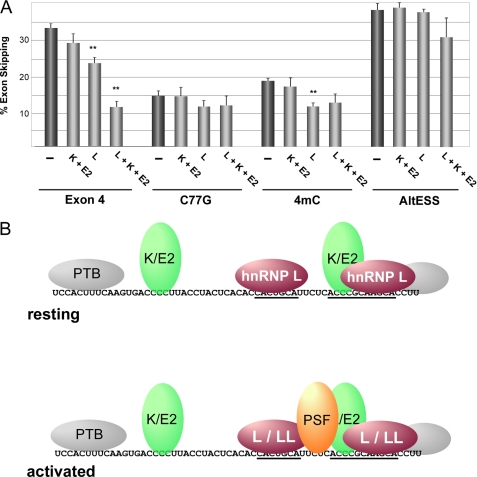
To test this hypothesis, we investigated whether the C77G mutation alters the dependence of silencing on hnRNPs L, K, and E2 in JSL1 cells (Fig. 7A). As expected, engineering the C77G mutation into the CD45 minigene used in Fig. 2 results in a notable loss of exon silencing relative to wild type (Fig. 7A, exon 4 versus C77G). Importantly, consistent with our model, there is no further loss of exon silencing of the C77G construct upon knockdown of hnRNPs K and E2. Moreover, knockdown of hnRNPs K and E2 shows no cooperative effect with knockdown of hnRNP L on the C77G construct (Fig. 7A). We do note that there is a small loss of silencing for the C77G construct upon knockdown of hnRNP L (Fig. 7A). This is consistent with our observation of some residual binding of hnRNP L to this mutant (Fig. 4), thereby allowing for partial silencing above baseline. The fact that the extent of silencing observed for C77G in the absence of hnRNP L alone is most similar to that observed for the wild-type control upon knockdown of hnRNPs K, E2, and L provides further evidence that hnRNPs K and E2 contribute nothing to the repression of the C77G construct.
FIGURE 7.
Presence of binding site for hnRNPs K and E2 is required for their ability to compensate for reduced hnRNP L function. A, the percentage of skipping of exon 4 variants containing the wild-type ESS1 (Exon 4), ESS1 with C77G mutation (C77G), or 4mC mutation (4mC) or substitution of the ESS1 with an unrelated silencer sequence (AltESS) in JSL1 cells upon knockdown of combinations of hnRNPs L, K, and E2. Experiments were performed and analyzed as described in the legend for Fig. 2. S.D. and p values are shown in supplemental Table 4. Double asterisks indicate p < 0.01. B, model for protein association with ESS1 in resting and stimulated T cells. The core ARS motifs are underlined. Colored proteins are those shown to be functional (hnRNPs L, K, and E2 in resting cells along with hnRNP LL and PSF in activated cells). PTB is added for completeness but is in gray to indicate its lack of contribution to ESS1-dependent silencing. Proteins shown to bind to the same or immediately overlapping sites are shown as a single oval (K/E2, L/LL) for simplicity.
We also observe a loss of sensitivity to depletion of hnRNPs K and E2 and weakening of sensitivity to hnRNP L, with the 4mC mutation similar to that observed for C77G (Fig. 7A), consistent with the similar in vitro studies for these mutants. By contrast, knockdown of hnRNP L, K, or E2 had no significant effect on the activity of an unrelated silencer used as a control (Fig. 7A, AltESS). Furthermore, consistent with the data in Fig. 2 and the effects of C77G and 4mC specifically on basal repression (Fig. 3), neither of these mutations nor knockdown of hnRNPs L, E2, or K had any effect on the response to phorbol 12-myristate 13-acetate stimulation (supplemental Fig. 3). Taken together, these data confirm that the C77G and 4mC mutations alter in vivo expression of CD45 exon 4 in resting cells by reducing hnRNP L function and abolishing the compensatory effect of hnRNPs K and E2.
DISCUSSION
Previous studies have shown that the regulation of CD45 isoform expression in resting T cells is controlled, in large part, by the binding of hnRNP L to exonic silencer sequences present in each of the variable exons. This binding of hnRNP L appears to be the sole determinant of repression for CD45 exon 5, and forced tethering of hnRNP L to exon 4 is likewise sufficient to cause silencing (19) (Fig. 2). However, a comparison of the regulation of CD45 exons 4 and 5 suggested that the natural silencer controlling exon 4 (ESS1) is more complex both in sequence and in recruited proteins. Here we delineate the functional involvement of additional ESS1-binding proteins and identify sequence determinants for the binding of each of the proteins that contribute to exon 4 silencing. Importantly, this detailed analysis uncovers the molecular mechanism of how the naturally occurring C77G polymorphism causes the disease-associated loss of exon silencing in humans.
Combinatorial Assembly of a Splicing Silencer Complex
In this study, we have found several previously unappreciated contributors to exon 4 silencing. First, we show that hnRNP K contributes, albeit weakly, to exon 4 repression under wild-type conditions. Secondly, we demonstrate that in addition to the core ARS motifs, two poly-cytosine tracts are required for maximal exon skipping in resting cells. Finally, we show that hnRNP K, together with hnRNP E2, play a redundant role in maintaining exon 4 silencing under conditions in which hnRNP L function is compromised. Interestingly, the newly identified poly-cytosine runs are necessary for this backup activity of hnRNPs K and E2.
This new description of a functional role of hnRNPs K and E2 and poly-cytosine tracts, together with previous data related to the binding determinants for the signal-specific proteins PSF and hnRNP LL (21, 24), allow us to form a comprehensive picture of the ESS1 silencing complex (Fig. 7B). Specifically, the new data we provide here indicate that in resting cells hnRNP L binds to the ARS core motifs, whereas hnRNPs K and E bind to the poly(C) regions (Fig. 7B, upper). Our data also suggest that PTB associates with the ends of the ESS1, although this interaction does not appear to contribute to silencing. Upon stimulation, PSF associates with the pyrimidine linker between the ARS repeats, whereas hnRNP LL associates with the repeats themselves (21, 24) (Fig. 7B, lower).
We do not yet know the stoichiometry of all the proteins bound to the ESS1. We have not observed cooperative binding interactions between any proteins tested; however, at least hnRNPs L and LL do appear to associate simultaneously with the ARS motifs based on the fact that we can detect binding of both proteins to ESS1 in UV cross-linking assays,4 and both contribute to exon silencing in stimulated cells (24). In sum, we conclude that the complexity of proteins associated with the ESS1 RNA ensures tight regulation of CD45 exon 4 by providing redundancy and contributing to silencing in a functionally combinatorial manner.
Functional Relevance of the C77G Polymorphism
Although CD45 activity is necessary for an initial T cell response to antigen (25, 26), subsequent repression of CD45 activity is an important mechanism to maintain homeostasis of the immune system (9). Exclusion of the highly glycosylated peptide sequences encoded by the three CD45 variable exons permits dimerization of the phosphatase domain of the molecule, which in turn inhibits catalytic activity and reduces cellular responsiveness to antigen (10, 12, 13). It follows then that a defect in alternative splicing that prevents the skipping of the CD45 variable exons would result in a hyperactive version of CD45 and therefore a hyperactive T cell compartment. Indeed, the C77G polymorphism has exactly this predicted phenotype in humans, in which a significant decrease in the expression of the smaller isoforms of CD45 correlates with immune dysfunction including increased susceptibility to autoimmune disease and HIV infection (16–18).
Our data here demonstrate, for the first time, the molecular consequence of the C77G mutation. Notably, this mutation does not have enough of an effect on the binding or function of the primary CD45 repressor protein hnRNP L to explain the dramatic in vivo phenotype. We show, however, that the C77G SNP also prevents hnRNPs K and E2 from performing a compensatory role to maintain silencing under conditions of compromised hnRNP L activity. This information is potentially of value in designing therapies to reverse the effect of the C77G SNP. Moreover, the fact that a mutation that does not significantly perturb hnRNP L activity has such a profound consequence on CD45 protein expression and function raises the specter of additional disease-causing mutations. Specifically, polymorphisms in the poly(C) region upstream of the ARS core motif would be predicted to have a similar functional outcome as C77G. Likewise, mutations at the C or A positions of the GCAs should have an even more severe phenotype. Finally, at a more global level, as C77G does not alter the coding potential of the CD45 mRNA, these results further emphasize the importance of understanding splicing determinants to better predict the physiological relevance of SNPs.
Supplementary Material
Acknowledgments
We thank Cara Boutte and Jason Nguyen for assistance with the early stages of this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM067719 and GM084034 (to K. W. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1–4.
S. A. Smith and K. W. Lynch, unpublished results.
- ARS
- activation-responsive sequence
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- ESS
- exonic splicing silencer
- mESS
- mutant ESS
- PTB
- polypyrimid tract binding protein
- PSF
- PTB-associated splicing factor.
REFERENCES
- 1. Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 2. Chen M., Manley J. L. (2009) Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nilsen T. W., Graveley B. R. (2010) Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 6. Modrek B., Resch A., Grasso C., Lee C. (2001) Nucleic Acids Res. 29, 2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evsyukova I., Somarelli J. A., Gregory S. G., Garcia-Blanco M. A. (2010) RNA Biol. 7, 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooper T. A., Wan L., Dreyfuss G. (2009) Cell 136, 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hermiston M. L., Xu Z., Majeti R., Weiss A. (2002) J. Clin. Invest. 109, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dornan S., Sebestyen Z., Gamble J., Nagy P., Bodnar A., Alldridge L., Doe S., Holmes N., Goff L. K., Beverley P., Szollosi J., Alexander D. R. (2002) J. Biol. Chem. 277, 1912–1918 [DOI] [PubMed] [Google Scholar]
- 11. Lynch K. W., Weiss A. (2000) Mol. Cell. Biol. 20, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Majeti R., Bilwes A. M., Noel J. P., Hunter T., Weiss A. (1998) Science 279, 88–91 [DOI] [PubMed] [Google Scholar]
- 13. Xu Z., Weiss A. (2002) Nat. Immunol. 3, 764–771 [DOI] [PubMed] [Google Scholar]
- 14. Majeti R., Xu Z., Parslow T. G., Olson J. L., Daikh D. I., Killeen N., Weiss A. (2000) Cell 103, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 15. Rothrock C., Cannon B., Hahm B., Lynch K. W. (2003) Mol. Cell 12, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 16. Zilch C. F., Walker A. M., Timón M., Goff L. K., Wallace D. L., Beverley P. C. (1998) Eur. J. Immunol. 28, 22–29 [DOI] [PubMed] [Google Scholar]
- 17. Jacobsen M., Schweer D., Ziegler A., Gaber R., Schock S., Schwinzer R., Wonigeit K., Lindert R. B., Kantarci O., Schaefer-Klein J., Schipper H. I., Oertel W. H., Heidenreich F., Weinshenker B. G., Sommer N., Hemmer B. (2000) Nat. Genet. 26, 495–499 [DOI] [PubMed] [Google Scholar]
- 18. Tchilian E. Z., Wallace D. L., Dawes R., Imami N., Burton C., Gotch F., Beverley P. C. (2001) AIDS 15, 1892–1894 [DOI] [PubMed] [Google Scholar]
- 19. Motta-Mena L. B., Heyd F., Lynch K. W. (2010) Mol. Cell 29, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rothrock C. R., House A. E., Lynch K. W. (2005) EMBO J. 24, 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melton A. A., Jackson J., Wang J., Lynch K. W. (2007) Mol. Cell. Biol. 27, 6972–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makeyev A. V., Liebhaber S. A. (2002) RNA 8, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan R. C., Black D. L. (1995) Mol. Cell Biol. 15, 6377–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Topp J. D., Jackson J., Melton A. A., Lynch K. W. (2008) RNA 14, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kishihara K., Penninger J., Wallace V. A., Kündig T. M., Kawai K., Wakeham A., Timms E., Pfeffer K., Ohashi P. S., Thomas M. L. (1993) Cell 74, 143–156 [DOI] [PubMed] [Google Scholar]
- 26. Byth K. F., Conroy L. A., Howlett S., Smith A. J., May J., Alexander D. R., Holmes N. (1996) J. Exp. Med. 183, 1707–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.