We report that the transacting factor CArG Box binding Factor A (CBF-A) binds the RNA trafficking sequences found in activity-regulated cytoskeleton-associated protein, calmodulin-dependent protein kinase II, and brain-derived neurotrophic factor mRNAs in an activity-dependent manner and accompanies the transcripts from gene to dendrites. CBF-A gene silencing impaired dendritic mRNA localization. We propose that CBF-A is important for trafficking of RNA trafficking sequence–containing neuronal mRNAs.
Abstract
In neurons certain mRNA transcripts are transported to synapses through mechanisms that are not fully understood. Here we report that the heterogeneous nuclear ribonucleoprotein CBF-A (CArG Box binding Factor A) facilitates dendritic transport and localization of activity-regulated cytoskeleton-associated protein (Arc), brain-derived neurotrophic factor (BDNF), and calmodulin-dependent protein kinase II (CaMKIIα) mRNAs. We discovered that, in the adult mouse brain, CBF-A has a broad distribution. In the nucleus, CBF-A was found at active transcription sites and interchromosomal spaces and close to nuclear pores. In the cytoplasm, CBF-A localized to dendrites as well as pre- and postsynaptic sites. CBF-A was found in synaptosomal fractions, associated with Arc, BDNF, and CaMKIIα mRNAs. Electrophoretic mobility shift assays demonstrated a direct interaction mediated via their hnRNP A2 response element (A2RE)/RNA trafficking sequence (RTS) elements located in the 3′ untranslated regions. In situ hybridization and microscopy on live hippocampal neurons showed that CBF-A is in dynamic granules containing Arc, BDNF, and CaMKIIα mRNAs. N-methyl-d-aspartate (NMDA) and α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) postsynaptic receptor stimulation led to CBF-A accumulation in dendrites; increased Arc, BDNF, and CaMKIIα mRNA levels; and increased amounts of transcripts coprecipitating with CBF-A. Finally, CBF-A gene knockdown led to decreased mRNA levels. We propose that CBF-A cotranscriptionally binds RTSs in Arc, BDNF, and CaMKIIα mRNAs and follows the transcripts from genes to dendrites, promoting activity-dependent nuclear sorting of transport-competent mRNAs.
INTRODUCTION
Transport and localization of mRNA transcripts to subcellular compartments occurs in many cell types and provides temporal and spatial control of gene expression. These mechanisms are dependent on the interplay between cis-acting elements located in the 3′ or 5′ untranslated regions (UTRs) of the transported mRNAs with a set of transacting factors (Rodrigues et al., 2008; Martin and Ephrussi, 2009; Percipalle et al., 2009). In fibroblasts and neurons, transport of the β-actin mRNA to leading edge and synapses is mediated by the cis-acting element zipcode, which is recognized by ZBP1 (Eom et al., 2003; Tiruchinapalli et al., 2003; Condeelis and Singer, 2005; Huttelmayer et al., 2005). In oligodendrocytes, the myelin basic protein (MBP) mRNA is transported to the myelin compartment (Carson et al., 2001; Carson and Barbarese, 2005; Percipalle et al., 2009). In its 3′ UTR, the MBP mRNA has a specific cis-acting element termed A2RE (hnRNP A2 response element) or more generally RTS (RNA trafficking sequence). This sequence is specifically recognized by the heterogeneous nuclear ribonucleoproteins (hnRNP) A2 and CArG Box binding Factor A (CBF-A) (Ainger et al., 1993; Hoek et al., 1998; Raju et al., 2008). In mammals, CBF-A is also known as hnRNP A/B. Similarly to hnRNP A2, CBF-A belongs to the A/B-type hnRNP subfamily and it is closely related to hnRNP A1 and hnRNP D, being characterized by two conserved RNA binding motifs and a C-terminal auxiliary domain involved in protein–protein interactions (Dreyfuss et al., 2002). Sequence analysis shows that CBF-A and hnRNP A2 exhibit 40% sequence identity. Consistent with a role as transacting factor, we discovered that CBF-A is part of the same RNP complex as hnRNP A2, and, in mouse oligodendrocytes, CBF-A gene silencing led to impairment of MBP mRNA trafficking. On the basis of these findings we proposed that A2RE/RTS recognition by CBF-A is important to sort transport-competent MBP mRNA to the final cytoplasmic destination (Percipalle et al., 2002, 2009; Raju et al., 2008).
In neurons, certain mRNA transcripts are transported to dendrites for localized translation (Bramham and Wells, 2007). In many cases, dendritic localization is dependent on the UTRs (Lisman et al., 2002; Bramham and Wells, 2007; Jambhekar and DeRisi, 2007; Andreassi and Riccio, 2009; Martin and Ephrussi, 2009). Even though these mechanisms are compatible with the establishment of excitatory synapses, it remains largely unclear how specific mRNAs are transported and localized to dendrites. RTS and RTS-like sequences were identified in a number of neuronal transcripts (Ainger et al., 1997), including Arc (activity-regulated cytoskeleton-associated protein) and CaMKIIα (calmodulin-dependent protein kinase II) mRNAs, which are believed to be dendritically transported via the A2RE/RTS pathway (An et al., 2008; Gao et al., 2008). CBF-A (or hnRNP A/B) is among the hnRNPs that are found in the RNA granules isolated from developing and adult mouse brains (Kanai et al., 2004; Elvira et al., 2006). Furthermore, CBF-A is a shuttling hnRNP component of pre-mRNP/mRNP particles and in neuron lineages we discovered that CBF-A–positive granules decorate dendrites (Percipalle et al., 2002; Raju et al., 2008). At this stage, however, it is not known whether CBF-A associates with the putative RTS-like sequences in the UTRs of dendritically transported transcripts and whether CBF-A has a role in trafficking of neuronal mRNAs (Bramham and Wells, 2007; Chua et al., 2010).
Here we report that CBF-A directly binds the RTSs found in Arc, CaMKIIα, and BDNF (brain-derived neurotrophic factor) mRNAs and accompanies the transcripts from gene to dendrites. Because in hippocampal neurons association is sensitive to postsynaptic receptor activation, we propose that CBF-A has an important role in the trafficking of RTS-containing neuronal mRNAs.
RESULTS
In vivo localization of CBF-A in the adult forebrain
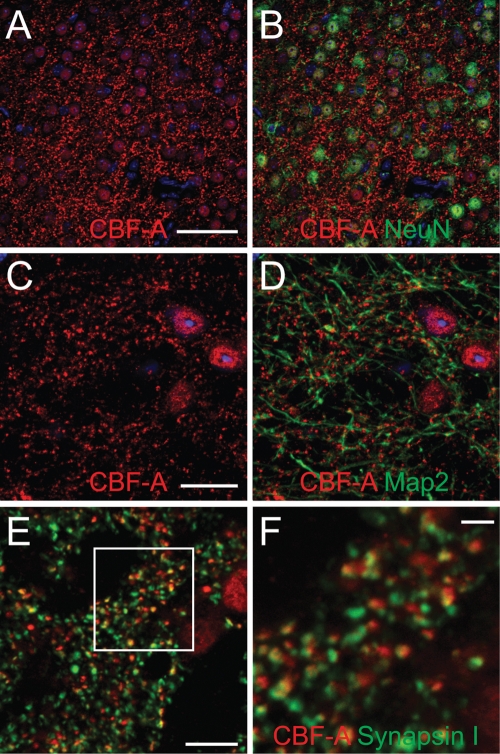
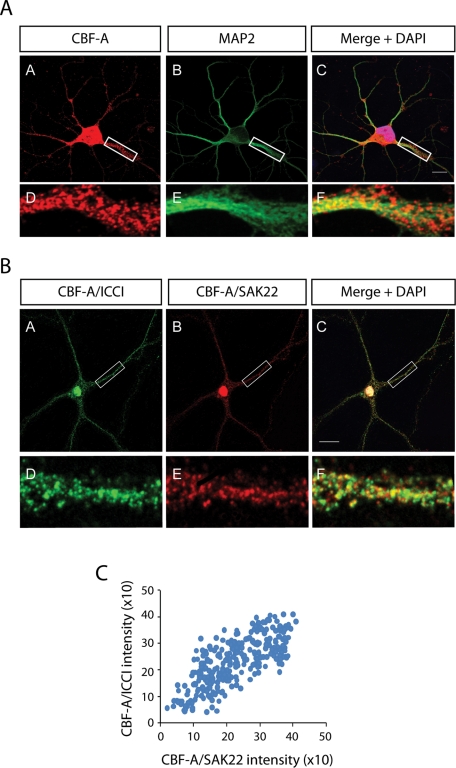
CBF-A is known to be present in two different splice variants termed p37 and p42 (Dean et al., 2002). Using the polyclonal anti–CBF-A antibody ICCI directed against the unique C-terminal epitope within the p42 variant we performed immunofluorescence staining of sections of adult mouse forebrain. Confocal microscopy confirmed that CBF-A is abundantly expressed in the mouse brain (Raju et al., 2008) and in neurons (Figure 1). In double immune labeling experiments we discovered that CBF-A has distinctive nuclear localization and that its distribution correlates with the neuronal nuclear marker NeuN (Figure 1B). A fraction of CBF-A was also found outside the cell nucleus in small clusters or discrete particles, reminiscent of dendritically transported mRNA granules, with different sizes and signal intensities depending on the brain regions analyzed (Figure 1). These clusters seem to be associated with microtubulin fibers of dendrites as revealed by costaining with the dendrite marker MAP2 (Figure 1D). Furthermore, coimmunostaining with the anti–CBF-A antibody ICCI and a mouse monoclonal antibody (mAb) against the presynaptic marker synapsin 1 revealed that CBF-A is located close to synapses (Figure 1, E and F). Distinctive nuclear staining as well as CBF-A–positive granules along dendrites were also revealed by double immunofluorescence staining performed on rat hippocampal neurons using ICCI and the MAP2 antibody (Figure 2A), as well as antibodies to synapsin 1 and postsynaptic density 95 (PSD95) (Supplemental Figure 1). These experiments were further validated with the anti–CBF-A antibody SAK22 raised against the CBF-A N terminus (Dean et al., 2002) and known to react with both p37 and p42 splice variants (Supplemental Figure 2). Interestingly, in double immunostainings of hippocampal neurons the signals obtained from ICCI and SAK22 displayed a linear correlation, and more than 80% overlap (Figure 2, B and C), supporting their specificity.
FIGURE 1:
In vivo localization of CBF-A in the adult mouse brain. (A–F) Coronal sections. (A and B) CBF-A is localized in neuronal nuclei and in small clusters in their proximity. (C and D) CBF-A clusters are associated with MAP2-positive dendrites in a synaptic pattern. (E and F) CBF-A clusters match the distribution of the presynaptic marker synapsin I but are not overlapping, suggesting preferential postsynaptic localization of CBF-A. The rectangle is shown in F. Scale bars: A and B = 50 μm, C and D = 20 μm, E = 10 μm, F = 2.5 μm.
FIGURE 2:
Dendritic localization of CBF-A in rat hippocampal neurons. (A) Consistent with the in vivo distribution, CBF-A is found in dendrites of hippocampal neurons, as revealed by coimmunostaining with anti-CBF-A (ICCI) and MAP2 antibodies. Scale bar, 10 μM. Magnifications of boxed areas are approximately fivefold in comparison to the corresponding overviews. (B) Double immunofluorescence staining with the anti–CBF-A antibodies ICCI and SAK22 reveals considerable overlap in nucleus and dendrites of rat hippocampal neurons. Scale bar, 20 μM. (C) Quantification of individual dendritic granules shows a linear correlation between the signals obtained with ICCI and SAK22 antibodies against CBF-A. More than 80% of the individual granules are positively labeled with both CBF-A antibodies.
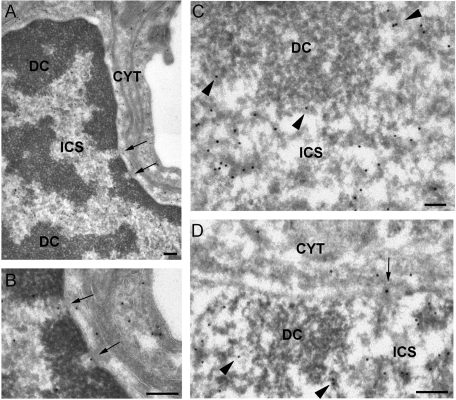
To further characterize the in vivo localization of CBF-A, we next carried out immuno-electron microcopy (IEM) experiments on thin sections of adult mouse brain. Sections of hippocampus incubated with the anti–CBF-A antibody SAK22 revealed that CBF-A has a widespread distribution, in agreement with the light microscopy results reported earlier in the text (Figure 1). Consistent with previous data on the subcellular distribution of CBF-A in oligodendrocytes (Raju et al., 2008), the anti–CBF-A antibodies revealed a high density of labeling in the nucleus of different types of brain cells, including neurons (see also Figures 1 and 2 as well as Supplemental Figures 1 and 2). The CBF-A antibodies decorated electrodense structures located in the interchromosomal space (ICS in Figure 3) and in the perichromatin area (arrowheads in Figure 2, C and D), where active transcription takes place (Fakan and Puvion, 1980). CBF-A was instead excluded from the patches of dense chromatin (DC in Figure 3). The location and morphology of the CBF-A–positive structures suggests that CBF-A is associated with (pre)-mRNP complexes at the sites of transcription and in the interchromosomal space. CBF-A was also found to be associated with electrodense structures, presumably mRNPs, at the nuclear pores and in transit to the cytoplasm (arrows in Figure 3, B and D).
FIGURE 3:
IEM localization of CBF-A in the cell nucleus. Thin sections of adult mouse brain were immunostained with the anti–CBF-A antibody SAK22. (A) Overview of a nucleus from a pericyte found wrapped around precapillary arterioles showing the typical appearance of dense chromatin regions (DC) and interchromatin space (ICS). The arrows point to two distinct nuclear pores. CYT, cytoplasm. (B) The same nuclear pores as in (A) at higher magnification. Note the presence of anti–CBF-A labeling in the central regions of the pores (arrows). (C and D) Examples of nuclear labeling in hippocampal neurons. The anti–CBF-A labeling is located in the interchromatin space (ICS) and in the perichromatin regions (arrowheads) but is excluded from the dense chromatin (DC). Note the labeling associated with a nuclear pore (arrow in D). The magnification bars represent 200 nm in A, B, and D and 100 nm in C.
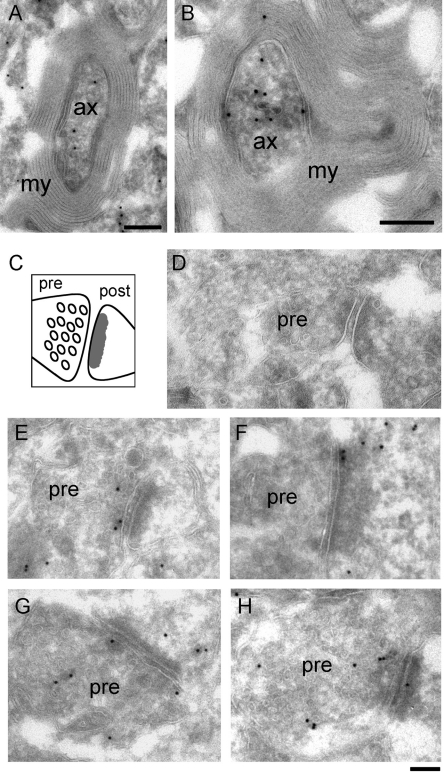
In the cytoplasm, significant immunolabeling was observed in myelinized axons (Figure 4, A and B). The SAK22 antibody also stained synapses, where immunogold markers were found on both presynaptic and postsynaptic compartments (Figure 4, E and H). The labeling density in synapses was relatively low compared to that observed in axons but was highly significant as judged by the absence of immunogold in sections processed in parallel with only secondary antibody (Figure 4D). We could also detect CBF-A in neuronal cytosol and over nonmyelinized processes. The labeling in axons and synapses was confirmed using the rabbit polyclonal anti–CBF-A antibody ICCI (Supplemental Figure 3).
FIGURE 4:
IEM labeling of CBF-A in axons and synapses. Thin sections of adult mouse brain were immunostained with the anti–CBF-A antibody SAK22. The antibody binding sites were detected with protein-A conjugated to colloidal gold particles. (A) and (B) show two examples of immunolabeling in myelinated hippocampal axons. The magnification bars represent 200 nm. Ax, axon; my, myelin sheath. A schematic representation of a synapse is provided in (C) and shows the vesicular appearance of the presynaptic compartment (pre) and the characteristic density of the postsynaptic button (post). (E–H) Examples of anti–CBF-A labeling in synapses in the hippocampus. CBF-A was revealed in both presynaptic and postsynaptic compartments. (D) A synapse taken from a negative control sample processed in parallel and incubated with only secondary antibody. The magnification bar in D–H represents 100 nm.
In synaptosomal fractions CBF-A associates with RTS-containing mRNA transcripts
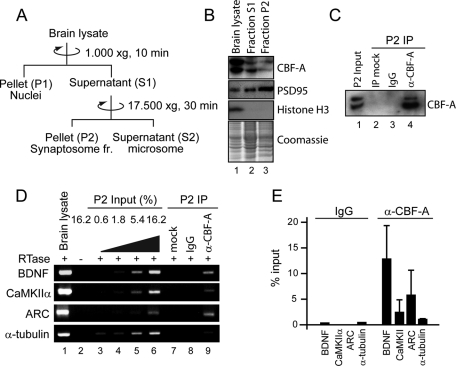
To confirm the subcellular localization of CBF-A in neurons, we fractionated brain lysates by ultracentrifugation on sucrose cushions (Figure 5A). The fractionated material was analyzed on immunoblots with antibodies against CBF-A as well as antibodies against the postsynaptic marker PSD95 and the nuclear marker histone H3. Consistent with the histological data and previous observations (Raju et al., 2008), CBF-A was found in nucleus and cytosol as well as in synaptosome-enriched fractions (Figure 5B).
FIGURE 5:
In synaptosome-enriched fractions, CBF-A specifically interacts with dendritically transported Arc, BDNF, and CaMKII mRNAs. (A) Preparation of synaptosome-enriched fractions. (B) P2 fractions were resolved by SDS–PAGE, stained with Coomassie, and analyzed on immunoblots with CBF-A (ICCI), PSD95, and histone H3 antibodies. (C) CBF-A can be specifically coprecipitated from synaptosomal preparations. P2 fractions were subjected to immunoprecipitation with the CBF-A (ICCI) antibody or nonspecific IgGs. Bound proteins were resolved by SDS–PAGE and CBF-A revealed on immunoblots with the ICCI antibody. Mock, protein A Sepharose incubated with P2 fraction in the absence of antibody. (D and E) In synaptosomal preparations, CBF-A is associated with BDNF, CaMKIIα, and Arc mRNAs. Total mRNA precipitated with anti–CBF-A (ICCI) antibody or nonspecific IgGs from synaptosomal preparations was isolated. The cDNA was analyzed with primers amplifying BDNF, CaMKIIα, BDNF, and α-tubulin mRNAs. Quantification of precipitated mRNA species was by densitometric analysis in at least three successful experiments.
We next applied RNA immunoprecipitation assays to examine whether in synaptosome-enriched P2 fractions CBF-A associates with synaptic mRNAs. Synaptosomal preparations were incubated with the CBF-A antibody ICCI. Total RNA was isolated from immunoprecipitated samples of mouse brain P2 fraction (see Figure 5C) using the TRI reagent and reverse-transcribed with oligo(dT) primers. The resulting cDNA was analyzed by semiquantitative RT-PCR using specific primers amplifying BDNF, CaMKIIα, and Arc mRNAs. As can be seen, we revealed a specific enrichment in the levels of Arc, BDNF, and CaMKIIα mRNAs coprecipitated with CBF-A (Figure 5D). Densitometric quantification revealed a considerable 3- to 10-fold increase in the levels of BDNF, CaMKIIα, and Arc mRNA coprecipitated with CBF-A in comparison to α-tubulin mRNA and control immunoprecipitations carried out with nonspecific immunoglobulin (Ig) Gs (Figure 5, D and E). We conclude that, in synaptosomal preparations, CBF-A associates with Arc, BDNF, and CaMKIIα mRNAs.
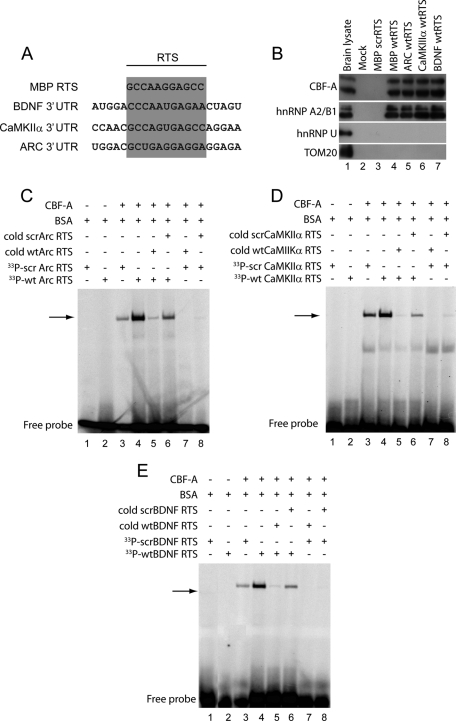
The 3′ UTRs of CaMKIIα and Arc mRNAs contain RTS-like sequences that are closely related to the MBP mRNA RTS (Gao et al., 2008), and we identified a similar RTS-like sequence in the 3′ UTR of BDNF mRNA (Figure 6A). To evaluate whether CBF-A bound any of these sequences, we synthesized biotinylated RNA oligonucleotides encompassing the wild-type (wt)RTS-like sequences present in Arc, BDNF, and CaMKIIα mRNAs and in the MBP mRNA RTS. As control, we used an RNA oligonucleotide encompassing a scrambled version (scrRTS) with identical nucleotide composition as the MBP mRNA RTS but different primary sequence (see Raju et al., 2008). These RNA oligonucleotides were coupled to streptavidin-coated Sepharose beads, and the beads were incubated with total mouse brain lysates. The blots in Figure 6B show that wtRTS sequences precipitated endogenous CBF-A (Figure 6B, lanes 4–7). Control scrRTS or mock beads that were not conjugated with any RNA oligonucleotides did not precipitate CBF-A (Figure 6B, lanes 2 and 3). As expected, similar results were obtained with endogenous hnRNP A2. On the contrary, hnRNP U, which is also found in RNA granules isolated from developing and adult mouse brains and the mitochondrial protein Tom20, were not coprecipitated with any of the beads used in the RNA affinity chromatography assays (Figure 6B), altogether supporting the specificity of the assay.
FIGURE 6:
CBF-A directly binds to Arc, BDNF, and CaMKIIα mRNA RTSs. (A) Sequence analysis of 3′ UTRs shows considerable homologies between MBP mRNA RTS and RTS-like sequences in BDNF, CaMKIIα, and ARC mRNAs. (B) CBF-A binds the RTS sequences found in BDNF, CaMKIIα, and ARC mRNAs. Biotinylated wtRTSs from BDNF, CaMKIIα, and MBP mRNA as well as a MBP scrRTS were conjugated to streptavidin Sepharose. Beads were incubated with mouse brain lysates. Bound proteins were resolved by SDS–PAGE and analyzed on immunoblots with antibodies to CBF-A (SAK22) and hnRNP A2B1 and with antibodies against hnRNP U, and the mitochondrial protein Tom20 was used as negative control. (C–E) RTS-binding EMSAs using 33P-labeled Arc wtRTS and scrRTS (C), BDNF wtRTS and scrRTS (D), and CaMKIIα wtRTS and scrRTS probes (E). To perform EMSAs, wtRTS or scrRTS probes were incubated with purified recombinant CBF-A without affinity tag. Incubations were also performed in the presence or absence of a 50-fold excess of unlabeled competitor RNA oligonucleotides as indicated.
We next applied electrophoretic mobility shift assays (EMSAs) to determine whether CBF-A directly binds the RTS motifs. We incubated purified recombinantly expressed glutathione S-transferase-tagged CBF-A (p37 isoform) where the affinity tag had been proteolytically removed with [33P]-labeled RNA oligonucleotides encompassing Arc, CaMKIIα, and BDNF wtRTSs. In parallel, CBF-A was also incubated with control RNA oligonucleotides encompassing scrambled versions of the wtRTS sequences. Figure 6 shows that incubation with CBF-A retarded more efficiently the electrophoretic mobility of wtRTS-containing oligonucleotides in comparison to the scrambled sequences (Figure 6, C–E). In addition, gel retardation of wtRTS-containing RNA oligonucleotides incubated with CBF-A was competed in the presence of corresponding unlabeled wtRTS oligonucleotides but not in the presence of unlabeled scrRTS-containing RNA oligonucleotides when added in large amounts to the reaction mixture (Figure 6, C–E). Finally, incubation of CBF-A with labeled scrRTS oligonucleotides was sensitive to competition with unlabeled wtRTS oligonucleotides but was not sensitive to competition with unlabeled scrRTS oligonucleotides (Figure 6, C–E).
Altogether these results indicate that CBF-A specifically associates with Arc, BDNF, and CaMKIIα mRNAs through direct interactions with their RTS sequences.
In dendrites CBF-A localizes to dynamic mRNA granules
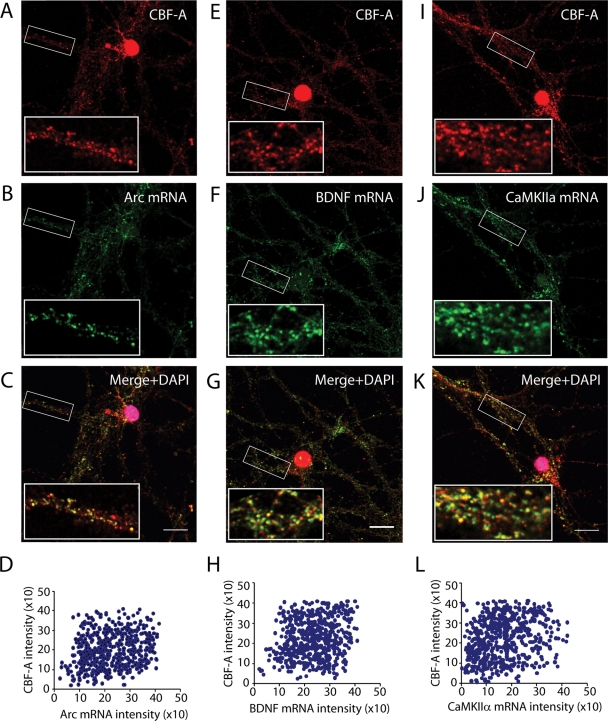
Immunofluorescence staining performed on hippocampal neurons demonstrated that CBF-A is present in dendrites (Figure 2A). If CBF-A associates with dendritic mRNAs via their RTS elements, it is likely that CBF-A is found in transported granules. To evaluate the possibility, hippocampal neurons were analyzed by immunofluorescence in situ hybridization (immuno-FISH) with the anti–CBF-A antibody ICCI and specific digoxigenin-labeled RNA probes hybridizing with endogenous Arc, BDNF, and CaMKIIα mRNAs. Confocal microscopy revealed that CBF-A–positive granules distributed along dendrites and their distributions correlated with the granular structures identified by the FISH signals using antisense probes specific for Arc, BDNF, and CaMKIIα mRNA (Figure 7). Notably, no FISH signal was detected with sense RNA probes against the corresponding transcripts just mentioned, supporting the specificity of the assay (Supplemental Figure 4). To evaluate colocalizations of endogenous CBF-A with Arc, BDNF, and CaMKIIα mRNAs, we performed unbiased statistical analysis on the fluorescence intensity levels from individual granules derived from the corresponding confocal images, as previously described (Ma et al., 2002; Raju et al., 2008). In all cases the results revealed linear correlations between the signals from CBF-A and the RNA transcripts analyzed (Figure 7, D–L). Quantification of the number of colocalization events occurring in individual particles showed that 60% to 70% of the granules simultaneously contain CBF-A and the RTS-containing Arc, BDNF, and CaMKIIα mRNA. These observations suggest that CBF-A is likely to be incorporated in endogenous mRNA granules containing Arc, BDNF, and CaMKIIα transcripts.
FIGURE 7:
In dendrites of hippocampal neurons CBF-A exhibits a granular distribution which correlates with transported Arc, BDNF, and αCaMKII mRNAs. Endogenous CBF-A and Arc (A–C), BDNF (E–G), or CaMKII mRNAs (I–K) were simultaneously monitored by immuno-FISH and confocal microscopy. Scale bar, 10 μm. Magnifications of boxed areas are approximately fivefold in comparison to corresponding overviews. Unbiased statistical quantification of individual CBF-A and Arc (D), BDNF (H), or CaMKIIα (L) RTS-positive granules are based on the immuno-FISH analysis in the top panels. In all cases, linear correlations between the fluorescence intensity levels of CBF-A and RTS-containing transcripts were revealed.
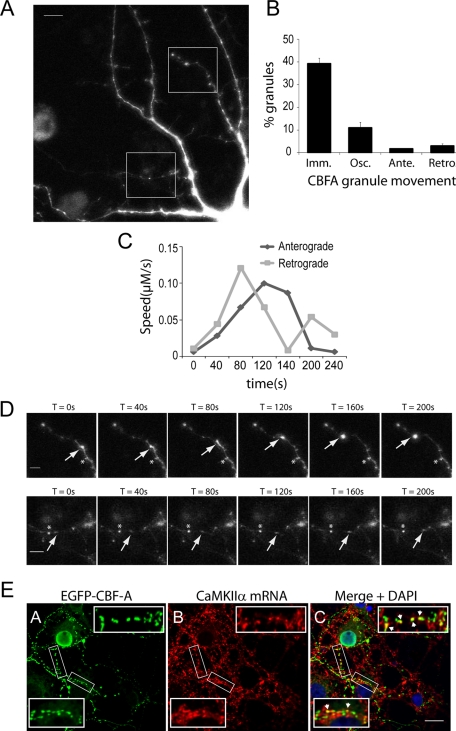
To evaluate whether in dendrites CBF-A–positive granules are dynamically transported, we transiently expressed full-length CBF-A (p37 isoform) tagged with enhanced green fluorescent protein (EGFP) in rat hippocampal neurons. Confocal microscopy showed that the EGFP/CBF-A construct displayed both nuclear distribution and granular clusters in dendrites, overall matching the distribution observed in the case of the endogenous protein ( Figure 8A). We next performed time-lapse live cell microscopy on hippocampal neurons transiently expressing EGFP/CBF-A. As can be seen in Figure 8B, we found that a major fraction of EGFP/CBF-A–positive granules were immobile or stationary. A fraction of EGFP/CBF-A–containing granules exhibited an oscillatory movement. Finally, a subset of EGFP/CBF-A–positive granules displayed a single direction motion, anterograde or retrograde, with a calculated speed of 0.028–0.13 μm/s (Figure 8, B–D), similar to that measured for Staufen 1-, DDX3-, and CaMKIIα mRNA–containing granules (Kohrmann et al. 1999; Rook et al., 2000; Elvira et al., 2006). Interestingly, we found that in dendrites of hippocampal neurons EGFP/CBF-A–positive granules correlated with the FISH signal from digoxigenin-labeled RNA probes hybridizing with CaMKIIα mRNAs (Figure 8E) as well as Arc and BDNF mRNAs (unpublished data). Taken altogether, these observations suggest that in hippocampal neurons a fraction of CBF-A is incorporated in dynamically transported granules which are likely to contain dendritic mRNAs.
FIGURE 8:
In dendrites CBF-A is present in dynamic mRNA granules. (A) Hippocampal neurons (10 DIV) transfected with EGFP/CBF-A. Rectangular areas represent the regions followed by live cell microscopy. Scale bar, 10 μm. (B) The diagram summarizes the different categories of EGFP/CBF-A–positive granules identified in transiently transfected neurons in terms of their movement. Particles that do not move at least three steps (as evaluated over four frames) in a row in the same direction are scored as either anterograde or retrograde. Particles that move one or two steps and after a stop move backward or forward to stop again, are scored as oscillating. (C and D) Time-course analysis of two EGFP/CBF-A–positive granules traced by the arrows in the time-lapse microscopy experiments (see rectangular areas in panel A). (D) Top panels, EGFP/CBF-A–positive granules moving anterogradely are indicated by arrows. Bottom panels provide examples of EGFP/CBF-A–positive granules which move retrogradely (arrows). In all cases, the asterisks indicate an immobile reference point. Scale bar, 5 μm. (E) In transfected hippocampal neurons, the distribution of EGFP/CBF-A correlates with CaMKIIα mRNAs. EGFP/CBF-A and CaMKIIα mRNA were simultaneously monitored by immuno-FISH and confocal microscopy with antibodies against GFP and an RNA probe hybridizing with CaMKIIα mRNA. Scale bar, 15 μm. Magnifications of boxed areas are approximately fivefold in comparison to corresponding overviews (arrowheads point to examples of colocalizations).
Postsynaptic receptor activation leads to increased levels of CBF-A in dendrites
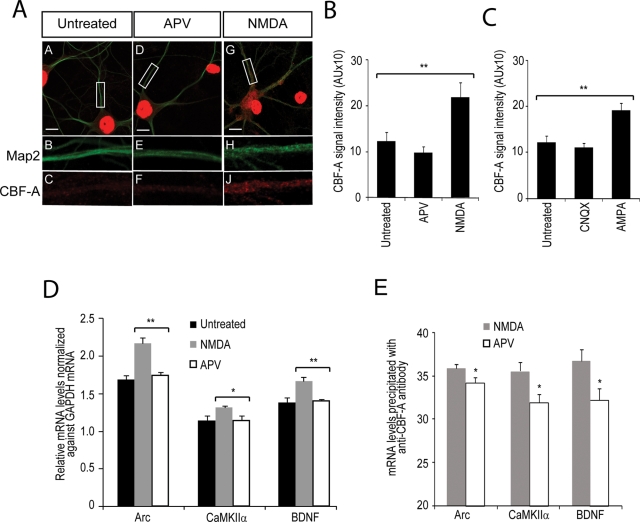
Treatment of hippocampal neurons with the selective agonists NMDA (N-methyl-d-aspartate) and AMPA (α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate) mimics the effects of the neurotransmitter glutamate, leads to synaptic activation, and upregulates dendritic mRNA transcripts, eventually leading to synaptic consolidation (Bramham and Wells, 2007; Tian et al., 2007). CBF-A is present in postsynaptic densities (Figure 4). Therefore, because CBF-A binds to Arc, BDNF, and CaMKIIα mRNAs, upon NMDA and AMPA treatment we expect to find increased CBF-A protein levels in dendrites. To test this possibility, we treated hippocampal neurons with the agonists NMDA or AMPA and the corresponding antagonists APV ((2R)-amino-5-phosphonovaleric acid)) and CNQX (6-cyano-7-nitroquinoxaline-2,3-dione) as negative controls. After a 4-h treatment, we monitored the distributions of CBF-A and dendrite marker MAP2 by immunofluorescence and confocal microscopy. We found that both NMDA (Figure 9A) and AMPA (unpublished data) induced increased levels of CBF-A along dendrites (Figure 8A). On the contrary, APV treatment (Figure 9A) or treatment with CNQX (unpublished data) did not induce any changes in the CBF-A distribution. To quantify the effects, we randomly selected multiple dendritic regions from 10 independent hippocampal neurons untreated or treated with antagonists or agonists, and in all cases we measured CBF-A signal intensities. We discovered that NMDA and AMPA induced an approximately twofold increase in the levels of CBF-A in dendrites in comparison to untreated cells (Figure 9, B and C). Only marginal effects were detected upon treatment with the respective agonists (Figure 9, B and C). We conclude that, in dendrites, trafficking and localization of endogenous CBF-A is sensitive to postsynaptic receptor stimulation.
FIGURE 9:
CBF-A association with RTS-containing Arc, BDNF, and CaMKIIα mRNAs is sensitive to postsynaptic receptor stimulation. CBF-A accumulates in dendrites of hippocampal neurons upon synaptic stimulation with the agonists NMDA or AMPA. (A) Merged images and fivefold magnification of rectangular areas obtained from (A–C) untreated neurons, (D–F) APV-treated neurons, and (G–J) NMDA-treated hippocampal neurons immunostained with antibodies to CBF-A and MAP2. (B and C) Quantification of NMDA and AMPA effects on the distribution of CBF-A. CBF-A intensities measured from randomly selected dendritic areas taken from neurons treated with the indicated reagents (NMDA or APV; AMPA or CNQX) were plotted in bar diagrams with standard deviations. CBF-A levels are specifically and significantly enriched in dendrites upon NMDA treatment in comparison to untreated or AMPA-treated cells (p values were calculated by Student's t test). AU, arbitrary units. (D) NMDA stimulation of hippocampal neurons induces an increased level of Arc, CaMKIIα, and BDNF mRNAs. Total RNA from untreated, NMDA-, or APV-treated neurons was reverse-transcribed with oligo(dT) primers and the cDNA analyzed by qRT-PCR with primers amplifying Arc, CaMKIIα, BDNF, and GAPDH mRNAs. The bar diagram shows relative amounts of the indicated RNAs toward GAPDH mRNA determined over three independent experiments. Error bars depict standard error estimated by Student's t test. (E) RNA coimmunoprecipitated with CBF-A from lysates of neurons treated with NMDA or APV was analyzed by qRT-PCR. Arc, BDNF, and CaMKIIα mRNA levels are specifically and significantly enriched following NMDA stimulation (p values were calculated by Student's t test). Error bars, standard deviations.
Association of CBF-A with dendritic mRNA transcripts is activity-dependent
During mRNA biogenesis, hnRNP proteins such as CBF-A are cotranscriptionally associated with nascent transcripts, facilitate RNP assembly, and in many cases accompany the mRNA from gene to polysomes (Visa et al., 1996; Daneholt, 2001; Dreyfuss et al., 2002; Percipalle et al., 2002).
CBF-A is an abundant nuclear protein that localizes to dendrites, and its distribution is activity dependent. Because the distribution of Arc, BDNF, and CaMKIIα mRNAs is sensitive to NMDA and AMPA treatment (Steward et al., 1998; Bramham and Wells, 2007), we reasoned that CBF-A association with transported mRNAs may be intrinsically sensitive to synaptic stimulation. To start proving this hypothesis, we stimulated rat hippocampal neurons with NMDA. Next total RNA was extracted and reverse-transcribed with oligo(dT) primers. The resulting cDNA was analyzed by quantitative real-time PCR (qRT-PCR) with primers amplifying Arc, BDNF, and CaMKIIα mRNAs. In at least three independent experiments, we found a 10–20% significant increase in the steady-state levels of Arc, BDNF, and CaMKIIα mRNAs, in all cases normalized against the levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA (Figure 9D). None of the transcripts were affected upon treatment with the antagonist APV (Figure 9D), altogether supporting the view that synaptic stimulation induces increased transcriptional rates of dendritically localized mRNAs (Bramham and Wells, 2007). We next tested whether NMDA stimulation correlates with increased binding of CBF-A to Arc, BDNF, and CaMKIIα mRNAs. For this purpose, total lysates from NMDA- or APV-treated rat hippocampal neurons were subjected to immunoprecipitations with anti–CBF-A antibodies. Total RNA was extracted and the cDNA analyzed by qRT-PCR. We found that the amounts of Arc, BDNF, and CaMKIIα mRNAs coprecipitated with CBF-A were significantly increased in comparison to APV-treated cells (Figure 9E) or untreated cells (unpublished data). As expected, nonspecific IgGs did not pull down any of the transcripts analyzed (unpublished data). Altogether these findings indicate that the specific binding of CBF-A to Arc, BDNF, and CaMKIIα mRNAs correlates with postsynaptic receptor stimulation and supports the view that CBF-A accompanies the transcripts from gene to dendrites.
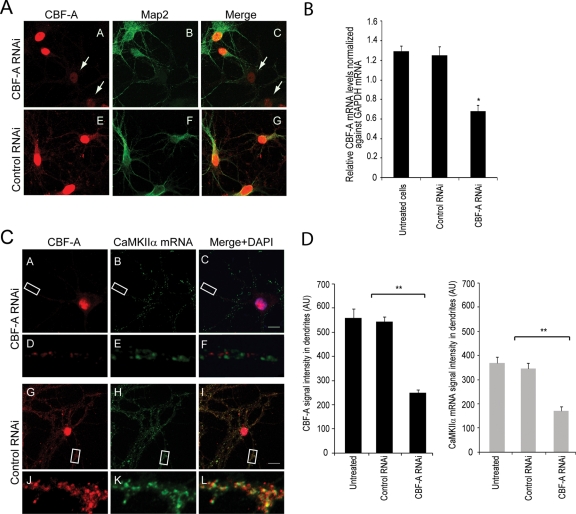
To test for an involvement of CBF-A in transport of dendritic transcripts, we next silenced the CBF-A gene by RNA interference (RNAi). Hippocampal neurons were transfected with RNA duplexes against target sequences on the CBF-A gene. Steady-state expression of endogenous CBF-A was monitored by immunofluorescence on hippocampal neurons anti–CBF-A antibodies and an anti-MAP2 antibody (Figure 10A), whereas the CBF-A mRNA levels were monitored by qRT-PCR (Figure 10B). A specific shutdown of the expression resulting in a drop in endogenous CBF-A steady-state level was observed approximately 3 d after transfection (Figure 10, A and B). To test the effect of CBF-A silencing on the distribution of dendritic mRNA, we performed immuno-FISH on CBF-A–silenced hippocampal neurons and monitored the distribution of CaMKIIα mRNA. We found that CBF-A gene silencing led to a drop in the levels of CaMKIIα mRNA in dendrites (Figure 10, C and D), whereas in control cells transfected with unrelated RNAi oligonucleotides, dendritic localization of CaMKIIα mRNA granules was not affected (Figure 10, C and D). For quantification, we randomly selected dendritic regions from nontransfected as well as control and CBF-A silenced hippocampal neurons and in all cases we measured CBF-A and FISH average signals intensities. Concomitantly, with an average 50% specific reduction in the steady-state expression of CBF-A, we observed a twofold drop in the levels of CaMKIIα mRNA in dendrites in comparison to controls (Figure 10D). Similar results were obtained for Arc and BDNF mRNA dendritic localization upon CBF-A gene silencing (unpublished data). We conclude that in neurons CBF-A gene knockdown specifically affects mRNA expression levels of transported mRNAs.
FIGURE 10:
CBF-A silencing impairs dendritic mRNA localization in hippocampal neurons. (A) Rat hippocampal neurons transfected with CBF-A specific or control siRNA oligonucleotides. Silencing is monitored by immunostaining with anti–CBF-A antibodies and with a mAb against MAP2. Arrows point to examples of cells in which CBF-A is specifically knocked down. Scale bar, 10 μm. (B) qRT-PCR was performed on cDNA derived from total RNA isolated from CBF-A–silenced or control hippocampal neurons. Significant down-regulation of relative CBF-A mRNA levels was observed upon CBF-A gene silencing in comparison to untreated or control siRNA treated cells (p values were calculated by Student's t test). Error bars represent standard deviations. (C) In CBF-A–silenced hippocampal neurons, there is a drop in the levels of CaMKIIα mRNA in dendrites as revealed by immuno-FISH with antibodies against CBF-A and an RNA probe hybridizing with CaMKIIα mRNA. Analysis is by confocal microscopy. Scale bars, 10 μm. (D–F) Fivefold magnifications of boxed areas in A–C. (J–L) Fivefold magnifications of boxed areas in G–I. Overall, these experiments were repeated in triplicate. (D) The bar diagrams display signal intensities for both CBF-A and CaMKIIα mRNA obtained in immuno-FISH experiments on untreated, control siRNA-, and CBF-A siRNA–treated hippocampal neurons. In all cases, signal intensities were measured over as many as 20–30 hippocampal neurons.
DISCUSSION
In the present study we found that the transacting factor CBF-A is abundantly expressed in neurons. In the cytosol, CBF-A is present in dendrites and synapses with a characteristic granular distribution. In synaptosomal preparations from total mouse brain lysates, CBF-A specifically associates with Arc, BDNF, and CaMKIIα mRNAs. A close look at the 3′ UTRs of the transcripts just mentioned shows the presence of RTS elements that are related to the MBP mRNA RTS. Consistent with previous work (Raju et al., 2008), these RTS elements are specifically and directly recognized by CBF-A. These observations and in situ evidence that in dendrites a fraction of CBF-A colocalizes with all three transcripts suggest that CBF-A is genuinely present in dendritic mRNA granules, presumably with hnRNP A2 (see also Raju et al., 2008), where CBF-A binds Arc, BDNF, and CaMKIIα mRNA through their RTS elements.
The RTS-containing 3′ UTRs of BNDF and CaMKIIα mRNAs play an essential role for localized translation. There is evidence that in mutated mice in which the protein-coding region of CaMKIIα is intact but lacks the 3′ UTR the mRNA is restricted to the soma. These animals show reduction of CaMKIIα transcripts in postsynaptic densities, a reduction in late-phase long-term potentiation, and impairments in spatial memory (Miller et al., 2002). For BDNF, the brain produces two different transcripts, with either short or long 3′ UTRs. In mutated mice with truncated long 3′ UTR, dendritic targeting of BDNF mRNAs is impaired, and these animals display selective impairment in long-term potentiation in dendrites of hippocampal neurons (An et al., 2008). Here we report that CBF-A–positive mRNA granules are dynamic in nature and that their localization in dendrites is activity-dependent. Similarly to Arc, BDNF, and CaMKIIα mRNAs, treatment of hippocampal neurons with NMDA and AMPA, which mimic the effect of the neurotransmitter glutamate, resulted in CBF-A accumulation in dendrites. CBF-A is also present in the characteristic postsynaptic densities where translation of Arc, BDNF, and CaMKIIα mRNAs is believed to take place (Bramham and Wells, 2007). Therefore it is conceivable that failure in correct localization and targeting of BDNF and CaMKIIα mRNAs upon truncation of their 3′ UTRs in mutated mice partly results from deletion of their RTS elements. Similarly to BDNF and CaMKIIα mRNAs, we predict that Arc mRNA lacking the RTS would not be transported to dendrites. On the basis of these considerations we propose that the RTS sequences in the 3′ UTRs of Arc, BDNF, and CaMKIIα mRNAs are bona fide cis-acting elements required for dendritic trafficking. Given that, in dendrites of CBF-A–silenced hippocampal neurons, we found a twofold drop in the levels of CaMKIIα mRNA, we suggest that CBF-A recognizes RTS elements and functions as transacting factor to facilitate mRNA transport and localization in dendrites. Because association of CBF-A with the RTS-containing mRNAs is sensitive to NMDA treatment, it is likely that the interplay between CBF-A and RTS elements correlates with the function of excitatory synapses.
IEM experiments demonstrated that in the cell nucleus CBF-A localizes in the perichromatin region where active transcription is believed to take place (Rouquette et al., 2010), presumably coupled to nascent pre-mRNPs as well as mature mRNPs in interchromatin space and passages through the nuclear pore complex. Because CBF-A is a shuttling hnRNP and core component of pre-mRNP/mRNPs (Percipalle et al., 2002), these observations suggest that CBF-A is assembled into mRNP complexes in the nucleus and exported from the nucleus to the cytoplasm as part of mature mRNPs. CBF-A is known to bind the MBP mRNA RTS already in the cell nucleus (Raju et al., 2008). Therefore, in view of the present findings, it is likely that RTS recognition by CBF-A is important during pre-mRNP assembly. We propose that CBF-A recognizes the RTS elements present in Arc, BDNF, and CaMKIIα mRNAs to facilitate cotranscriptional pre-mRNP assembly, a mechanism that promotes the establishment of transport-competent RNPs. We suggest that cotranscriptional binding of CBF-A to RTS elements provides de facto a way to sort transported Arc, BDNF, and CaMKIIα mRNPs at an early stage during RNP biogenesis.
At later stages, when granules have reached their final destinations, there is evidence that hnRNP A2 phosphorylation by Fyn kinase leads to unwinding of the MBP mRNA granule for localized translation (White et al., 2008), which indicates a structural role for hnRNP A2 in the maintenance of the RNP granule. Similar mechanisms may be envisaged in neurons for dendritic RTS-mediated mRNA targeting. In any case, we propose that in the cell nucleus RTS recognition by CBF-A mediates RNP packaging of Arc, BDNF, and CaMKIIα mRNAs through a mechanism that probably leads to nuclear remodeling of their 3′ UTRs. This in turn contributes to keep dendritic mRNAs in a state which is transport-competent and presumably translationally dormant. We speculate that these mechanisms are required for establishment of excitatory synapses and may have a long-term effect on the plasticity of neuronal contacts.
MATERIALS AND METHODS
Antibodies
The mouse mAbs against Map2, NeuN, synapsin I, and PSD95 were, respectively, obtained from Sigma (St. Louis, MO), Chemicon (now Millipore; Billerica, MA), Cell Signaling Technology (Danvers, MA), and Abcam (Cambridge, UK). The rabbit polyclonal anti–CBF-A antibody ICCI was raised against the peptide YQQGYGPGYGGYDY located in the C terminus of the p42 CBF-A splice variant as described by Raju et al. (2008). The guinea pig anti–CBF-A antibody SAK22 was designed against the peptide EEQPMETTGATEN located in the N terminus of both p37 and p42 CBF-A isoforms as previously described (Dean et al., 2002) and was purchased from Peptide Specialty Laboratories (Heidelberg, Germany). The mAbs against histone H3, hnRNP U (3G6), and hnRNP A2 were obtained from Abcam, whereas the anti-Tom20 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Nonspecific mouse IgGs were obtained from Dako (Carpinteria, CA). For immunofluorescence experiments, the primary antibodies were revealed with species-specific fluorophore-conjugated (Cy3, Cy5 from Jackson ImmunoResearch, West Grove, PA; Alexa 488 from Molecular Probes, Eugene, OR) secondary antibodies.
Immunohistochemistry on mouse forebrain
Adult C57BL/6 mice (Charles River Laboratories, Wilmington, MA) were transcardially perfused with phosphate-buffered saline (PBS) followed by 4% formaldehyde in PBS, and brains were dissected and postfixed overnight at 4°C. Coronal sections were made at 35 μm using a vibratome (Leica, Wetzlar, Germany). Staining was performed on free-floating sections. Sections were incubated with blocking solution (10% donkey serum in PBS with 0.2% Triton X-100) for 1 h at room temperature and then incubated at 4°C for 14 h with primary antibody diluted in blocking solution. Primary antibodies against CBF-A (ICCI), Map2, NeuN, Synapsin I, and PSD95 were used on the sections. Primary antibodies were revealed with Cy3-, Cy5-, or Alexa 488–conjugated secondary antibodies. Control sections were stained with secondary antibodies alone.
Primary neuronal cultures and cell stimulation
Hippocampal neurons were prepared as described in Ledda et al. (2007). Briefly, hippocampi were dissected from E18–20 rat embryos, and hippocampal neurons were cultured in neurobasal medium (Invitrogen, Carlsbad, CA) supplemented with B-27 (Gibco, now Invitrogen) on poly-d-lysine–coated coverslips or dishes. Neurons were fed once weekly by replacing half of the medium. Neuronal cell stimulation was carried out as described earlier (Tian et al., 2007). Where indicated, 18 d after in vitro (DIV) hippocampal neurons were incubated with the 20 μM NMDA (Sigma) or 20 μM APV (Sigma) or, alternatively, with 20 μM AMPA or 20 μM CNQX. In all cases treatment was performed for 4 h. Cells were subsequently fixed with the 3.7% formaldehyde in PBS and further processed for immunofluorescence.
Immunofluorescence microscopy and immuno-FISH
In situ hybridization and immunostaining were performed as described by Raju et al. (2008). Digoxigenin-labeled RNA probes were synthesized using DIG RNA labeling mix (Roche, Basel, Switzerland). PCR products obtained with primer pairs amplifying Arc mRNA (forward, AGC AGC AGA CCT GAC ATC CT; reverse, GGC TTG TCT TCA CCT TCA GC), BDNF mRNA (forward, TGG CCT AAC AGT GTT TGC AG; reverse, GGA TTT GAG TGT GGT TCT CC), CaMKIIα mRNA (forward, GAC ACC AAA GTG CGC AAA CAGG; reverse, GCG AAG CAA GGA CGC AGG) were cloned into the p-GEM-T vector (Promega, Madison, WI) and used as a template for in vitro transcription by T7 RNA polymerase. Briefly, hippocampal neurons grown on poly-d-lysine–coated coverslips were fixed with 3.7% formaldehyde and hybridized overnight at 42ºC. After hybridization, the cells were extensively washed three times for 30 min using 2× saline-sodium citrate buffer supplemented with 50% formamide. The cells were briefly fixed again, and immunostaining was performed with the rabbit polyclonal antibody against CBF-A (ICCI). Detection of the hybridized probe and endogenous CBF-A was performed with anti-DIG rhodamine (Roche) and donkey anti–rabbit Alexa 488–conjugated secondary antibody (Invitrogen). Where indicated, the same probes were used in FISH experiments performed on rat hippocampal neurons expressing EGFP/CBF-A (see next section) and in hippocampal neurons subjected to CBF-A gene silencing (see section on CBF-A gene silencing).
Transfection and live cell microscopy on cultured hippocampal neurons
Rat hippocampal neurons were grown on poly-d-lysine–coated glass coverslips in 60-mm dishes (Corning, Corning, NY) and transfected at DIV 9–10 with an EGFP/CBF-A expression plasmid (gift from Tomas Leanderson, Lund University, Sweden) using Lipofectamine 2000 as described in the manufacturer's handbook (Invitrogen). The following day, EGFP/CBFA-expressing neurons were imaged with a 40× objective using a Zeiss (Thornwood, NY) LSM510 META laser scanning confocal microscope equipped with heated chamber and humidity control unit at 37°C and 5% CO2. Images were acquired every 40 s, and the data were analyzed using ImageJ and the Volocity image analysis software.
IEM on sections of adult mouse brain
Adult mouse brains were fixed following Godsave et al. (2008) with 2% paraformaldehyde (PFA) and 0.2% glutaraldehyde in 0.1 M PHEM buffer (25 mM HEPES, 10 mM EGTA, 60 mM PIPES, 2 mM MgCl2, pH 7.2) for 20 h after intracardiac perfusion. Small brain pieces were postfixed for 7 d in 1% PFA and stored in 0.5% PFA in the same buffer, washed with 50 mM glycine in PHEM buffer, and embedded gradually in gelatin (2, 5, and 12% in PHEM) before infiltration overnight in 2.3 M sucrose. Brain pieces were frozen in liquid nitrogen and sectioned at –120°C in a cryoultramicrotome (Leica EM UC6/FC6; Leica, Vienna, Austria). The sections were retrieved in Formvar-coated gold EM grids of 200 mesh using a mixture 1:1 of 2.3 M sucrose and 2% methyl cellulose. For immunogold labeling, the grids were sequentially incubated at 37ºC in 2% gelatin for 20 min, in 50 mM glycine in PHEM, in 5% bovine serum albumin (BSA) in PHEM, and finally in 2% BSA in PHEM. Incubation with the anti–CBF-A antibody SAK22 (1:7 vol/vol) and protein A gold conjugated (1:100 vol/vol) were also done in 2% BSA in PHEM for 1 h and 30 min, respectively. Washes were then done in PHEM. Grids were finally postfixed with 1% glutaraldehyde in PHEM, washed in distilled water, and stained with 2% methyl cellulose/saturated uranyl acetate (9:1) for 10 min. The observations and images were made in a Tecnai Spirit 120 kv electron microscope (FEI, Eindhoven, The Netherlands).
Isolation of synaptosome-enriched fraction
Brain fractions were prepared as previously described ( Huttner et al., 1983; Zhou et al., 2007). Briefly, three adult mice were killed by cervical dislocation, and the forebrains were dissected out on ice. All of the following steps were performed at 4ºC. The tissues were quickly minced and homogenized in 10× volumes of 0.32 M sucrose buffer (0.32 M sucrose in 10 mM HEPES, pH.7.4) supplemented with protease inhibitors (Complete; Roche) with nine up-and-down strokes of a glass homogenizer. The homogenate was centrifuged at 1000 × g for 10 min, and supernatant (S1) was centrifuged again at 17,500 × g for 30 min. The pellet (P2) was dissolved into lysis buffer (0.4% NP-40, protease inhibitors in 1× PBS) and used for further experiments. For the characterization of each fraction, the same amount of proteins (20 μg/lane) was resolved by SDS–PAGE, detected by Coomassie staining, and analyzed on immunoblots with antibodies against CBF-A (ICCI), PSD-95, and H3.
Protein and RNA immunoprecipitation
Mouse brain P2 fractions were incubated overnight at 4ºC with anti–CBF-A antibody (ICCI), no antibody (mock), or nonspecific mouse IgGs (Dako). Where indicated, protein extracts were treated with the RNase inhibitor (100 μg/ml, RNaseout; Fermentas, Glen Burnie, MD) for 15 min before incubation with antibodies. In all cases the reaction mixes were incubated for 1 h with Protein G-Sepharose 4B conjugate (Zymed, Invitrogen). The precipitated samples were resolved by SDS–PAGE and analyzed on immunoblot with antibodies against CBF-A. For analysis of the RNA species coprecipitated with the CBF-A antibodies, the immunoprecipitated RNA was extracted using the TRI reagent (Sigma), and reverse-transcribed using Superscript II and oligo(dT) primers, as described in the manufacturer's protocols (Invitrogen). An equal volume of RNA incubated without Superscript II was used as negative control (RT–). The samples were then analyzed by semiquantitative PCR with primers specific to BDNF mRNA with long 3′ UTR (BDNF forward, TGG CCT AAC AGT GTT TGC AG; BDNF reverse, GGA TTT GAG TGT GGT TCT CC), CaMKIIα mRNA (CaMKIIα forward, GAC ACC AAA GTG CGC AAA CAGG; CaMKIIα reverse, GCG AAG CAA GGA CGC AGG), Arc mRNA (Arc forward, AGC AGC AGA CCT GAC ATC CT; Arc reverse, GGC TTG TCT TCA CCT TCA GC), and α-tubulin mRNA (forward, TTC GTA GAC CTG GAA CCC AC; reverse, TGG AAT TGT AGG GCT CAA CC). Quantification of PCR products was performed over three independent experiments using ImageJ software.
RNA–protein interaction assays
For RNA affinity chromatography, the experiments were performed essentially as described ( Hoek et al., 1998; Raju et al., 2008). The following RTS-containing RNA oligonucleotides (Thermo Fisher Scientific, Walldorf, Germany) were used in the binding assays: Arc wtRTS, GCU GAG GAG GAG GAG AUC AUU; CaMKIIα wtRTS, AAC GCC AGU GAG CCA GGA ACU; and BDNF wtRTS, AUG GAC CCA AUG AGA ACU AGU, as well as the previously described MBP wtRTS and scrRTS sequences (Raju et al., 2008). In all cases RNA oligonucleotides were coupled to streptavidin-coated Sepharose (GE Healthcare, Little Chalfont, UK) and incubated with total mouse brain lysates. Bound proteins were resolved by SDS–PAGE and analyzed on immunoblots with antibodies to CBF-A, hnRNP A2, hnRNP U, or Tom20. EMSAs were performed as described by Raju et al. (2008). Briefly, RNA oligonucleotides encompassing Arc wtRTS, CaMKIIα wtRTS, or wtRTS BDNF (see all sequences earlier in the text) and the RNA oligonucleotides Arc scrRTS (GGCUGAGAAUGCGGAAUAGUG), CaMKIIα scrRTS (GGCAAGGUAGCGCCUAAACA), and BDNF scrRTS (GUGAACCAGGGCACUAAAUAU) encompassing scrambled versions of the RTSs were 5′ end-labeled using [γ-33P]ATP (GE Healthcare) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Purified recombinant CBF-A was incubated with 50 fmol of 33P-labeled wtRTS or scrRTS RNA oligonucleotides in EMSA buffer (20 mM HEPES, pH 7.6, 5 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, 5% glycerol) containing heparin (5 μg/μl) and BSA (100 μg/ml) for 30 min at room temperature. Protein–RNA complexes were resolved by native gel electrophoresis and analyzed with a Fuji-BAS 2000 phosphorimager (Tokyo, Japan).
CBF-A gene silencing
Rat hippocampal neurons (2 or 5 DIV) were transfected with 50 nM predesigned siGENOME smart pool (Dharmacon, now Thermo Scientific) against the hnRNP CBFA or control siRNA against EGFP (for the sequences, see Raju et al., 2008) using the Lipofectamine RNAiMAX reagent according to the manufacturer's protocols (Invitrogen). Transfection was allowed for 72–96 h. Silencing was monitored by immunofluorescence staining with anti–CBF-A antibodies and confocal microscopy as mentioned earlier in the text. CBF-A mRNA levels were monitored by qRT- PCR (forward primer 5′ CCG AACACTGGTCGATCA AGAG 3′, reverse primer 5′ ACACGACCATCCAGTCTGTGCT 3′) (also see next section).
qRT-PCR
For analysis of steady-state mRNA levels, total RNA was extracted from hippocampal neurons treated with NMDA, AMPA, APV, or CNQX and from hippocampal neurons subjected to CBF-A gene silencing (see preceding section). In all cases total RNA was reverse-transcribed and the cDNA analyzed by qRT-PCR on an ABI 7500 (Applied Biosystems, Sweden) as recently described (Sahlgren et al., 2008). Individual amplification reactions were carried out using the Fast SYBR® Green Master Mix (Applied Biosystems) and specific primers against Arc mRNA (forward primer, CCCTGCAGCCCAAGTTCAAG; reverse primer, GAAGGCTCAGCTGCCTGCTC), BDNF mRNA (forward primer, AGCTGAGCGTGTGTGACAGT; reverse primer, ACCCATGGGATTACACTTGG), CaMKIIα mRNA (forward primer, CCATCCTCACCACTATGCTG; reverse primer, ATCGATGAAAGTCCAGGCCC), and GAPDH mRNA (forward primer, GCATCCTGCACCACCAACTG; reverse primer, ACGCCACAGCTTTCCAGAGG). In all cases GAPDH is used as internal control, and all the values are normalized accordingly over five independent experiments.
Supplementary Material
Acknowledgments
We thank Carlos Ibanez for comments and Malte Wittman, Maurice Perrinjaquet, Annalisa Vicario, and Debashish Das for help with rat embryos and qRT-PCR. We also thank Gema Martínez and Nieves Hernández for EM cryo-immunogold. This work was supported by grants from the Swedish Research Council (Vetenskapsrådet) and the Swedish Cancer Society (Cancerfonden) to P.P. and N.V. N.F. is supported by a postdoctoral grant from the Wenner-Gren Foundations, Sweden.
Abbreviations used:
- A2RE
hnRNP A2 response element
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- APV
(2R)-amino-5-phosphonovaleric acid
- Arc
activity-regulated cytoskeleton-associated protein
- BDNF
brain-derived neurotrophic factor
- BSA
bovine serum albumin
- CaMKIIα
calmodulin-dependent protein kinase II
- CBF-A
CArG Box binding Factor A
- CNQX
6-cyano-7-nitroquinoxaline-2,3-dione
- DIV
days after in vitro
- EGFP
enhanced green fluorescent protein
- EMSA
electrophoretic mobility shift assay
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IEM
immuno-electron microcopy
- Ig
immunoglobulin
- immuno-FISH
immunofluorescence in situ hybridization
- MBP
myelin basic protein
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PSD95
postsynaptic density-95
- qRT-PCR
quantitative real-time PCR
- RNAi
RNA interference
- RTS
RNA trafficking sequence
- scr
scrambled
- UTR
untranslated region
- wt
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0904) on April 6, 2011.
REFERENCES
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, et al. Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3’ UTR. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Carson JH, Barbarese E. Systems analysis of RNA trafficking in neural cells. Biol Cell. 2005;97:51–62. doi: 10.1042/BC20040083. [DOI] [PubMed] [Google Scholar]
- Carson JH, Cui H, Krueger W, Schlepchenko B, Brumwell C, Barbarese E. RNA trafficking in oligodendrocytes. Results Probl Cell Differ. 2001;34:69–81. doi: 10.1007/978-3-540-40025-7_5. [DOI] [PubMed] [Google Scholar]
- Chua JJE, Kindler S, Boyken J, Jahn R. The architecture of an excitatory synapse. J Cell Sci. 2010;123:819–823. doi: 10.1242/jcs.052696. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Daneholt B. Assembly and transport of a pre-messenger RNP particle. Proc Natl Acad Sci USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JLE, Sully G, Wait R, Rawlinson L, Clark AR, Saklatvala J. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem J. 2002;366:709–719. doi: 10.1042/BJ20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messenger they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;4:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond S-J, Serban A, Prusiner SB, Peters PJ. Cryo-immunogold electron microscopy for prions: Toward identification of a conversion site. J Neurosci. 2008;28:12489–12499. doi: 10.1523/JNEUROSCI.4474-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Paratcha G, Sandoval-Guzmán T, Ibáñez CF. GDNF and GFRα1 promote formation of neuronal synapses by ligand-induced cell adhesion. Nat Neurosci. 2007;10:293–300. doi: 10.1038/nn1855. [DOI] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Ma ASW, Moran-Jones K, Shan J, Munro TP, Snee MJ, Hoek KS, Smith R. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem. 2002;277:18010–18020. doi: 10.1074/jbc.M200050200. [DOI] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIa impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Jonsson A, Nashchekin D, Karlsson C, Bergman T, Guialis A, Daneholt B. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 2002;30:1725–1734. doi: 10.1093/nar/30.8.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Raju C, Fukuda N. Actin-associated hnRNP proteins as transacting factors in the control of mRNA transport and localization. RNA Biol. 2009;6:171–174. doi: 10.4161/rna.6.2.8195. [DOI] [PubMed] [Google Scholar]
- Raju CS, Goritz C, Nord Y, Hermanson O, Lopez-Iglesias C, Visa N, Castelo-Branco G, Percipalle P. In Cultured Oligodendrocytes the A/B-type hnRNP CBF-A Accompanies MBP mRNA Bound to mRNA Trafficking Sequences. Mol Biol Cell. 2008;19:3008–3019. doi: 10.1091/mbc.E07-10-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Cremer C, Cremer T, Fakan S. Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol. 2010;282:1–90. doi: 10.1016/S1937-6448(10)82001-5. [DOI] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tian L, Stefanidakis M, Ning L, Van Lint P, Nyman-Huttunen H, Libert C, Itohara S, Mishina M, Rauvala H, Gahmberg CG. Activation of NMDA receptors promotes dendritic spine development through MMP-mediated ICAM-5 cleavage. J Cell Biol. 2007;178:687–700. doi: 10.1083/jcb.200612097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- White R, Gonsior C, Krämer-Albers EM, Stöhr N, Hüttelmaier S, Trotter J. Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J Cell Biol. 2008;181:579–586. doi: 10.1083/jcb.200706164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Martinez SJ, Haber M, Jones EV, Bouvier D, Doucet G, Corera AT, Fon EA, Zisch AH, Murai KK. EphA4 signaling regulates phospholipase Cgamma1 activation, cofilin membrane association, and dendritic spine morphology. J Neurosci. 2007;27:5127–5138. doi: 10.1523/JNEUROSCI.1170-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.