DHHC2 regulates dynamic palmitoylation of PSD95, a process that appears to be linked to changes in DHHC2 localization. The precise distribution and membrane dynamics of DHHC2 are not clear. We demonstrate that DHHC2 cycles between the plasma membrane and recycling endosomes (RE) and that this dynamic localization requires an intact C-terminal domain.
Abstract
Intracellular palmitoylation dynamics are regulated by a large family of DHHC (Asp-His-His-Cys) palmitoyl transferases. The majority of DHHC proteins associate with endoplasmic reticulum (ER) or Golgi membranes, but an interesting exception is DHHC2, which localizes to dendritic vesicles of unknown origin in neurons, where it regulates dynamic palmitoylation of PSD95. Dendritic targeting of newly synthesized PSD95 is likely preceded by palmitoylation on Golgi membranes by DHHC3 and/or DHHC15. The precise intracellular distribution of DHHC2 is presently unclear, and there is very little known in general about how DHHC proteins achieve their respective localizations. In this study, membrane targeting of DHHC2 in live and fixed neuroendocrine cells was investigated and mutational analysis employed to define regions of DHHC2 that regulate targeting. We report that DHHC2 associates with the plasma membrane, Rab11-positive recycling endosomes, and vesicular structures. Plasma membrane integration of DHHC2 was confirmed by labeling of an extrafacial HA epitope in nonpermeabilized cells. Antibody-uptake experiments suggested that DHHC2 traffics between the plasma membrane and intracellular membranes. This dynamic localization was confirmed using fluorescence recovery after photo-bleaching analysis, which revealed constitutive refilling of the recycling endosome (RE) pool of DHHC2. The cytoplasmic C-terminus of DHHC2 regulates membrane targeting and a mutant lacking this domain was associated with the ER. Although DHHC2 is closely related to DHHC15, these proteins populate distinct membrane compartments. Construction of chimeric DHHC2/DHHC15 proteins revealed that this difference in localization is a consequence of divergent sequences within their C-terminal tails. This study is the first to highlight dynamic cycling of a mammalian DHHC protein between clearly defined membrane compartments, and to identify domains that specify membrane targeting of this protein family.
INTRODUCTION
S-palmitoylation, the attachment of long-chain fatty acids (predominantly palmitate, C16:0) onto cysteine residues, is a posttranslational modification occurring on a diverse array of cellular proteins (Kang et al., 2008). Our understanding of the cell biology of palmitoylation is advancing rapidly, with research on distinct cells and systems converging to highlight key regulatory mechanisms attributable to this lipid modification (for reviews, see Greaves and Chamberlain, 2007; Linder and Deschenes, 2007; Greaves et al., 2009; Fukata and Fukata, 2010; Salaun et al., 2010). Protein palmitoylation has been shown to regulate 1) membrane binding; 2) the precise membrane micro-localization of proteins; 3) protein traffic and sorting; 4) protein–protein interactions; and 5) protein stability.
Protein palmitoylation is a reversible process and many proteins undergo constitutive or regulated changes in palmitoylation status. A small number of depalmitoylating enzymes have been identified, although the contribution made by these proteins to cellular palmitoylation dynamics is not clear (Duncan and Gilman, 1998; Dekker et al., 2010). In contrast to this sketchy understanding of depalmitoylation, recent studies identified a family of 23 mammalian DHHC (Asp-His-His-Cys) palmitoyl transferases (Fukata et al., 2004; Huang et al., 2004; Keller et al., 2004). Genetic analyses in yeast have found that DHHC proteins are responsible for the bulk of palmitoylation reactions that occur in cells (Roth et al., 2006). The defining feature of DHHC proteins is the presence of a 51 amino acid cysteine-rich domain, which includes a largely invariant DHHC motif (Putilina et al., 1999; Lobo et al., 2002; Roth et al., 2002). The DHHC domain is critical for palmitoyl transferase activity, and is present within a cytoplasmic loop of these polytopic membrane proteins (Mitchell et al., 2006). The palmitoylation reaction appears to involve a two-step process that proceeds via a DHHC-palmitate intermediate (Mitchell et al., 2010), and the integrity of the DHHC motif is also important for autopalmitoylation of DHHC proteins (Greaves and Chamberlain, 2011).
Recent studies have shown that many peripheral proteins undergo dynamic cycling between the Golgi and the plasma membrane, which is regulated by palmitoylation–depalmitoylation dynamics (Goodwin et al., 2005; Rocks et al., 2005, 2010). Palmitoylation of these proteins is proposed to occur exclusively at the Golgi, facilitating stable membrane interaction and traffic to post-Golgi compartments. Subsequent depalmitoylation results in membrane release and cytosolic exchange, and is followed by repalmitoylation at the Golgi. Thus it is proposed that Golgi DHHC proteins play a major function in directing anterograde traffic of peripheral palmitoylated proteins. In contrast to the central function performed by Golgi DHHC proteins in palmitoylation of peripheral membrane proteins, far less is known about the role of specific DHHC proteins in regulating palmitoylation dynamics at post-Golgi compartments.
A recent study reported that DHHC2 localizes to mobile vesicles in dendrites, which respond to changes in synaptic activity by translocating toward the plasma membrane, regulating dynamic palmitoylation of the postsynaptic scaffold protein PSD-95 (Noritake et al., 2009). However, the identity and nature of these vesicles was not determined, and it is not clear whether DHHC2 remains associated with vesicles or if these vesicles undergo fusion with the plasma membrane; fusion would result in integration of DHHC2 into the plasma membrane. The delivery of newly synthesized PSD-95 to dendritic regions likely requires prior palmitoylation on Golgi membranes, which would facilitate stable membrane binding and synaptic targeting. PSD-95 is a substrate for Golgi enzymes, including DHHC3, DHHC7, and DHHC15 (Fukata et al., 2004). DHHC3 was reported to contribute to constitutive palmitoylation of PSD-95 in hippocampal neurons (Noritake et al., 2009). In neuroendocrine PC12 cells, DHHC2 decorates the plasma membrane and an unidentified intracellular compartment visualized by confocal imaging (Greaves et al., 2010); possible targets of DHHC2 in this cell type include the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) membrane fusion synaptosomal-associated proteins (SNAP) SNAP25 and SNAP23 (Greaves et al., 2010). To fully delineate the functions of DHHC2 and to understand regulated trafficking of this protein, it is essential to characterize the membrane interaction dynamics of this protein. Furthermore, understanding how DHHC2 localization is achieved is an important step toward uncovering the mechanisms that define intracellular patterning of the DHHC protein family. These questions are particularly relevant, as recent work suggested that DHHC2 is one of the major DHHC isoforms expressed in neuroendocrine cells and in mouse brain at the mRNA level (Jia et al., 2011).
RESULTS
DHHC2-EGFP localizes to the plasma membrane, recycling endosomes, and vesicles
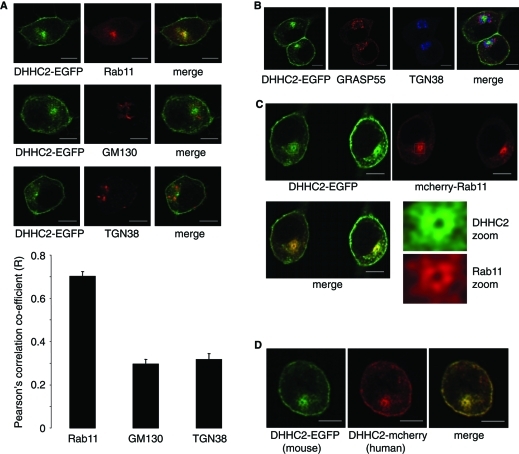
We recently reported that murine DHHC2 expressed in PC12 cells associates with the plasma membrane and an unidentified intracellular compartment (Greaves et al., 2010). This localization was unaffected by the size (hemagglutinin [HA] or enhanced green fluorescent protein [EGFP]) or the location (N- or C-terminal) of the tag. To identify the compartment(s) with which DHHC2 associates, we examined codistribution with markers of recycling endosomes (Rab11), Golgi (GM130), and trans-Golgi network (TGN38) (Figure 1A). Clear overlap of DHHC2 was observed with Rab11 but not with either GM130 or TGN38; this was confirmed by quantitative analysis of fluorescence intensity covariance (Figure 1A). Triple labeling experiments also revealed the physical separation of DHHC2-EGFP from both Golgi (Grasp55) and trans-Golgi (TGN38) membranes (Figure 1B), whereas cotransfection with mCherry-Rab11 highlighted association of DHHC2-EGFP with recycling endosomes (Figure 1C). In addition to the plasma membrane and recycling endosomes, DHHC2 was also associated with vesicular structures in the cytoplasm. Human DHHC2 displayed the same localization as murine DHHC2 (Figure 1D).
FIGURE 1:
Intracellular localization of DHHC2-EGFP in PC12 cells. (A) PC12 cells transfected with DHHC2-EGFP were labeled with antibodies against Rab11, GM130, or TGN38, and then with Alexa Fluor 633–conjugated secondary antibodies. Representative images are shown in the top three panels. Pearson's r values denoting covariance of fluorescence signals were calculated using Image J software and results show average values ± SEM (n = 6 or 7 cells for each condition). The correlation coefficient for DHHC2-EGFP and Rab11 was significantly greater than values obtained for DHHC2-EGFP against GM130 and TGN38 (p < 0.001, one-way analysis of variance [ANOVA]). (B) PC12 cells transfected with DHHC2-EGFP were labeled with antibodies against Grasp55 (sheep) and TGN38 (mouse), and then with anti-sheep(543)and anti–mouse(633) secondary antibodies. (C) PC12 cells were cotransfected with DHHC2-EGFP and mCherry-Rab11 and representative confocal images are shown. (D) Localization of mouse DHHC2-EGFP and human DHHC2-mCherry cotransfected in PC12 cells. Scale bars on all figure panels represent 5 μm.
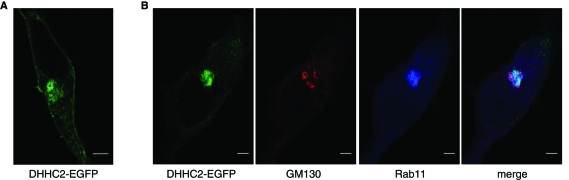
To determine whether the plasma membrane/endosomal localization of DHHC2 in PC12 cells is also observed in other cell types, DHHC2-EGFP was transfected into HEK293 cells. Figure 2A shows that DHHC2 associated with the plasma membrane and an intracellular compartment in HEK293 cells. Staining of the Golgi (GM130) and recycling endosomes (Rab11) revealed that DHHC2 was indeed associated with a Rab11-positive compartment in this cell type (Figure 2B).
FIGURE 2:
Intracellular localization of DHHC2-EGFP in HEK293 cells. (A) Representative image showing association of DHHC2-EGFP with the plasma membrane and an intracellular compartment in HEK293 cells. (B) HEK293 cells transfected with DHHC2-EGFP were labeled with mouse GM130 and rabbit Rab11 antibodies and subsequently with anti-mouse(543) and anti-rabbit(633). Scale bars on all figure panels represent 5 μm.
Plasma membrane integration of DHHC2
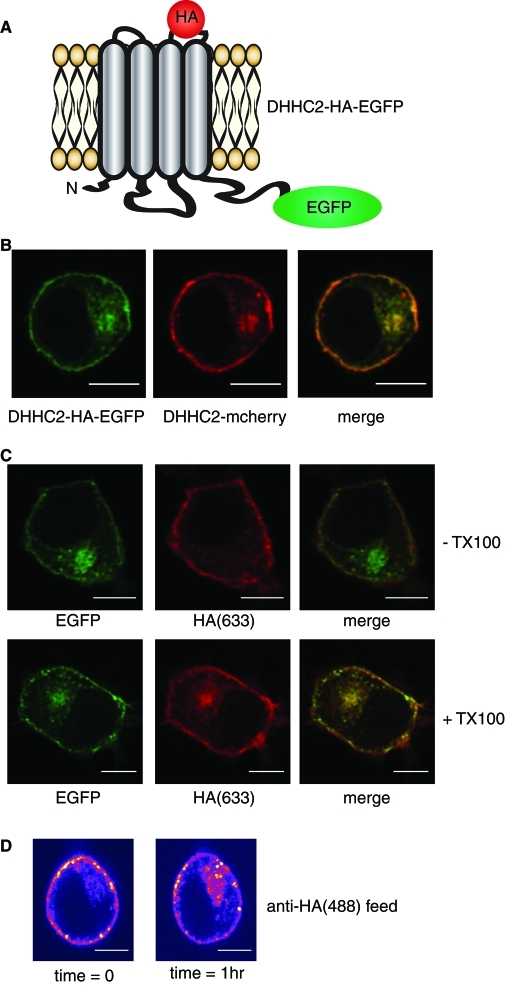
To ensure that the plasma membrane signal detected for DHHC2-EGFP reflected the true integration of DHHC2 into this membrane, we created a DHHC2 construct with an HA epitope inserted into the predicted extrafacial loop between transmembrane helices 3 and 4 (Figure 3A). The presence of the HA epitope did not affect the localization of DHHC2-EGFP as judged by colocalization with DHHC2-mCherry (Figure 3B). If DHHC2 is properly inserted into the plasma membrane, the HA epitope should be accessible to antibody added to intact cells. Thus cells expressing DHHC2-HA-EGFP were fixed and incubated in the presence or absence of 0.2% Triton X-100, and then incubated with a monoclonal HA antibody. In nonpermeabilized cells, the HA antibody decorated the plasma membrane but was absent from the recycling endosome (RE) compartment (Figure 3C, Top). However, in permeabilized cells, the antibody labeled both the plasma membrane and recycling endosomes (Figure 3C, Bottom). This analysis clearly shows that the HA epitope of plasma membrane-associated DHHC2 is accessible on nonpermeabilized cells and, therefore, that the protein is integrated into the plasma membrane. In addition, this result provides experimental support to the predicted membrane topology for DHHC2 that is illustrated inFigure 3A.
FIGURE 3:
Integration of DHHC2 into the plasma membrane and evidence of recycling. (A) Illustration of the predicted membrane topology of DHHC2 at the plasma membrane, highlighting the position of the inserted HA epitope and EGFP fluorescent tag. (B) Comparison of the intracellular localization of DHHC2-HA-EGFP and DHHC2-mCherry constructs cotransfected into PC12 cells. (C) PC12 cells transfected with the DHHC2-HA-EGFP construct were fixed and incubated in the absence (− TX100) or presence (+ TX100) of Triton X-100 for 6 min. The cells were then incubated with a mouse monoclonal HA antibody and subsequently with anti-mouse(633). (D) PC12 cells expressing DHHC2-HA-mCherry were incubated with anti-HA conjugated to Alexa Fluor 488 for 30 min at room temperature. The cells were then imaged directly, incubated on the microscope stage at 37°C for a further 60 min, and imaged again using identical settings. Scale bars on all figure panels represent 5 μm.
Membrane cycling of DHHC2
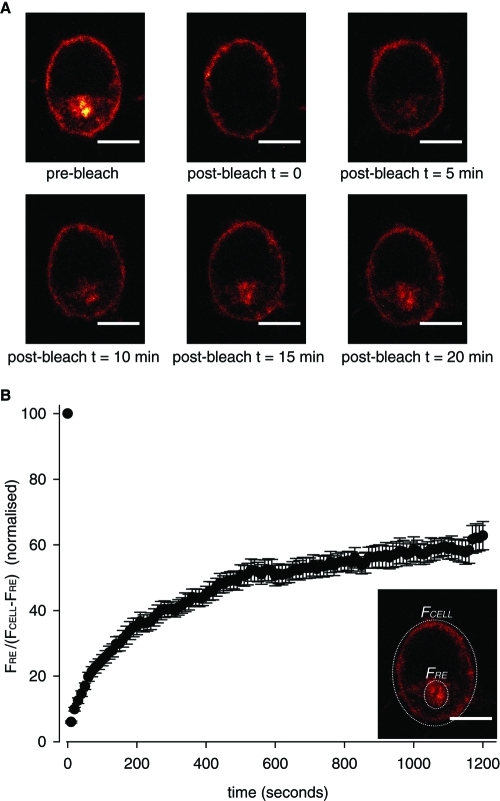
When live PC12 cells expressing DHHC2-HA-mCherry were incubated with an Alexa Fluor 488–conjugated HA antibody, we observed a time-dependent accumulation of the antibody on intracellular membranes (Figure 3D), suggesting that DHHC2 might cycle between the plasma membrane and endosomes. To further investigate whether the localization of DHHC2 is dynamically regulated, we employed fluorescence recovery after photo-bleaching (FRAP). For FRAP analysis, the intracellular fluorescence was selectively photo-bleached, and fluorescence recovery of the RE compartment was monitored over time (Figure 4). Measuring the ratio of fluorescence of the RE pool (FRE) against the remainder of the total cellular fluorescence (FCELL − FRE) revealed that the RE pool of DHHC2-EGFP recovered from 6 ± 0.3% to 62.8 ± 4.3% of the starting fluorescence intensity value within 20 min following photo-bleach. This analysis shows the localization of DHHC2-EGFP is dynamic, and there most likely is cycling of the protein between the plasma membrane and endosomes.
FIGURE 4:
Evidence for plasma membrane-to-endosome cycling of DHHC2-EGFP. (A) DHHC2-EGFP fluorescence present within the cytosolic space was photo-bleached as described in Materials and Methods. Top, comparison of the left and middle image panels shows the fluorescence signal that was photo-bleached. Fluorescence recovery was monitored by acquiring images every 10 s after photo-bleaching, and representative images are shown prebleach, immediately following photo-bleaching (t = 0) and after 5, 10, 15, and 20 min of recovery. Scale bars represent 5 μm. (B) The fluorescence intensities of the RE compartment (FRE) and the whole cell (FCELL) were calculated using Image J software, and presented as a normalized ratio of FRE/(FCELL − FRE) (n = 10 cells). The figure inset depicts example regions that would be selected for analysis.
Intracellular localization of DHHC2 is regulated by a C-terminal domain
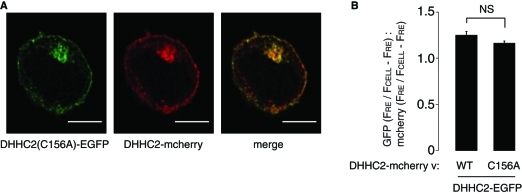
There has been little previous analysis of the targeting features that govern DHHC protein localization. Given our reported localization of DHHC2 to a dynamic membrane cycling pathway in PC12 cells, we next investigated how targeting to this pathway is achieved. Palmitoylation of many DHHC substrate proteins regulates their intracellular targeting. Thus, as a starting point, we examined the possibility that intramolecular autopalmitoylation of DHHC2 might be important for targeting to the plasma membrane and recycling endosomes. To test this, we examined a DHHC2-EGFP construct containing a substitution of the cysteine in the DHHC domain with an alanine (C156A); mutation of the DHHC motif in this manner blocks autopalmitoylation of DHHC proteins. Our previous work showed that mutation of the DHHC motif does not have a gross effect on DHHC2 localization (Greaves et al., 2010). To detect more subtle changes in the localization of inactive DHHC2, the DHHC2(C156A)-EGFP construct was cotransfected into PC12 cells with wild-type DHHC2-mCherry. The representative image shown in Figure 5 demonstrates that the localization of wild-type and C156A mutant DHHC2 are similar, suggesting that intramolecular autopalmitoylation of DHHC2 is not having a major impact on targeting of the protein. To provide a quantitative comparison of wild-type and C156A mutant targeting to the RE compartment, z projections were generated from image stacks of cells expressing either wild-type or C156A DHHC2-EGFP together with wild-type DHHC2-mCherry. The fluorescence intensity of the RE compartment (FRE) was then expressed as a ratio against the remaining cellular fluorescence (FCELL − FRE) for both EGFP and mCherry fluorescence in the same cell. The graph presented in Figure 5 demonstrates that there was no significant difference in targeting of DHHC2-EGFP wild-type or C156A mutant to the RE compartment compared with DHHC2-mCherry.
FIGURE 5:
Effect of mutation of the DHHC motif on localization of DHHC2. (A) DHHC2-mCherry was cotransfected into PC12 cells with either DHHC2-EGFP (not depicted) or with DHHC2-EGFP containing a C156A mutation (C156 is the cysteine of the DHHC motif). Scale bars represent 5 µm. (B) The fluorescence intensity values of the RE compartment and of the whole cell were calculated as described in Materials and Methods for both EGFP and mCherry. The graph shows the mean ratio ± SEM of endosomal fluorescence of the EGFP-tagged constructs relative to DHHC2-mCherry (n = 7 cells for DHHC2-EGFP, and 5 cells for DHHC2(C156A)-EGFP). A Student's t test revealed no significant difference between RE targeting of wild-type and mutant DHHC2.
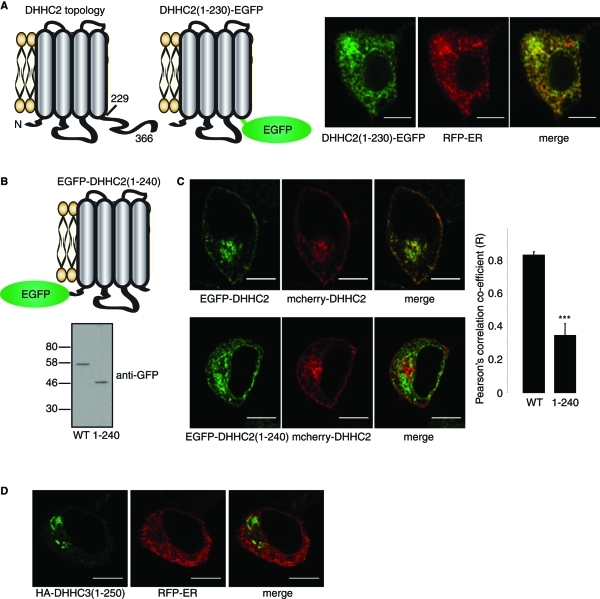
DHHC2 is predicted to have a short 16 amino acid N-terminal domain, a large intracellular loop containing the DHHC domain, and a relatively long cytoplasmic C-terminus (residues 229–366 for mouse DHHC2) (Figure 6A). As palmitoylation did not appear to regulate intracellular targeting of DHHC2, we next examined whether the large C-terminal domain played a role in DHHC2 targeting. Replacing the entire C-terminal domain of DHHC2 with EGFP (DHHC2(1–230)-EGFP) led to a redistribution of the protein to a compartment that overlapped with the endoplasmic reticulum (ER) marker, RFP-ER (Figure 6A). To confirm that this redistribution was a result of removal of the C-terminal domain and not caused by the EGFP tag, we constructed an additional mutant with an N-terminal EGFP tag (EGFP-DHHC2(1–240)); this mutant also contained a short amino acid sequence (residues 230–240) downstream of the transmembrane domain (TMD) to limit any disruption of the membrane interaction of the TMD (Figure 6B). This truncation mutant expressed at similar levels to full-length DHHC2 and did not show signs of aggregation (Figure 6B), suggesting that the protein is folded and stable. The EGFP-DHHC2(1–240) mutant displayed a similar ER-like localization as the mutant with a C-terminal EGFP tag, and its distribution was quantifiably distinct from full-length mCherry-DHHC2 in cotransfection experiments (Figure 6C). Finally, we checked whether similar C-terminal truncations in other DHHC proteins also cause ER retention. Truncation of DHHC3 15 amino acids downstream of the final predicted TMD (DHHC3(1–250)) had no effect on the targeting of this protein to Golgi membranes and did not cause ER retention; thus an intact C-terminal domain is not required for ER exit of all DHHC proteins.
FIGURE 6:
The C-terminus of DHHC2 regulates intracellular targeting. (A) Schematic of DHHC2 membrane topology, and representation of the topology of a DHHC2(1–230)-EGFP construct. Numbers represent amino acid positions. The images show PC12 cells cotransfected with DHHC2(1–230)-EGFP and RFP-ER. (B) Schematic of the membrane topology of an EGFP-DHHC2(1–240) construct (Top). EGFP-DHHC2 and EGFP-DHHC2(1–240) were transfected into HEK293 cells. Lysates prepared from transfected cells were resolved by SDS–PAGE and analyzed by immunoblotting with a monoclonal GFP antibody (Bottom). Position of molecular weight markers (kDa) are indicated on the left. (C) EGFP-DHHC2 (Left, Top) or EGFP-DHHC2(1–240) (Left, Bottom) were cotransfected into PC12 cells with mCherry-DHHC2. Pearson's r values showing covariance of the fluorescence intensity of GFP-tagged constructs relative to mCherry-DHHC2 were calculated using Image J software and results show mean values ± SEM (n = 5 cells for each construct). The data were analyzed using a Student's t test, which revealed a significant difference between DHHC2 and DHHC2(1–240) (*** denotes p < 0.001). (D) Confocal images showing the localizations of HA-DHHC3(1–250) stained with HA antibody and a mouse secondary antibody conjugated to Alexa Fluor 488, and RFP-ER. Scale bars on all images represent 5 μm.
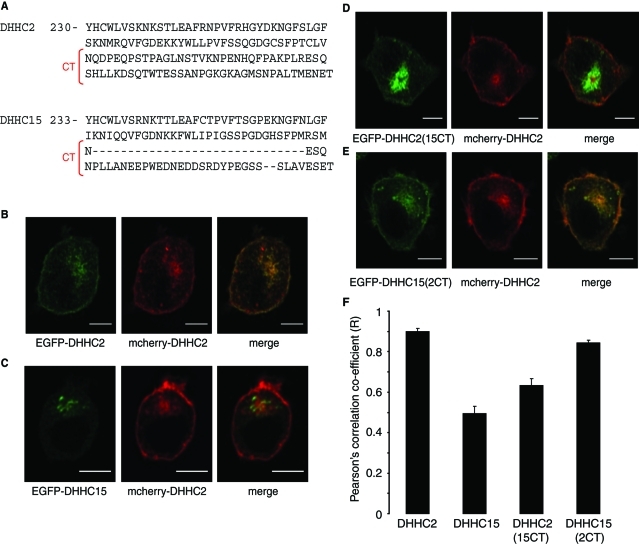
DHHC2 shares high sequence identity with DHHC15, which is nevertheless localized to Golgi-like membranes (Greaves et al., 2008). It is interesting to note that despite the similarity of DHHC2 and DHHC15, the protein sequences diverge in the final half of the C-terminal tail (Figure 7A). We therefore reasoned that the distinct localizations of DHHC2 and DHHC15 might be due to these divergent regions in the C-termini of the proteins. To test this idea, we swapped the C-terminal regions of DHHC2 and DHHC15 downstream of amino acid N298 in DHHC2 and N301 in DHHC15 (as highlighted in Figure 7A), generating DHHC2(15CT) and DHHC15(2CT) mutants. All proteins examined in this experiment had the EGFP tag placed at the N-terminal side of DHHC2/DHHC15, and the localizations of these proteins were compared to mCherry-DHHC2 expressed in the same cell. As expected, EGFP-DHHC2 and mCherry-DHHC2 displayed a similar distribution (Figure 7B), whereas EGFP-DHHC15 was clearly present on distinct intracellular membranes from DHHC2 and largely absent from the plasma membrane (Figure 7C). Interestingly, the DHHC2(15CT) mutant displayed an intracellular localization that was distinct from mCherry-DHHC2 (Figure 7D). Conversely, adding the C-terminal region of DHHC2 onto DHHC15 resulted in a localization of DHHC15(2CT) that was similar to DHHC2 (Figure 7E). These results imply that the distinct C-terminal domains of DHHC2 and DHHC15 are responsible for the different intracellular localizations of these two related DHHC proteins. The observed changes in localization of the chimeric proteins were confirmed by calculating Pearson's correlation coefficient values from multiple transfected cells (Figure 7F).
FIGURE 7:
Analysis of the intracellular localizations of DHHC2 and DHHC15 chimeric proteins. (A) Comparison of the amino acid sequences of the C-terminal domains of DHHC2 and DHHC15. The regions of DHHC2 and DHHC15 that were swapped to make the DHHC2(15CT) and DHHC15(2CT) chimeras are highlighted (CT). PC12 cells were cotransfected with mCherry-DHHC2 together with EGFP-DHHC2 (B), EGFP-DHHC15 (C), EGFP-DHHC2(15CT) (D), or EGFP-DHHC15(2CT) (E). F) Pearson's r values showing covariance of fluorescence intensities of the EGFP-tagged constructs relative to mCherry-DHHC2 were calculated using Image J software and results show mean values ± SEM (n = 4–6 cells for each construct). The data were analyzed using a one-way ANOVA, which revealed significant differences between DHHC2 and DHHC15 (p < 0.001), DHHC2 and DHHC2(15CT) (p < 0.001), DHHC15 and DHHC15(2CT) (p < 0.001), DHHC15 and DHHC2(15CT) (p = 0.015), and DHHC2(15CT) and DHHC15(2CT) (p = 0.001). Scale bars on images represent 5 μm.
DISCUSSION
The majority of active mammalian DHHC proteins characterized to date are localized to Golgi membranes (see Fernandez-Hernando et al., 2006; Greaves et al., 2008, 2010). Indeed, a recent study highlighted the Golgi as a super-reaction center for palmitoylation of peripheral membrane proteins (Rocks et al., 2010). Thus, following synthesis, such proteins are palmitoylated at the Golgi, leading to stable membrane association and facilitating transport to more distal membrane compartments. Palmitoylation is a reversible process, with many proteins undergoing rapid and continuous cycles of palmitoylation and depalmitoylation. For proteins such as H- and N-Ras, dynamic palmitoylation is thought to involve cytosolic diffusion and repalmitoylation on Golgi membranes (Goodwin et al., 2005; Rocks et al., 2005). However, many proteins may not undergo complete depalmitoylation (and hence cytosolic exchange) and dynamic palmitoylation of these proteins might be regulated by DHHC proteins beyond the confines of the Golgi complex. Two mammalian DHHC proteins that display post-Golgi sorting have been characterized: DHHC5 associates with the plasma membrane and the postsynaptic density in neurons (Ohno et al., 2006; Li et al., 2010), whereas DHHC2 is present on mobile vesicles in the dendrites of hippocampal neurons and at the plasma membrane in neuroendocrine PC12 cells (Noritake et al., 2009; Greaves et al., 2010). DHHC2 substrates include PSD95, and DHHC2 depletion leads to a loss of activity-dependent changes in palmitoylation of this protein. DHHC2 is thought to play a specific role in dynamic palmitoylation of PSD95 occurring at synaptic regions, whereas synaptic targeting of freshly synthesized PSD95 may be regulated by DHHC3 present on the somatic Golgi (Noritake et al., 2009). SNAP25 is also a substrate of DHHC2 in HEK293T cells (Greaves et al., 2010), although if and how DHHC2 contributes to palmitoylation dynamics of endogenous SNAP25 is not currently known.
A key finding to emerge from the analysis of DHHC2 regulation of PSD95 is that DHHC2 activity toward this substrate is affected by changes in membrane trafficking of the DHHC protein rather than changes in intrinsic activation (Noritake et al., 2009). Because of this, it is essential to delineate the precise steady-state and dynamic localizations of DHHC2. The observation that DHHC2 cycles between the plasma membrane and endosomes is of particular relevance to the observed movement of DHHC2 in hippocampal neurons (Noritake et al., 2009). Total internal reflection fluorescence (TIRF) microscopy analysis revealed that DHHC2 vesicles move closer to the membrane following synaptic blockade. However, it was not clear whether this increased fluorescence reflected insertion of DHHC2 into the plasma membrane or if vesicles were just accumulating closer to the cytosolic face of the membrane. The results of this present study suggest that the observed increase of DHHC2 at the membrane is likely to arise due to increased insertion of the protein into the plasma membrane, where it can effectively regulate PSD95 palmitoylation dynamics. It is particularly interesting that DHHC2 localized to Rab11-positive recycling endosomes, as this compartment has previously been reported to supply α-amino-3-hydroxy-5-methyl-4- isoxazole-propionic acid (AMPA) receptors during long-term potentiation (Park et al., 2004). It is thus possible that neuronal DHHC2 cycles together with AMPA receptors between the dendritic plasma membrane and endosomes.
The functions of DHHC2 are not restricted to neuronal cells, and this protein has also been shown to regulate tetraspanins and cytoskeleton-associated protein 4 (Sharma et al., 2008; Planey et al., 2009). Furthermore, a previous study examining palmitoyl transferase activity against the cation-dependent mannose-6-phosphate receptor concluded that the protein that palmitoylates this particular substrate is cycling between the plasma membrane and endosomes (Stockli and Rohrer, 2004). Thus, given the results reported in the current study, we believe that this activity might represent DHHC2, and this possibility clearly merits further investigation.
Previous studies examining DHHC2 localization suggested that the protein is associated with Golgi membranes (Ohno et al., 2006; Sharma et al., 2008). However, it is important that proteins present in a pericentriolar localization are thoroughly examined using quantitative colocalization with markers of multiple intracellular compartments. This is particularly true when defining localizations with Golgi, trans-Golgi network (TGN), and recycling endosomes, as these compartments are all present in a similar intracellular location. Indeed, a recent study claimed that the major steady-state localization of H- and N-Ras is on recycling endosomes (Misaki et al., 2010) and not on Golgi membranes, as was previously reported.
There is no information available on how mammalian DHHC proteins target their respective intracellular compartments, and this study is the first to highlight trafficking signals in this class of protein. Intriguingly, the C-terminal 68 amino acids of DHHC2 and the corresponding (albeit shorter) region of DHHC15 were shown to play an important role in defining the intracellular localizations of these proteins. It will be interesting to determine whether the C-terminus of DHHC15 contains Golgi retention signals and/or if the corresponding region of DHHC2 contains active information for targeting to the plasma membrane and recycling endosomes. Work is currently ongoing in our laboratory to more precisely define the targeting signals present within this region of DHHC2/DHHC15 and in other members of the DHHC protein family.
MATERIALS AND METHODS
Cell culture and transfection
PC12 cells were maintained in RPMI 1640 medium (Life Technologies) containing 10% horse serum and 5% fetal calf serum (FCS). HEK293 cells were maintained in DMEM supplemented with 10% FCS. Both cell lines were cultured at 37°C in a humidified atmosphere containing 5% (HEK293) or 7.5% (PC12) CO2. Cells growing on poly-d-lysine-coated glass coverslips were transfected with 0.5 μg of each plasmid using Lipofectamine 2000 (Invitrogen, Paisley, UK).
Plasmids
Mouse DHHC2 and DHHC15 were amplified from previously described constructs (Fukata et al., 2004) and cloned into pEGFPC2 or pEGFPN1 vectors between HindIII and Sal1 sites (Clontech, Oxford, UK). DHHC2(1–240) was generated by inserting a stop codon at the appropriate position in EGFP-DHHC2 using site-directed mutagenesis. Human DHHC2 was amplified from HEK293 cell cDNA using the following primers: GATCAAGCTTATGGCGCCCTCGGGCCCGGG and GATCGTCGACGCAGTCTCATTTTCCATGGTTAATGCAG, and cloned into HindIII and Sal1 restriction sites upstream of an mCherry tag in the pEGFPN1 vector backbone. The DHHC2-HA-EGFP vector was made by inserting an HA epitope (KYPYNVPNYA) between amino acids F196 and W197 of DHHC2-EGFPN1 using site-directed mutagenesis. The DHHC2(15CT) and DHHC15(2CT) chimeras were generated within the HA-tagged constructs as follows: site-directed mutagenesis was used to insert Nhe1 and Sal1 restriction sites at the boundaries of the domains that were swapped (upstream of N298 in DHHC2 and N301 in DHHC15, and downstream of the stop codon of each DHHC protein). These regions were then swapped by restriction/ligation, and the Nhe1/Sal1 restriction sites removed using site-directed mutagenesis. The chimeric constructs were amplified by PCR and cloned into pEGFPC2 between HindIII and Sal1 restriction sites. All mCherry-tagged constructs were generated by excising EGFP using Age1 and BsrG1 restriction enzymes and replacing with mCherry. A GFP-Rab11 construct kindly provided by Giampietro Schiavo (Cancer Research UK London Research Institute) was used to generate mCherry-Rab11. RFP-ER was from Clontech (San Diego, CA).
Antibodies
Francis Barr (Cancer Research Centre, University of Liverpool, United Kingdom) kindly provided a sheep Grasp55 antibody. Monoclonal HA antibody (16B12) was purchased from Covance (Emeryville, CA). Monoclonal anti-HA (16B12) conjugated to Alex Fluor 488, rabbit antibody–recognizing Rab11, and Alexa Fluor–conjugated secondary antibodies were purchased from Invitrogen. Monoclonal TGN38 and GM130 antibodies were from Becton Dickinson (Oxford, UK). The GFP antibody (JL8) used for immunoblotting was from Clontech.
Cell fixation, labeling, and confocal microscopy
Cells were prepared for confocal microscopy or directly analyzed ∼40 h posttransfection. For most experiments, cells were washed in phosphate-buffered saline (PBS) and fixed in 4% formaldehyde for 30 min at room temperature. For analysis of EGFP/mCherry fluorescence, the coverslips were mounted in Mowiol and dried overnight. For antibody staining, the cells were washed in PBS containing 0.3% bovine serum albumin (BSA) (PBSB) and then permeabilized in PBSB containing 0.25% Triton X-100 for 6 min at room temperature. The cells were washed and incubated with primary antibodies (1:50) in PBSB for 1 h and then with the appropriate Alexa Fluor–conjugated secondary antibodies (1:400) for 1 h. The specific antibodies that were used for each experiment are indicated in the figure legends. Imaging was performed on Zeiss Axiovert and Leica SP5 laser-scanning confocal microscopes. Image data were acquired at Nyquist sampling rates and deconvolved using Huygens software (Scientific Volume Imaging, Hilversum, The Netherlands). The data were analyzed using Image J software (http://imagej.nih.gov/ij/download.html).
For comparison of targeting of DHHC2-EGFP (wild-type of C156A mutant) and DHHC2-mCherry constructs to recycling endosomes, summed z projections were generated from image stacks and the fluorescence intensities of the RE compartment (FRE) and the whole cell (FCELL) were calculated from identical regions of interest for the EGFP and mCherry images. A value of FRE/(FCELL − FRE) was calculated for both EGFP and mCherry images and a ratio of [FRE/(FCELL − FRE)]EGFP:[FRE/(FCELL − FRE)]mCherry was calculated.
For FRAP analysis, live PC12 cells expressing DHHC2-EGFP were maintained at 37°C and 5% CO2. Regions of interest encompassing the fluorescent signal from the cell cytosol were selected and photo-bleached for 50 iterations using 100% laser power. The bleached cells were then imaged for 20 min at 10-s intervals. Image J software was used to calculate fluorescence intensities of the RE compartment (FRE) and whole cell (FCELL) for every time point. These values were then converted to a ratio of RE fluorescence against the remaining cell fluorescence by calculating FRE/(FCELL − FRE). The data were normalized by setting the prebleach value to 100, and all other values were expressed relative to this.
Antibody-uptake experiments
PC12 cells expressing DHHC2-HA-mCherry were incubated with HA-488 antibody for 30 min at room temperature. The cells were then washed and imaged directly; following incubation for 1 h at 37°C/5% CO2 on the microscope stage, cells were imaged again using identical microscope settings.
Acknowledgments
Confocal imaging was performed with the help of Trudi Gillespie at the IMPACT imaging facility at Edinburgh University and also at the CALM imaging facility headed by Rolly Wiegand. We thank Giampietro Schiavo for the gift of GFP-Rab11 and Francis Barr for the sheep Grasp55 antibody. Work in the authors’ laboratory is funded by an MRC senior fellowship award to LHC.
Abbreviations used:
- AMPA
α-amino-3-hydroxy-5-methyl-4- isoxazole-propionic acid
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- FCS
fetal calf serum
- FRAP
fluorescence recovery after photo-bleaching
- HA
hemagglutinin
- PBS
phosphate-buffered saline
- RE
recycling endosome
- RFP-ER
endoplasmic reticulum marker
- SNAP
synaptosomal-associated protein
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- TGN
trans-Golgi network
- TIRF
total internal reflection fluorescence
- TMD
transmembrane domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0924) on April 6, 2011.
REFERENCES
- Dekker FJ, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21RAS. J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS, Sessa WC. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J Cell Biol. 2006;174:369–377. doi: 10.1083/jcb.200601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting 10.1083/jcb.200610151. J Cell Biol. 2007;176:249–254. doi: 10.1083/jcb.200610151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Greaves J, Gorleku OA, Salaun C, Chamberlain LH. Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases. J Biol Chem. 2010;285:24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Prescott GR, Gorleku OA, Chamberlain LH. The fat controller: roles of palmitoylation in intracellular protein trafficking and targeting to membrane microdomains (review) Mol Membr Biol. 2009;26:67–79. doi: 10.1080/09687680802620351. [DOI] [PubMed] [Google Scholar]
- Greaves J, Salaun C, Fukata Y, Fukata M, Chamberlain LH. Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J Biol Chem. 2008;283:25014–25026. doi: 10.1074/jbc.M802140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Jia L, Linder ME, Blumer KJ. Gi/o signaling and the palmitoyltransferase DHHC2 regulate palmitate cycling and shuttling of RGS7-family binding protein. J Biol Chem. 2011;286:13695–13703. doi: 10.1074/jbc.M110.193763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The γ2 subunit of GABAA receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu J, Höfer K, Wong AMS, Cooper JD, Birnbaum SG, Hammer RE, Hofmann SL. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem. 2010;285:13022–13031. doi: 10.1074/jbc.M109.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, Matsuda M, Taguchi T. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–29. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Thematic review series: lipid posttranslational modifications. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- Noritake J, et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Planey SL, Keay SK, Zhang C-O, Zacharias DA. Palmitoylation of cytoskeleton associated protein 4 by DHHC2 regulates antiproliferative factor-mediated signaling. Mol Biol Cell. 2009;20:1454–1463. doi: 10.1091/mbc.E08-08-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putilina T, Wong P, Gentleman S. The DHHC domain: a new highly conserved cysteine-rich motif. Mol Cell Biochem. 1999;195:219–226. doi: 10.1023/a:1006932522197. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PIH. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Rocks O, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, III, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Yang XH, Hemler ME. DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol Biol Cell. 2008;19:3415–3425. doi: 10.1091/mbc.E07-11-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockli J, Rohrer J. The palmitoyltransferase of the cation-dependent mannose 6-phosphate receptor cycles between the plasma membrane and endosomes. Mol Biol Cell. 2004;15:2617–2626. doi: 10.1091/mbc.E03-11-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]