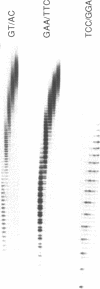
Abstract
The analysis of slippage synthesis of simple sequence DNA in vitro sheds some light on the question of how simple sequences arise in vivo. We show that it is possible to synthesize all types of repetitious di- and trinucleotide motifs starting from short primers and a polymerase in vitro. The rate of this synthesis depends on a sequence specific slippage rate, but is independent of the length of the fragments being synthesized. This indicates that only the ends of the DNA fragments are involved in determining this rate and that slippage is accordingly a short range effect. Slippage synthesis occurs also on a fixed template where only one strand is free to move, a situation which resembles chromosome replication in vivo. It seems therefore likely that slippage during replication is the cause of the observed length polymorphism of simple sequence stretches between individuals of a population.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Campbell J. L. Eukaryotic DNA replication. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- Epplen J. T. On simple repeated GATCA sequences in animal genomes: a critical reappraisal. J Hered. 1988 Nov-Dec;79(6):409–417. doi: 10.1093/oxfordjournals.jhered.a110544. [DOI] [PubMed] [Google Scholar]
- Fresco J. R., Alberts B. M. THE ACCOMMODATION OF NONCOMPLEMENTARY BASES IN HELICAL POLYRIBONUCLEOTIDES AND DEOXYRIBONUCLEIC ACIDS. Proc Natl Acad Sci U S A. 1960 Mar;46(3):311–321. doi: 10.1073/pnas.46.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- KORNBERG A., BERTSCH L. L., JACKSON J. F., KHORANA H. G. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID, XVI. OLIGONUCLEOTIDES AS TEMPLATES AND THE MECHANISM OF THEIR REPLICATION. Proc Natl Acad Sci U S A. 1964 Feb;51:315–323. doi: 10.1073/pnas.51.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987 Jul 10;15(13):5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Love J. M., Knight A. M., McAleer M. A., Todd J. A. Towards construction of a high resolution map of the mouse genome using PCR-analysed microsatellites. Nucleic Acids Res. 1990 Jul 25;18(14):4123–4130. doi: 10.1093/nar/18.14.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Coulter M. B., Flintoff W. F., Paetkau V. H. Enzymatic synthesis of deoxyribonucleic acids with repeating sequences. A new repeating trinucleotide deoxyribonucleic acid, d(T-C-C)n-d(G-G-A)n. Biochemistry. 1974 Apr 9;13(8):1596–1603. doi: 10.1021/bi00705a007. [DOI] [PubMed] [Google Scholar]
- Tautz D. Genomic finger printing goes simple. Bioessays. 1990 Jan;12(1):44–46. doi: 10.1002/bies.950120111. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986 Aug 14;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Wells R. D., Jacob T. M., Narang S. A., Khorana H. G. Studies on polynucleotides. LXIX. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: DNA-like polymers containing repeating trinucleotide sequences. J Mol Biol. 1967 Jul 28;27(2):237–263. doi: 10.1016/0022-2836(67)90018-6. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Ohtsuka E., Khorana H. G. Studies on polynucleotides. L. Synthetic deoxyribopolynucleotides as templates for the DNA polymerase of Escherichia coli: a new double-stranded DNA-like polymer containing repeating dinucleotide sequences. J Mol Biol. 1965 Nov;14(1):221–237. doi: 10.1016/s0022-2836(65)80242-x. [DOI] [PubMed] [Google Scholar]