Summary
Inhibitor of growth 4 (ING4) is a candidate tumor suppressor gene that was shown to be deleted in 10% to 20% of breast cancers by array comparative genome hybridization analysis. We developed fluorescent in situ hybridization to detect the ING4 gene directly in the tissue samples on tumor tissue microarrays. We evaluated the ING4 gene status in 1033 breast cancer tissue samples and observed that ING4 was deleted in 16.5% (170/1033) of all breast cancers. ING4 deletion was significantly associated with Her2 overexpression: of the tumors with ING4 deletion, 23.8% (39/164) were human epidermal growth factor 2 (HER2) positive, as compared with 14.1% (115/814) of the tumors without ING4 deletion (P = .002). In addition, the tumors with ING4 deletion were more likely to belong to the HER2 molecular subtype (estrogen receptor negative/progesterone receptor negative/human epidermal growth factor positive) of breast cancer, compared with the other subtypes (28.4% HER2 versus 15.7% all, P = .002). ING4 deletion did not affect survival outcome of all patients with breast cancer (P = .797) or of the patients with HER2-positive tumors (P = .792). We conclude that ING4 deletion in breast cancer is relatively common, as 1 in 6 breast cancer harbors ING4 deletion. Furthermore, ING4 deletion is more prevalent in HER2-positive tumors, suggesting a functional antagonistic relationship between the ING4 tumor suppressor and the HER2 oncogene. These results sustain the view that ING4 is a tumor suppressor in breast cancer and suggest that ING4 deletion may contribute to the pathogenesis of HER2-positive breast cancer.
Keywords: ING4, Tumor suppressor gene, Breast cancer, FISH, HER2/neu
1. Introduction
ING4 is a member of the inhibitor of growth (ING) tumor suppressor family (ING1-5) and has been shown to play a role in cancer-related cellular processes, including cell proliferation, apoptosis, contact inhibition, tumor angiogenesis, DNA damage response, cell migration, and hypoxia [1–7]. The ING4 gene is mapped to 12p13 in the short arm of chromosome 12. Loss of heterozygosity at 12p13 has been reported in cancers such as hematologic malignancies, ovarian cancer, and prostate cancer [8–10]. In addition, loss of heterozygosity and single-locus deletion of ING4 have been found in head and neck squamous cell carcinoma and breast cancer, respectively [3,11]. Low levels of ING4 expression have been correlated with high-grade tumors and poor patient outcome in malignant neoplasias, including glioma, melanoma, gastric adenocarcinoma, and hepatocellular carcinoma [4,12–14]. Moreover, inactivating point mutations of ING4 have been found in several cancer cell lines and glioma [3,15]. Thus, ING4 appears to be disabled by various mechanisms in cancer and may play a role as a tumor suppressor in various cancers that arise from diverse tissue types.
In a previous study, ING4 deletion has been estimated to occur in 10% to 20% of breast cancers by array comparative genomics hybridization (aCGH) analysis using 2 DNA probes flanking the ING4 gene [3]. In this study, we used fluorescent in situ hybridization (FISH) using a single bacterial artificial chromosome (BAC) clone that contains the ING4 gene. We evaluated ING4 deletion in more than 1500 breast cancer specimens using tissue microarray (TMA) and correlated ING4 deletion with clinicopathologic parameters in breast cancer.
2. Materials and methods
2.1. Breast cancer TMA
TMAs contained 2020 tumor breast cancer tissue punches from 1579 independent formalin-fixed and paraffin-embedded tumor samples collected from patients with breast cancer diagnosed between the years 1985 and 2007 at the Institute for Pathology, University Hospital of Basel, and the Institute of Viollier in Basel, Switzerland. Of 2020 tissue spots on TMAs, 1566 tissue spots came from the same tumor samples used in the TMA studies previously described [16,19]. The median age of patients was 63 years ranging from 27 to 101 years. The mean follow-up time was 80.8 months ranging from 1 to 263 months. Raw patient survival data were obtained from the Cancer Registry of Basel or from the patients’ attending physicians. Tumor data regarding histologic subtype, TNM classification, Bloom-Richardson-Elston-Ellis (BRE) grade, and diameter were obtained from pathology reports. Tissue samples and data were used according to the ethical standards of the University Hospital of Basel, Switzerland. TMAs were constructed as described previously [16]. In brief, tissue cylinders with a diameter of 0.6 mm were punched out from the “donor” tumor tissue blocks and transferred into a “recipient” paraffin block using a semiautomated tissue arrayer (Institute for Pathology, University Hospital of Basel, Basel, Switzerland). Each TMA contained a number of tumor punch spots ranging from 159 to 522. Three hundred thirty-six “double tissue spots” were included on TMAs from 168 samples by obtaining tissue punches from the tumor center and periphery.
2.2. Fluorescent labeling of the DNA probe
The BAC clone, RP11-433J6, was purchased from the Children’s Hospital Oakland Research Institute (Oakland, CA). BAC DNA was purified from Escherichia coli using a plasmid purification kit (Qiagen, Valencia, CA). Eight hundred eighty nanograms of BAC DNA was digested with AluI restriction enzyme (Invitrogen, Carlsbad, CA). AluI-digested BAC DNA was labeled with Cy3-dUTP (GE Healthcare USA, Piscataway, NJ) using BioPrime Array CGH kit (Invitrogen) for 2 hours at 37°C. Cy3-labeled BAC DNA (BAC-ING4) was purified using Vivaspin column (Sartorius Biolab, Göttingen, Germany). spectrum green–labeled chromosome 12 centromere probe (CEP12) was purchased from Vysis (Downers Grove, IL).
2.3. Fluorescent in situ hybridization
Metaphase spread of T47D cells was prepared using a standard method. In brief, cells were grown to 80% confluent and treated with colcemid (GIBCO, Grand Island, NY) at 10 μL/mL for 2 hours. Cells were harvested and incubated in prewarmed hypotonic solution (0.2% KCl, 0.2% sodium citrate) at 37°C for 7 minutes. Cells were washed, fixed in cold methanol/acetic acid (3:1 vol/vol), and dropped on a slide. Lymphocyte metaphase spread was purchased from Vysis. Paraffin-embedded normal breast tissue sections were obtained from the Institute for Pathology, University Hospital of Basel, Switzerland. Five-micrometer tissue and TMA sections were deparaffinized and pretreated with a paraffin pretreatment kit (Vysis).
Fluorescent-labeled DNA probes were hybridized to metaphase spread and tissue sections on slides at 37°C overnight. The slides were washed with wash solutions (Vysis; 0.4X saline-sodium citrate [SSC], 0.3% Nonidet P40, pH 7–7.5) and counterstained with 4′,6-diaminidino-2-phenylindole (DAPI) (Vysis) before mounting. The slides were visualized with a Zeiss Axiophot 2 epifluorescence microscope (Zeiss, Jena, Germany) using filter sets for DAPI, spectrum orange, spectrum green and DAPI/spectrum green/spectrum orange filter (Abbott Molecular, Abbott Park, IL). The FISH probe signals were counted in 10 nonoverlapping nuclei per tissue spot on TMA, and the BAC-ING4/CEP12 ratio was calculated. We used the ratio less than 0.8 to define a gene deletion, as described previously [17,18].
2.4. Immunohistochemistry
Immunochemical staining of TMA sections was performed using iView DAB Detection Kit (Ventana, Tucson, AZ). Antibodies used were anti–estrogen receptor α (anti-ER) monoclonal (1:40; Novocastra, Newcastle, UK), anti–progesterone receptor (anti-PR) monoclonal (1:100; Novocastra), and anti–HER2/neu polyclonal (no dilution; Ventana). The staining intensity of ER, PR, and HER2 was scored as described previously [19]. In brief, tumors were considered positive for ER or PR for nuclear staining in more than 10% of tumor cells, with an intensity score between 1 and 3. HER2 expression was scored as 0 for no staining, 1+ for faint and partial membranous staining, 2+ for weak complete staining of the membrane in more than 10% of tumor cells, and 3+ for intense complete staining of the membrane in more than 10% of tumor cells. All slides were scored manually by at least one pathologist (C.T. or E.K.).
2.5. Statistical analysis
Relationship between clinicopathologic features and ING4 deletion in tumors was analyzed using Pearson χ2 test. Wilcoxon rank sum test was used for the analysis of age and tumor size distribution. Kaplan-Meier method and log-rank test were used to assess survival time differences in univariate analysis and in subgroup analysis. All analyses were carried out using SAS V9.1 (SAS Institute, Cary, NC). The Bonferroni correction was used to adjust the level of significance for multiple comparisons, and P values less than .004 were considered statistically significant.
3. Results
3.1. FISH detection of the ING4 gene
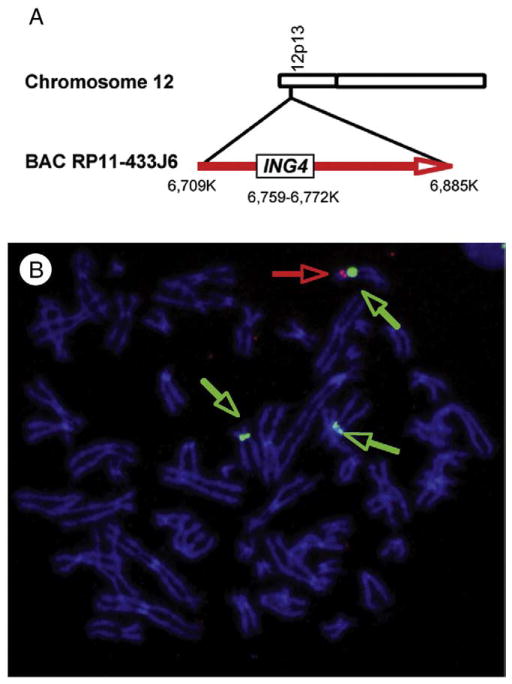
In a previous study, ING4 was shown to be deleted in 10% to 20% of breast cancers, suggesting a tumor-suppressive role of ING4 in breast cancer [3]. The deletion was estimated by aCGH analysis using 2 BAC probes flanking the ING4 gene [3]. In this study, we used FISH using a single BAC clone that contains the ING4 gene to determine the prevalence of ING4 deletion in breast cancer. The ING4 gene maps to the short arm of chromosome 12 and is located 6.7 mb from the telomere (www.ncbi.nlm.nih.gov). We chose BAC RP11-433J6 to detect the ING4 gene (www.genome.ucsc.edu). The BAC is 176 kb in size and contains the ING4 gene that spans 12.8 kb (Fig. 1A). First, we used 2-color FISH to determine the chromosomal location of the BAC on a metaphase spread of normal lymphocytes, using Cy3-labeled BAC (red) and spectrum green–labeled chromosome 12 centromere probe (green). We observed that Cy3-labeled BAC was hybridized to the short arm of chromosome 12, verifying the chromosomal location of the BAC (data not shown). We refer to the fluorescent-labeled BAC RP11-433J6 and chromosome 12 centromere probe as BAC-ING4 and CEP12, respectively.
Fig. 1.
FISH detection of the ING4 gene. A, Schematic diagram of chromosome 12, BAC RP11-433J6, and the ING4 gene. B, T47D cells have deletions in the 2 copies of the ING4 gene. FISH shows 3 CEP12 (green) and one copy of BAC-ING4 (green). DNA is stained with DAPI (blue).
Next, we hybridized BAC-ING4 to a metaphase spread of T47D breast cancer cells. T47D breast cancer cells were characterized as hypotriploid and contain 3 copies of chromosome 12 (www.ATCC.org). In a previous study, it was determined by aCGH that at least 2 copies of the ING4 gene locus were deleted in T47D cells [3]. Using FISH, we detected 3 green signals of CEP12, indicating that T47D cells contain 3 copies of chromosome 12 (Fig. 1B). In contrast, only one red signal of BAC-ING4 was detected (Fig. 1B). These results confirmed that T47D breast cancer cells contain 3 copies of chromosome 12 with 2 copies of the ING4 gene deleted, retaining only 1 copy of the gene.
To evaluate the FISH probes on interphase chromosomes, we hybridized CEP12 and BAC-ING4 to a 5-μm section of paraffin-embedded normal breast tissue. We detected 1 or 2 CEP12 signals and 1 or 2 BAC-ING4 signals per nucleus. After counting 100 nuclei, the average ratio between BAC-ING4 and CEP12 (BAC-ING4/CEP12) was approximately 1 (data not shown).
3.2. ING4 deletion in breast tumor tissue samples on TMAs
We next hybridized CEP12 and BAC-ING4 to TMAs containing breast cancer tissues. The probe signals were counted in 10 tumor cell nuclei per tissue spot. To avoid misinterpretation due to technical variability, we only counted the tissue areas that showed at least 1 signal for either BAC-ING4 or CEP12. In addition, all TMA sections were hybridized twice to clarify uneven or insufficient hybridization of the probes. After excluding ambiguous and discrepant FISH scores, 1033 tissue spots (51.1%) among 2020 tissue spots were used for data evaluation.
We found that the BAC-ING4/CEP12 ratio ranged from 0.28 to 2.18 between the tissue samples on TMAs. A tumor with an overall ratio of 1 showed 2 BAC-ING4 and 2 CEP12 signals per nucleus (Fig. 2A), indicating no deletion of ING4. A tumor that contained 0 to 3 BAC-ING4 signals and 3 to 7 CEP12 signals per nucleus resulted in an overall ratio of 0.38 (Fig. 2B), showing underrepresentation of the ING4 gene copy number compared with the number of chromosome 12 centromeres, thus indicating deletion of ING4. Another example of tumor with ING4 deletion is shown in Fig. 2C, with 0 to 2 BAC-ING4 signals and 2 to 7 CEP12 signals, resulting in an overall BAC-ING4/CEP12 ratio of 0.55 (Fig. 2C).
Fig. 2.
FISH on breast cancer TMA. A, Infiltrating ductal carcinoma with no ING4 deletion shows 2 CEP12 (green; green arrows) and 2 BAC-ING4 (BAC, red; red arrows) signals, resulting in a ratio of 1 BAC-ING4/CEP12. B, Ductal carcinoma with pleomorphic nuclei with ING4 deletion shows 3 to 7 CEP12 (green; green arrows) and 0 to 3 BAC-ING4 (red; red arrows) signals with an overall ratio of 0.38 BAC-ING4/CEP12. C, High-grade infiltrating ductal carcinoma with nuclear polymorphy shows an overall ratio of 0.55 BAC-ING4/CEP12. The nuclei contained up to 7 CEP12 (green arrows) and 0 to 2 BAC-ING4 (red arrows). The section shows cancer cells (DAPI, blue) infiltrating the surrounding soft tissue (autofluorescent, light green).
We used a ratio of 0.8 as a gene deletion reference point, as described previously [17,18]. We detected 170 tumor tissues with the BAC-ING4/CEP12 ratio of 0.8 or less, which made up 16.5% (170/1033) of all breast tumors. We concluded that ING4 is deleted in 16.5% of breast cancer. This indicated that ING4 deletion is relatively common in breast cancer.
3.3. Clinicopathologic correlation of ING4 deletion
We then compared the clinicopathologic features of 170 breast tumors harboring ING4 deletion to 863 tumors with no ING4 deletion. We calculated the percentage of tumors with or without ING4 deletion in each parameter and determined statistical significance. The results are tabulated in Table 1.
Table 1.
Relationship between ING4 deletion and clinicopathologic features
| Clinicopathologic feature |
ING4 (n = 1033) |
P | |
|---|---|---|---|
| Deletion (ratio, ≤0.8) (n = 170) | No deletion (ratio, >0.8) (n = 863) | ||
| Histologic subtype | |||
| Ductal carcinoma | 132 (77.7%) | 619 (71.7%) | .113 |
| Other | 38 (22.4%) | 244 (28.3%) | |
| Size | |||
| pT1 | 51 (30.0%) | 295 (34.8%) | .258 |
| pT2 | 96 (56.5%) | 410 (48.3%) | |
| pT3 | 8 (4.7%) | 57 (6.7%) | |
| pT4 | 15 (8.8%) | 87 (10.3%) | |
| Lymph node status | |||
| N0 | 81 (53.6%) | 395 (51.6%) | .652 |
| >N0 | 70 (46.4%) | 370 (48.4%) | |
| BRE | |||
| Grade 1 | 26 (15.3%) | 183 (21.5%) | .187 |
| Grade 2 | 78 (45.9%) | 358 (42.0%) | |
| Grade 3 | 66 (38.8%) | 311 (36.5%) | |
| ER | |||
| Positive | 126 (76.8%) | 627 (74.9%) | .603 |
| Negative | 38 (23.2%) | 210 (25.1%) | |
| PR | |||
| Positive | 60 (47.2%) | 273 (42.1%) | .281 |
| Negative | 67 (52.8%) | 376 (57.9%) | |
| HER2/neu receptor | |||
| 0 + 1 | 125 (76.2%) | 699 (85.9%) | .002 |
| 2 + 3 | 39 (23.8%) | 115 (14.1%) | |
| Age at diagnosis (y), median (minimum-maximum) | 61.5 (28–88) | 63 (27–93) | .174 |
| Tumor size (mm), median (minimum-maximum) | 24 (4–110) | 25 (0–140) | .854 |
| Survival rate | |||
| 5 y (95% CI) | 81.4% (74%–87%) | 83.0% (80%-86%) | .591* |
| 6 y (95% CI) | 80.4% (72%–86%) | 81.4% (78%-84%) | |
| 10 y (95% CI) | 72.6% (62%-81%) | 71.8% (67%-76%) | |
Abbreviation: CI, confidence interval.
No difference in survival time throughout the duration of follow-up.
First, we found that most of the clinicopathologic features were in concordance with each other between the 2 types of tumors with no statistical differences: most tumors were ductal carcinoma (ING4 deletion versus no deletion, 77% versus 71%), T2 (56% versus 48%), node negative (53% versus 51%), and BRE grade 2 (45% versus 42%). The median ages between breast cancer patients with and without ING4 deletion were comparable (61 years versus 63 years). The average tumor sizes were also comparable (24 mm versus 25 mm). In addition, most tumors with ING4 deletion were ER positive (76%), as were the tumors without ING4 deletion (74%). The percentages of PR-positive tumors were also comparable between the 2 tumor types (47% versus 42%). Survival information was available for 956 patients of the corresponding tumor tissues evaluated by FISH (956/1033; 92.5%). We did not observe any significant differences in survival time between patients with breast cancer with or without ING4 deletion (Table 1 and also see Fig. 3).
Fig. 3.
ING4 deletion does not influence a 5-year patient survival. A, Patient survival with breast cancer with (black open circle) and without (pink open circle) ING4 deletion. B, Patient survival with HER2+ breast cancer with (black open circle) and without (pink open circle) ING4 deletion.
One distinct feature of ING4 deleted tumors was a significant association with HER2 overexpression. Thirty-nine (23.8%) of 164 of tumors with ING4 deletion were HER2 positive, as compared with 115 (14.1%) of 814 of tumors with no deletion (Table 1). Moreover, 25.3% (39/154) of HER2-positive tumors harbored ING4 deletion compared with 15.1% (125/824) of HER2-negative tumors. These data showed that 1 of 4 HER2-positive tumors harbors ING4 deletion compared with 1 of 7 HER2-negative tumors, indicating that ING4 deletion is more prevalent in HER2-positive tumors. We compared clinicopathologic features between 39 HER2-positive ING4-deleted tumors with 115 HER2-positive tumors with no ING4 deletion and observed no discernable features correlating with ING4 deletion among HER2-positive tumors (data not shown).
3.4. ING4 deletion is more prevalent in the HER2-positive molecular subtype of breast cancer
We examined whether ING4 deletion was associated with any molecular subtypes of breast cancer. The molecular subtypes of breast cancer were initially defined by distinct gene expression signatures [20–22]. Subsequently, the subtypes have also been defined by the presence or absence of 3 correlative surrogate markers: ER, PR, and HER2/neu receptor (HER2) [23]: luminal A (ER+ PR+ HER2−), luminal B (ER+ PR+ HER2+), HER2 (ER− PR− HER2+), and basal-like (ER− PR− HER2−). Among our TMA tumor tissue samples, we had information regarding the status of all 3 markers in 465 tumors. Our study cohort consisted of 37.2% luminal A (173/465), 1.1% luminal B (5/465), 17.4% HER2 (81/465), and 44.3% basal-like (206/465) subtype (Table 2). These indicated a markedly lower percentage of the luminal B subtype and a higher percentage of the basal-like subtype in our study cohort, compared with other studies [24]. The reason for this skewed distribution of the subtypes in our study samples is not known.
Table 2.
ING4 deletion in the molecular subtypes of breast cancer
| Molecular subtype | n = 465 | ING4 deletion, 73 (15.7%) | No deletion, 392 (84.3%) | P |
|---|---|---|---|---|
| Luminal A (ER+ PR+ HER2−) | 173 | 25 (14.5%) | 148 (85.5%) | <.001 |
| Luminal B (ER+ PR+ HER2+) | 5 | 1 (20%) | 4 (80%) | NS |
| HER2 (ER− PR− HER2+) | 81 | 23 (28.4%) | 58 (71.6%) | .002 |
| Basal-like (ER− PR− HER2−) | 206 | 24 (11.7%) | 182 (88.3%) | .001 |
Abbreviation: NS, not significant.
We then determined the prevalence of ING4 deletion in each molecular subtype of breast cancer by calculating the percentage of tumors that harbor ING4 deletion. The results are tabulated in Table 2. In all subtypes, 15.7% contained ING4 deletion (73/465). In the luminal A subtype, 14.5% of tumors contained ING4 deletion (25/173), comparable with the average ING4 deletion rate of 15.7%. In the luminal B subtype, we could not assess the prevalence of ING4 deletion with any statistical significance because of the small number of cohort. In the HER2 subtype, a significantly higher percentage of tumors contained ING4 deletion (23/81; 28.4%). In contrast, only 11.7% (24/206) of the basal-like subtype tumors contained ING4 deletion. We conclude that ING4 deletion is more prevalent in the HER2 molecular subtype, whereas it is less prevalent in the basal-like subtype.
3.5. ING4 deletion does not affect patient survival
We determined whether the ING4 deletion status influenced patient survival, using subgroup analyses. The results showed no difference in overall survival rate between the patients with and without ING4 deletion (Table 1 and Fig. 3A). ING4 deletion did not affect the survival rate of the patients with HER2-positive tumors (Fig. 3B). We conclude that ING4 deletion does not affect the survival rate of patients with breast cancer.
4. Discussion
We have found that ING4 is deleted in 16.5% of breast cancer by evaluating a cohort of 1033 patient samples. This result is consistent with the previous estimate of ING4 deletion in 10% to 20% of breast cancer [3]. We also found that ING4 deletion was more prevalent in HER2-positive tumors and in the HER2 molecular subtype (ER− PR− HER2+) of breast cancer. Although we did not observe any effect of ING4 deletion in patient survival either in all patients with breast cancer or in patients with HER2-positive tumors, we do not know if ING4 deletion influences other clinical features such as response to therapy.
Breast cancer is a heterogeneous disease with the subtypes defined by distinct molecular characteristics, clinical features, and survival outcome [21,22]. HER2-positive tumors with HER2 gene amplification or HER2/neu overexpression make up approximately 20% of all breast cancers [25]. The presence of HER2 has been correlated with high-grade tumors and associated with poor prognosis [21,22,26]. Therapy targeting HER2 such as trastuzumab has been shown effective as did monotherapy in less than 35% of patients with HER2-positive tumors [27,28]. Genetic factors that determine the responsiveness to the HER2-target therapy are unknown. It is possible that ING4 deletion may be one of the factors that affect responses to therapy in patients with HER2-positive breast cancer. However, our clinical data regarding therapeutics were limited so that we could not address such a hypothesis.
Our results showing that ING4 deletion is more prevalent in HER2-positive tumors suggest a suppressive role of ING4 in the HER2-driven oncogenesis. Molecular mechanism of ING4 antagonizing HER2 is not known. The molecular mechanism of ING4 characterized to date involves transcription regulation via chromatin remodeling [15,29,30]. Consistently, ING4 copurifies with chromatin remodeling complexes containing histone acetyl transferases and histone deacetylases [1,29]. In addition, ING4 could directly bind to methylated histone H3 [30,31]. Therefore, molecular mechanism of the ING4 tumor suppressor in HER2-positive breast cancer may involve gene regulation downstream of the HER2 receptor signal.
HER2 receptor signaling has been shown to activate the PI3K/Akt/mTOR pathways leading to the activation of the nuclear factor κB (NF-κB) transcription factor [32,33]. In glioma, ING4 was shown to modulate downstream targets genes of NF-κB [4,15]. Taken together, ING4 may suppress HER2-driven breast cancer by modulating HER2-activated NF-κB. In this case scenario, we would expect frequent deletion of ING4 in HER2-positive tumors, which would result in constitutive activation of NF-κB. Our results showing that ING4 deletion is more prevalent in the HER2-positive tumors are consistent with the antagonistic relationship between HER2/NF-κB and ING4.
NF-κB activation has been detected predominantly in ER-negative and HER2-positive breast cancer [34]. Biswas et al [34] also reported that NF-κB activation was rare in the basal-like molecular subtype of breast cancer (ER negative and HER2 negative). These observations are also consistent with our findings that ING4 deletion is more prevalent in the HER2 subtype but less prevalent in the basal-like subtype of breast cancer. Thus, it appears that ING4 deletion may correlate with NF-κB activation in breast cancer. A direct functional relationship between HER2, NF-κB, and ING4 will require further investigation.
In conclusion, we have developed a FISH assay that can assess ING4 deletion in breast cancer. Our results showing ING4 deletion in 1 of 6 breast cancer reenforce a tumor-suppressive role of ING4 in breast cancer. Furthermore, we showed that ING4 deletion is twice more prevalent in HER2-positive tumors and the HER2 molecular subtype, suggesting that ING4 deletion may contribute to the pathogenesis of HER2-driven breast cancer. ING4 deletion may be used as a molecular marker to further delineate molecular subtypes in breast cancer with distinct characteristics and therapeutic implications.
Acknowledgments
We greatly appreciate Tom Dennis, Chris Gooden, and Dr Mike Bittner for their expert advice on developing FISH. We thank Drs Hans-Jörg Spichtin and H. Boss for breast tissue samples. We also thank Dr Galen Hostetter for helpful discussions on the manuscript.
Footnotes
This study was supported by a postdoctoral fellowship award from the Novartis Foundation (Basel, Switzerland), formerly the Ciba-Geigy Jubilee Foundation (to C.T.) and National Cancer Institute (Bethesda, MD) 5K01CA115681 Howard Temin Award (to S.K.).
References
- 1.Shiseki M, Nagashima M, Pedeux RM, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–8. [PubMed] [Google Scholar]
- 2.Zhang X, Xu LS, Wang ZQ, et al. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci U S A. 2004;101:16251–6. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garkavtsev I, Kozin SV, Chernova O, et al. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–32. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 5.Shen JC, Unoki M, Ythier D, et al. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–8. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc Natl Acad Sci U S A. 2005;102:7481–6. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colla S, Tagliaferri S, Morandi F, et al. The new tumor-suppressor gene inhibitor of growth family member 4 (ING4) regulates the production of proangiogenic molecules by myeloma cells and suppresses hypoxia-inducible factor-1 alpha (HIF-1alpha) activity: involvement in myeloma-induced angiogenesis. Blood. 2007;110:4464–75. doi: 10.1182/blood-2007-02-074617. [DOI] [PubMed] [Google Scholar]
- 8.Cavé H, Gérard B, Martin E, et al. Loss of heterozygosity in the chromosomal region 12p12–13 is very common in childhood acute lymphoblastic leukemia and permits the precise localization of a tumor-suppressor gene distinct from p27KIP1. Blood. 1995;86:3869–75. [PubMed] [Google Scholar]
- 9.Hatta Y, Takeuchi S, Yokota J, Koeffler HP. Ovarian cancer has frequent loss of heterozygosity at chromosome 12p12.3–13.1 (region of TEL and Kip1 loci) and chromosome 12q23-ter: evidence for two new tumour-suppressor genes. Br J Cancer. 1997;75:1256–62. doi: 10.1038/bjc.1997.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawana Y, Ichikawa T, Suzuki H, et al. Loss of heterozygosity at 7q31.1 and 12p13–12 in advanced prostate cancer. Prostate. 2002;53:60–4. doi: 10.1002/pros.10131. [DOI] [PubMed] [Google Scholar]
- 11.Gunduz M, Nagatsuka H, Demircan K, et al. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene. 2005:356. doi: 10.1016/j.gene.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–9. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Jin Y, Sun WJ, Yu Y, Bai J, Tong DD, et al. Reduced expression and novel splice variants of ING4 in human gastric adenocarcinoma. J Pathol. 2009;219:87–95. doi: 10.1002/path.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang F, Luo LB, Tao YM, Wu F, Yang LY. Decreased expression of inhibitor of growth 4 correlated with poor prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:409–16. doi: 10.1158/1055-9965.EPI-08-0575. [DOI] [PubMed] [Google Scholar]
- 15.Nozell S, Laver T, Moseley D, et al. The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol Cell Biol. 2008;28:6632–45. doi: 10.1128/MCB.00697-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Kuraya K, Novotny H, Bavi P, et al. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60:768–72. doi: 10.1136/jcp.2006.038281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Leo A, Gancberg D, Larsimont D, et al. HER-2 amplification and topoisomerase IIalpha gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res. 2002;8:1107–16. [PubMed] [Google Scholar]
- 18.Knoop AS, Knudsen H, Balslev E, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23:7483–90. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Tapia C, Schraml P, Simon R, et al. HER2 analysis in breast cancer: reduced immunoreactivity in FISH non-informative cancer biopsies. Int J Oncol. 2004;25:1551–7. [PubMed] [Google Scholar]
- 20.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 21.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen PL, Taghian AG, Katz MS, et al. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26:2373–8. doi: 10.1200/JCO.2007.14.4287. [Epub 2008 Apr 2314] [DOI] [PubMed] [Google Scholar]
- 24.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [Epub 2003 Jun 8426] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5:63–9. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 26.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. 1987;235:4785. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 27.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–26. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 28.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26:3637–43. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 29.Doyon Y, Cayrou C, Ullah M, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Hung T, Binda O, Champagne KS, et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–56. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palacios A, Muñoz IG, Pantoja-Uceda D, et al. Molecular basis of histone H3K4me3 recognition by ING4. J Biol Chem. 2008;283:15956–64. doi: 10.1074/jbc.M710020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou BP, Hu MC, Miller SA, et al. HER-2/neu blocks tumor necrosis factor–induced apoptosis via the Akt/NF-kappaB pathway. J Biol Chem. 2000;275:8027–31. doi: 10.1074/jbc.275.11.8027. [DOI] [PubMed] [Google Scholar]
- 33.Pianetti S, Arsura M, Romieu-Mourez R, Coffey RJ, Sonenshein GE. Her-2/neu overexpression induces NF-kappaB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IkappaB-alpha that can be inhibited by the tumor suppressor PTEN. Oncogene. 2001;20:1287–99. doi: 10.1038/sj.onc.1204257. [DOI] [PubMed] [Google Scholar]
- 34.Biswas DK, Shi Q, Baily S, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004;101:10137–42. doi: 10.1073/pnas.0403621101. [Epub 12004 Jun 10125] [DOI] [PMC free article] [PubMed] [Google Scholar]