Abstract
Folate coenzymes function as one-carbon group carriers in intracellular metabolic pathways. Folate-dependent reactions are compartmentalized within the cell and are catalyzed by two distinct groups of enzymes, cytosolic and mitochondrial. Some folate enzymes are present in both compartments and are likely the products of gene duplications. A well-characterized cytosolic folate enzyme, FDH (10-formyltetrahydrofolate dehydrogenase, ALDH1L1), contains a domain with significant sequence similarity to aldehyde dehydrogenases. This domain enables FDH to catalyze the NADP+-dependent conversion of short-chain aldehydes to corresponding acids in vitro. The aldehyde dehydrogenase-like reaction is the final step in the overall FDH mechanism, by which a tetrahydrofolate-bound formyl group is oxidized to CO2 in an NADP+-dependent fashion. We have recently cloned and characterized another folate enzyme containing an ALDH domain, a mitochondrial FDH. Here the biological roles of the two enzymes, a comparison of the respective genes, and some potential evolutionary implications are discussed. The phylogenic analysis suggests that the vertebrate ALDH1L2 gene arose from a duplication event of the ALDH1L1 gene prior to the emergence of osseous fish >500 millions years ago.
Keywords: Folate metabolism, mitochondria, 10-formyltetrahydrofolate dehydrogenase, aldehyde dehydrogenase, domain structure, evolution
1. Introduction
In the cell, folate coenzymes participate in numerous reactions of one-carbon transfer (Fig. 1, reviewed in [1–3]), including de novo nucleotide biosynthesis, conversions of several amino acids and the incorporation of formate-derived carbon into folate pool. Another group of biochemical reactions of folate involve interconversions of different forms of the coenzyme, in which one-carbon groups remain folate-bound but alter their oxidation state. Additional folate reactions, which do not involve the conversion of one-carbon groups, are: (i) the reduction of folic acid to dihydrofolate and then to tetrahydrofolate, and (ii) the addition of glutamic acid residues to folate monoglutamate to form folate polyglutamates. These reactions are required to produce the active form of the coenzyme (via dihydrofolate reductase) and to retain folate within the cell (via folylpolyglutamate synthetase). Finally, a reaction catalyzed by 10-formyltetrahydrofolate dehydrogenase (ALDH1L1, FDH) irreversibly removes carbon groups from the folate pool in the form of CO2 [4]. This pathway is distinct from other folate-dependent reactions in terms of the utilization of a one-carbon group: instead of being used in a biosynthetic pathway it is diverted toward energy production coupled with the final step of carbon oxidation, from the level of formate to the level of CO2.
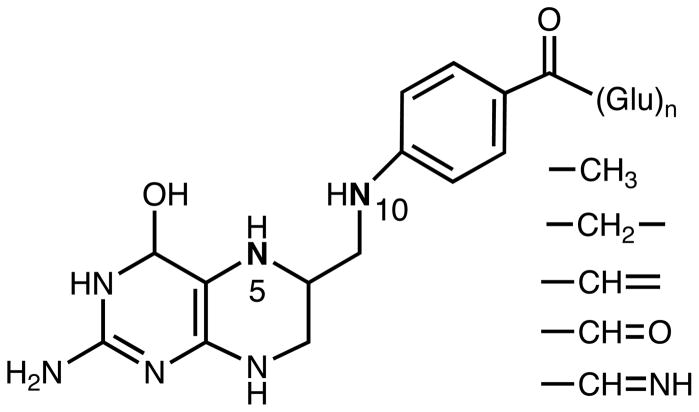
Fig. 1.
Structure of tetrahydrofolate (THF) and transferred one-carbon groups. The folate molecule consists of pteridine, p-aminobenzoic acid and glutamate. Inside the cell, reduced folates exist as polyglutamates with up 8 glutamate residues bound through α-amino and γ-carboxyl groups. One-carbon groups can be bound to N5, N10 or both of these positions in THF molecule. CH3, methyl; CH2, mehtylene; CH, methenyl; CHO, formyl, CHNH, formimino.
Folate metabolism is highly compartmentalized in the cell with the major pathways being localized to either the cytoplasm or mitochondria [3]. Mitochondrial folate metabolism is generally viewed as a supplier of one-carbon groups for cytosolic folate-dependent biosynthetic reactions [5, 6]. In addition, recent studies have indicated a nuclear compartmentation for some folate-dependent reactions as well [2, 3, 7, 8]. The nucleus-related aspects of folate metabolism, however, are less studied than the cytosolic and mitochondrial folate pathways. The cytosolic and mitochondrial compartmentation of folate metabolism occurs at two levels. First, there are two different folate pools, which are not easily interchangeable since folate cannot freely traverse mitochondrial membrane [3]. Transport of folate into mitochondria is carried out by a specific transporter, which is homologous to several inner mitochondrial wall transporters [9]. Second, there are separate sets of folate enzymes residing in the cytosol or mitochondria that define the specificity of folate pathways in each compartment [2, 3]. Some folate enzymes and corresponding reactions are unique to a single compartment, either cytosolic or mitochondrial. Several enzymes, however, are present in both compartments, and FDH belongs to this group (Table 1).
Table 1.
Subcellular localization and function of folate-related enzymes.
| Enzyme (genes are listed in case of multiple enzymes) | Localization | E.C. number* | Function | References |
|---|---|---|---|---|
| SHMT (SHMT1 and 2) | Cytosol/mitochondria | 2.1.2.1 | Ser/Gly interconversion | [10, 11] |
| C1-Synth (MTHFD1, MTHFD2, MTHFD1L, MTHFD2L) | Cytosol/mitochondria | 1.5.1.5/3.5.4.9/6.3.4.3 | Incorporation of formate into folate pool, interconversion of folates | [3, 12–14] |
| FDH (ALDH1L1 and 1L2) | Cytosol/mitochondria | 1.5.1.6 | Conversion of folate-bound one-carbon groups to CO2 | [4, 15, 16] |
| FPGS | Cytosol/mitochondria | 6.3.2.17 | Polyglutamylation of folate | [17, 18] |
| MTHFS** | Cytosol/mitochondria | 6.3.3.2 | 5-formyl-THF to 5,10-methenyl-THF conversion | [19] |
| GARFT | Cytosol | 2.1.2.2 | De novo purine biosynthesis | [20, 21] |
| AICARFT | Cytosol | 2.1.2.3 | De novo purine biosynthesis | [20, 21] |
| Thymidylate synthase | Cytosol | 2.1.1.45 | TMP biosynthesis | [2] |
| DHFR | Cytosol | 1.5.1.3 | Reduction of DHF to THF*** | [2] |
| MTHFR | Cytosol | 1.5.1.20 | Methylene-THF to methyl-THF conversion | [22, 23] |
| Methionine synthase | Cytosol | 2.1.1.13 | Biosynthesis of Met from Hcys | [24] |
| GNMT**** | Cytosol | 2.1.1.20 | Conversion of Gly to sarcosine | [25] |
| FTCD | Cytosol | 2.1.2.5/4.3.1.4 | Histidine degradation | [26–28] |
| Aminomethyltransferase | Mitochondria | 2.1.2.10 | Glycine oxidation (glycine cleavage system) | [29] |
| FMT | Mitochondria | 2.1.2.9 | Biosynthesis of fmet-tRNA (mitochondrial translation initiation) | [30] |
| DMGDH | Mitochondria | 1.5.99.2 | Conversion of dimethylglycine to sarcosine | [1, 31, 32] |
| SarDH | Mitochondria | 1.5.99.1 | Conversion of sarcosine to glycine | [1, 33] |
Enzymes listed are: SHMT, serine hydroxymethyltransferase; C1-Synth (C1-synthase), a trifunctional enzyme containing 10-formyl-THF synthetase, 5,10-mehtylene-THF dehydrogenase and 5,10-methenyl-THF cyclohydrolase activities (only MTHFD1 possesses all three activities); FDH, 10-formyl-THF dehydrogenase; FPGS, folylpolyglatamate synthetase; MTHFS, 5,10-methenyl-THF synthetase; GARFT, glycinamide ribonucleotide formyltransferase; AICARFT, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase; DHFR, dihydrofolate reductase; MTHFR, 5,10-methylenetetrahydrofolate reductase; GNMT, glycine N-methyltransferase; FTCD, bifunctional formiminotransferase/cyclodeaminase; FMT, methionyl-tRNA formyltransferase; DMGDH, dimethylglycine dehydrogenase; SarDH, sarcosine dehydrogenase.
C1-synthase and FTCD are multifunctional enzymes; GARFT is a part of a trifunctional enzyme; AICART is a part of a bifunctional enzyme.
Only MTHFS activity has been observed in a mitochondrial fraction, the presence of the enzyme in mitochondria is still an open question.
DHFR also converts folic acid (a component of multivitamin supplementation/food fortification) to dihydrofolate.
GNMT does not use folate as a coenzyme but is regulated by 5-methyl-THF.
FDH converts 10-formyl-THF to THF and CO2 in a NADP+-dependent dehydrogenase reaction (Fig. 2 inset). The cytosolic form of this enzyme, ALDH1L1, has been known for a long time and is a well-characterized protein [4, 15]. ALDH1L1 appears to be a natural fusion of three unrelated genes that determines a complex domain structure of the protein. The functional domains, which compose the protein, are an aldehyde dehydrogenase (carboxyl-terminal), a folate-binding/hydrolase (amino-terminal), and an acyl carrier protein-like intermediate domain [34–36]. The conversion of 10-formyl-THF to THF and CO2 includes three steps, two catalytic and one transfer step. In the first step, the formyl group is removed from the folate molecule in a hydrolase reaction; in the second step, this group, covalently attached to a 4′-phosphopantetheine moiety of the intermediate domain, is transferred to the carboxyl-terminal domain where it undergoes oxidation through an ALDH-like mechanism as the third step [4]. The presence of the ALDH domain enables the enzyme to perform the aldehyde dehydrogenase catalysis as well [15]. It is not clear, however, whether this reaction has an independent physiological significance and what would be a substrate for the enzyme in such a reaction in vivo.
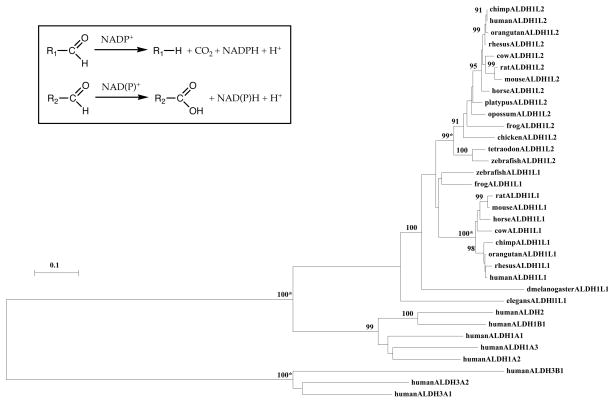
Fig. 2.
Phylogenetic tree of vertebrate and invertebrate ALDH1L and human ALDH proteins. The tree is labeled with the ALDH name and the name of the animal. Note the 5 major clusters corresponding to the vertebrate ALDH1L family; the vertebrate and invertebrate ALDH1L1 family; human class 1 ALDHs (ALDH1A1, ALDH1A2 and ALDH1A3); human ALDH1B1 and ALDH2; and human class 3 ALDHs (ALDH3A1, ALDH3A2 and ALDH1B1). A genetic distance scale is shown. The numbers of times a clade (sequences common to a node or branch) occurred in the bootstrap replicates are shown. Only replicate values of 90 or more which are highly significant are shown with 100 bootstrap replicates performed in each case. The asterisks (*) refer to significant gene duplication events leading to the divergence of ALDH1L1 and ALDH1L2 genes, and of ALDH1L and ALDH genes. Inset shows reactions catalyzed by ALDH1L enzymes (top reaction) and aldehyde dehydrogenases (bottom reaction). R1, THF (the aldehyde group is bound to THF through a C-N bond at N5); R2, broad spectra of moieties (the aldehyde group is bound to R2 through a C-C bond).
We have recently identified and characterized a homolog of ALDH1L1, ALDH1L2, which is the product of a separate gene. ALDH1L2 is a mitochondrial enzyme with high sequence similarity to ALDH1L1 [16]. An additional sequence at its amino-terminus is unique to ALDH1L2 and is a functional mitochondrial leader sequence which is absent in ALDH1L1. In the present study, we have evaluated the presence of both ALDH1L1 and ALDH1L2 genes in the genomes of several species and examined the organization and appearance of these genes during vertebrate and invertebrate evolution.
2. Materials and Methods
2.1. ALDH1L1 and ALDH1L2 gene and protein identification
BLAST (Basic Local Alignment Search Tool) studies were undertaken using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [37]. Protein BLAST analyses used ALDH1L1 amino acid sequences. Non-redundant protein sequence databases for several vertebrate and invertebrate genomes were examined using the BLASTP algorithm, including human (Homo sapiens), chimpanzee (Pan troglodytes), orangutan (Pongo abelii), rhesus monkey (Macaca mulata), cow (Bos Taurus), horse (Equus caballus), mouse (Mus musculus), rat (Rattus norvegicus), opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), frog (Xenopus tropicalis), zebrafish (Danio rerio), tetraodon fish (Tetraodon nigroviridis), fruit fly (Drosophila melanogaster) and nematode (Caenorhabditid elegans). This procedure produced multiple BLAST ‘hits’ for each of the protein databases which were individually examined and retained in FASTA format, and a record was kept of the sequences for predicted mRNAs and encoded ALDH1L-like proteins. These records were derived from annotated genomic sequences using the gene prediction method: GNOMON and predicted sequences with high similarity scores. Predicted ALDH1L-like protein sequences were obtained in each case and subjected to analyses of predicted protein and gene structures. BLAT analyses were subsequently undertaken for each of the predicted ALDH1L amino acid sequences using the UC Santa Cruz genome browser [http://genome.ucsc.edu/cgi-bin/hgBlat] [38] with the default settings to obtain the predicted locations for each of the ALDH1L genes, including predicted exon boundary locations and gene sizes. BLAT analyses were similarly undertaken for other human ALDH genes (Table 2).
Table 2.
Vertebrate and Invertebrate ALDH1L and Human ALDH Genes and Proteins
| Animal | Species | Gene | Chromosome Coordinates (NCBI) | Exon No (strand) | Gene Size (bps) | AA1 | NCBI Gene ID4 | Ensemble Transcript ID5 |
|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | ALDH1L1 | 3: 125,822,408 – 125,899,485 | 23 (−ve) | 77,078 | 902 | 10840 | ENST00000393434 |
| ALDH1L2 | 12: 105,418,202 – 105,478,236 | 23 (−ve) | 60,035 | 923 | 160428 | ENST00000258494 |
||
| Chimp | Pan troglodytes | ALDH1L1 | 3: 130,482,081 – 130,561,624 | 24 (−ve) | 79,544 | 1144 | 460655 | ENSPTRT00000028630 |
| ALDH1L2 | 12: 106,168,092 – 106,236,200 | 23 (−ve) | 68,109 | 923 | 452195 | ENSPTRT00000009886 |
||
| Orangutan | Pongo abelii | ALDH1L1 | 3: 6,608,523 – 6,684,098 | 22 (+ve) | 75,576 | 811 | 100172380 | ENSPPYT00000015651 |
| ALDH1L2 | 12: 106,775,750 – 106,845,415 | 24 (−ve) | 69,666 | 921 | 100459691 | ENSPPYT00000005804 | ||
| Rhesus | Macaca mulatta | ALDH1L1 | 2: 45,883,053 – 45,964,675 | 23 (−ve) | 81,623 | 912 | 716977 | ENSMMUT00000017512 |
| ALDH1L2 | 11: 106,144,801 – 106,206,822 | 23 (−ve) | 62,022 | 923 | 701242 | ENSMMUT00000045704 | ||
| Mouse | Mus musculus | ALDH1L1 | 12: 90,500,842 – 90,549,165 | 23 (+ve) | 48,324 | 902 | 107747 | ENSMUST00000130418 |
| ALDH1L2 | 10: 82,950,195 – 82,996,807 | 23 (−ve) | 46,613 | 923 | 216188 | ENSMUST00000020497 | ||
| Rat | Rattus norvegicus | ALDH1L1 | 4: 20,327,151 – 20,373,257 | 2 | 46,107 | 902 | 64392 | 2 |
| ALDH1L2 | 7: 22,398,067 – 22,450,381 | 25 (+ve) | 52,315 | 923 | 299699 | ENSRNOT00000059639 | ||
| Horse | Equus caballus | ALDH1L1 | Un3: 198,300 – 259,293 | 21 (−ve) | 60,994 | 905 | 100061108 | ENSECAT00000022380 |
| ALDH1L2 | 12: 28,298,287 – 28,351,588 | 23 (−ve) | 53,302 | 923 | 100053705 | ENSECAT00000026071 | ||
| Cow | Bos taurus | ALDH1L1 | 3: 127,196,357 – 127,224,313 | 24 (+ve) | 27,957 | 902 | 505677 | ENSBTAT00000019240 |
| ALDH1L2 | 5: 73,448,754 – 73,507,074 | 24 (−ve) | 58,321 | 923 | 516864 | ENSBTAT00000008298 | ||
| Opossum | Monodelphis domestica | ALDH1L2 | 8: 84,727,305 – 84,811,901 | 23 (−ve) | 84,597 | 933 | 100022528 | ENSMODT00000002576 |
| Platypus | Ornithorhynchus anatinus | ALDH1L2 | Un3: 22,773 – 60,939 | 23 (−ve) | 38,167 | 1010 | 100074128 | ENSOANT00000002867 |
| Chicken | Gallus gallus | ALDH1L2 | 1: 56,337,623 – 56,365,874 | 23 (+ve) | 28,252 | 922 | 418078 | ENSGALT00000020714 |
| Frog | Xenopus tropicalis | ALDH1L1 | 2 | 22 (+ve) | 2 | 902 | 496436 | ENSXETT00000025284 |
| ALDH1L2 | Un3: 349,398 – 378,431 | 22 (+ve) | 29,034 | 922 | 100127737 | ENSXETT00000003988 | ||
| Zebrafish | Danio rerio | ALDH1L1 | 6: 23,886,064 – 23,919,577 | 24 (+ve) | 33,514 | 903 | 798292 | ENSDART00000112636 |
| ALDH1L2 | 4: 8,500,281 – 8,520,437 | 22 (−ve) | 20,157 | 923 | 100333269 | ENSDART00000102893 | ||
| Tetraodon | Tetraodon nigroviridis | ALDH1L1 | Un3:14,197,867 – 14,204,712 | 21 (+ve) | 6,845 | 904 | NA | ENSTNIT00000005042 |
| Fruit fly | Drosophila melanogaster | ALDH1L1 | 2L: 21,371,232 – 21,377,995 | 3 (+ve) | 6,764 | 913 | 35407 | FBtr0081517 |
| Nematode | Caenorhabditis elegans | ALDH1L1 | IV: 11,022,573 – 11,025,841 | 7 (+ve) | 3,269 | 908 | 177999 | F36H1.6 |
| Human | Homo sapiens | ALDH1A1 | 9: 75,515,587 – 75,567,969 | 13 (−ve) | 52,383 | 501 | 216 | ENST00000297785 |
| ALDH1A2 | 15: 58,245,627 – 58,357,906 | 13 (−ve) | 112,280 | 518 | 8854 | ENST00000249750 | ||
| ALDH1A3 | 15: 101,420,009 – 101,456,831 | 13 (+ve) | 36,823 | 512 | 220 | ENST00000329841 | ||
| ALDH1B1 | 9: 38,392,702 – 38,398,658 | 2 (+ve) | 5,957 | 517 | 219 | ENST00000377698 | ||
| ALDH2 | 12: 112,204,346 – 112,247,784 | 13 (+ve) | 43,439 | 517 | 217 | ENST00000261733 | ||
| ALDH3A1 | 17: 19,641,297 – 19,651,746 | 11 (−ve) | 10,450 | 453 | 218 | ENST00000225740 | ||
| ALDH3A2 | 17: 19,552,064 – 19,580,908 | 10 (+ve) | 28,845 | 485 | 224 | ENST00000176643 | ||
| ALDH3B1 | 11: 67,777,790 – 67,796,743 | 10 (+ve) | 18,954 | 468 | 221 | ENST00000434449 |
Predicted NCBI amino acid sequence;
not available;
unknown chromosome;
Gene IDs were obtained from NCBI web sources http://www.ncbi.nlm.nih.gov/genbank/
Ensembl ID was derived from Ensembl genome database http://www.ensembl.org; bps refers to base pairs of nucleotide sequences as annotated by NCBI; the number of exons are listed as predicted by Ensembl.
2.2. Predicted Mitochondrial Targeting Sequences for Vertebrate ALDH1L2 Proteins
MitoProt web tools were used to predict the N-terminal protein region that can support a mitochondrial targeting sequence and the cleavage site for each of the predicted vertebrate ALDH1L2 sequences (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html) [39].
2.3. Phylogeny Studies and Sequence Divergence
Alignments of vertebrate ALDH-like protein sequences for ALDH1L (eg. residues 417-902 for human ALDH1L1 and residues 428-923 for human ALDH1L2) and human ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, ALDH3A1, ALDH3A2 and ALDH3B1 sequences (see Table 2 for sources) were assembled using BioEdit v.5.0.1 with the default settings [40]. Alignment of ambiguous regions, including the amino and carboxyl termini, were excluded prior to phylogenetic analysis yielding alignments of 396 residues for comparisons of vertebrate ALDH1L and human ALDH sequences with the fruit fly (Drosophila melanogaster) and nematode (Caenorhabditis elegans) ALDH1L1 sequences (Table 2). Evolutionary distances were calculated using the Kimura option [41] in TREECON [42]. Phylogenetic trees were constructed from evolutionary distances using the neighbor-joining method [43]. Tree topology was reexamined by the boot-strap method (100 bootstraps were applied) of resampling and only values that were highly significant (≥90) are shown [44].
3. Results
3.1. Gene Locations and Exonic Structures for Vertebrate and Invertebrate ALDH1L Genes
Table 2 summarizes the predicted locations for vertebrate and invertebrate ALDH1L-like genes based upon BLAT interrogations of several genomes using the reported sequences for human/mouse [45] and rat [15, 46] and the predicted sequences for other vertebrate genes and the UC Santa Cruz genome browser [38]. Predicted primate ALDH1L1 and ALDH1L2 genes were predominantly transcribed on the negative strand, with the exception of the orangutan (Pongo abelii) ALDH1L1 gene, which was transcribed on the positive strand. Vertebrate ALDH1L1 genes examined contain between 21 and 25 exons and ALDH1L2 genes contain between 22 and 25 exons (Table 2). Within the same species, the number of exons between the two genes can be equal or not. While this variability could be attributed to incomplete annotation of some genomes at present, in most genomes the first exon encodes the 5′-non-translatable mRNA region in ALDH1L1 genes and for the N-terminal mitochondrial targeting sequence in ALDH1L2 genes. The invertebrate genomes examined (fruit fly and nematode) exhibited only a single ALDH1L-like sequence which lacked the N-terminal mitochondrial targeting sequence in each case. Fewer exons were observed for the invertebrate ALDH1L1 genes examined, with the fruit fly (Drosophila melanogaster) and nematode (Caenorhabditid elegans) genes exhibiting 2 and 7 exons respectively. It is apparent however that the fused nature of ALDH1L1 and ALDH1L2 genes and proteins, previously reported for mammalian enzymes [4], have been retained for all of the invertebrate and other vertebrate genes and enzymes examined.
3.2. Phylogeny and Divergence of ALDH1L and Human ALDH1, ALDH2 and ALDH3 Sequences
A phylogenetic tree (Figure 2) was calculated by the progressive alignment of 24 vertebrate ALDH1L1 and ALDH1L2 amino acid sequences with human ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, ALDH3A1, ALDH3A2 and ALDH3B1 sequences with the fruit fly (Drosophila melanogaster) and nematode (Caenorhabditis elegans) ALDH1L1 sequences (Table 2). The phylogram showed clustering of the ALDH sequences into groups which were consistent with their evolutionary relatedness as well as groups for vertebrate ALDL1L1 and ALDH1L2 sequences, which were distinct from the human ALDH1-, ALDH2- and ALDH3-like sequences. The ALDH1L1 and ALDH1L2 groups were significantly different from each other (with bootstrap values of 99–100/100) supporting a hypothesis that these are distinct but related family groups. It is apparent from this study of vertebrate ALDH1L genes and proteins that this is an ancient protein for which a proposed common gene ancestor has predated the appearance of osseous fish > 500 million years ago [47]. In addition, the ALDL1L1 gene, which encodes the cytoplasmic form of this enzyme, may have served as the ancestral gene, given that both fruit fly and nematode genomes exhibited only a single ALDH1L1-like gene. Genetic distances for human, cow, mouse and rat ALDH1L1 and ALDH1L2 sequences calculated from the corresponding zebrafish sequences were 0.846±0.006 and 0.858±0.008, respectively, which suggests that these sequences are diverging at similar rates during vertebrate evolution.
4. Discussion
FDH, a multidomain enzyme, is the product of a fusion of three unrelated genes [4, 15, 36]. The two catalytic modules of the enzyme, the amino-terminal hydrolase and the carboxyl-terminal aldehyde dehydrogenase, retain their respective catalytic activities when expressed as individual proteins [46, 48]. The third module, an acyl carrier-like domain, couples the two catalytic domains together that produces a new catalytic activity, the 10-formyltetrahydrofolate dehydrogenase [36]. Several molecular mechanisms, including exon shuffling and gene duplication/fusion, must underlie the origin of such new chimeric genes from more simple ancient ones [49–51]. In fact, the complex domain organization seen in the FDH molecule is a common phenomenon in nature [52–54]. The presence of different functionalities within one protein molecule can be beneficial for several reasons. In the case of multifunctional enzymes, the combination of catalytic activities from the same metabolic pathway allows for substrate channeling, a process protecting unstable short-living intermediates, preventing the loss of a substrate due to diffusion and eliminating side-reactions [53, 55]. The expected effect of substrate channeling would be more efficient catalysis. Such multifunctional enzymes encoded by a single gene are found in several biochemical pathways including lipid metabolism [56], de novo purine and pyrimydine biosynthesis [20, 57], and folate metabolism [2, 58]. In some other proteins resulting from gene fusion or exon shuffling, the combination of domains could create a new function, and FDH is an outstanding example of this phenomenon.
The presence of ALDH1L1 gene obviously provided a selective advantage for higher organisms since it has been retained throughout their evolution. The importance of the new reaction for the cell, as well as its precise evolutionary advantage, is not completely understood. More ancient organisms (e.g. bacteria, plants and fungi) do not have a corresponding enzyme. The loss of Aldh1l1 in mice, while affecting the distribution of reduced folate pools, is not lethal and did not produce a distinct phenotype when animals are kept on a folate-rich diet [59]. These mice, however, demonstrated decreased reproductive efficiency [60]. We suggest that FDH controls the overall flux of one-carbon groups through the folate pool. In agreement with this hypothesis, it has been shown that the enzyme regulates the major folate-dependent biosynthetic processes, de novo purine pathways and regeneration of methionine from homocysteine [61–63]. The key role of FDH is associated with the fact that it functions as a catabolic enzyme with regard to the one-carbon group conversion: the enzyme removes these groups, in the form of CO2, from the folate pool thus counteracting biosynthetic processes. In this sense, FDH could serve to limit excessive proliferation, which is an unwanted process for most tissues in an adult organism. In support of this possibility, it has been observed that FDH is strongly and ubiquitously down-regulated in cancers [61].
It has also been suggested that FDH is a crucial component of the methanol detoxification pathway [64]. Methanol toxicity is primarily caused by its metabolite, formic acid, which is responsible for the metabolic acidosis and ocular toxicity observed in methanol-intoxicated humans [64]. Thus, on a more general note, the enzyme should be considered as a component of the formate degradation pathway. This pathway converts formate to neutral CO2, through 10-formyltetrahydrofolate as an intermediate. The two steps of this pathway are catalyzed by cytosolic C1-synthase and ALDH1L1, correspondingly. In the cell, formate is directly produced in pathways involving the degradation of 3-methyl-branched fatty acids and the shortening of 2-hydroxy long chain fatty acids [65]. In addition, methanol is produced during fermentation from the hydrolysis of fruit pectin and thus is present in juices and alcoholic beverages [66]. Interestingly, the artificial sweetener aspartame also generates a small amount of methanol [67]. Importantly, it has been demonstrated that the ALDH1L1 pathway is more prominent for the clearance of lower, physiological doses of formate [68]. Bacteria, yeast and plants possess an enzyme, formate dehydrogenase (EC 1.2.1.2), which directly oxidizes formate to CO2 [69], and this enzyme is not found in higher animals. Thus, it can be speculated that FDH-catalyzed reaction evolved as a compensatory pathway to clear formate.
The mitochondrial FDH (mtFDH, ALDH1L2) is structurally very similar to the cytosolic enzyme [16]. While ALDH1L1 was obviously the natural product of the gene fusion, mtFDH most likely was the result of a duplication of the ALDH1L1 gene. This point of view is supported by the fact that mtFDH is seen later on the evolutionary tree than cytosolic FDH and that the two ALDH1L genes have higher similarity to each other than to the potential ALDH ancestors. For instance, for the ALDH domain, the similarity between the two proteins is about 79% while the closest member of ALDH family, retinaldehyde dehydrogenase, is only about 50% similar to either of the FDH isoforms. Of note, gene duplication is not uncommon for folate enzymes with at least two other examples known, MTHFD and SHMT [2, 3, 70]. Interestingly, another genetic mechanism that can create mitochondrial and cytosolic isoforms is through alternative splicing. In this mechanism, the exon encoding for a mitochondrial leader sequence is spliced out which produces a protein localized to the cytosol. The two isoforms of FPGS, mitochondrial and cytosolic, are the result of the alternative splicing of a single gene [18]. Moreover, this mechanism is also possible for the gene encoding mitochondrial SHMT [8]. Such a mechanism, however, has not been seen in the ALDH gene family.
In the case of the ALDH1L genes, it appears that an opposing mechanism was responsible for the creation of the mitochondrial enzyme: instead of losing a mitochondrial leader sequence, its acquisition took place. This has apparently occurred without significant changes in the ALDH1L1 gene organization. Thus, both genes, ALDH1L1 and ALDH1L2, have a similar number of exons in all species examined, with some species having identical number of exons in both genes and others demonstrating the difference of only one or two exons (Table 2). The first exon of ALDH1L1 is non-translatable, but it encodes for the mitochondrial leader sequence in the case of ALDH1L2. Evidently, alterations within this exon allowed for the acquisition of a mitochondrial leader sequence.
It is not clear whether the presence of two ALDH1L genes, and thus two FDH isoforms, provides a selective advantage for a species or whether it was a random act of a gene duplication. C. elegans and insects have only the ALDH1L1 gene. Our recent analysis of the zebrafish genome revealed two FDH-like genes, for which protein products were predicted to reside in mitochondria. In addition, the combination of these proteins corresponded to a full-length FDH [16]. However, these two genes have been deleted from the most recent annotation of the zebrafish genome, and a new annotation of the ALDH1L2 gene encoding for full-length mitochondrial FDH has been included in the database. Thus, the duplication of ALDH1L1 gene took place prior to appearance of osseous fish. The present study identified only a single FDH gene in birds, ALDH1L2, which may be due to the incomplete annotation of ALDH1L genes and proteins in these species. Alternatively, the two ALDH1L genes may be redundant in bird species and similar to mitochondrial SHMT in mice [8], avian ALDH1L2 could produce a cytosolic enzyme through an alternative splicing mechanism. Whether alternative splicing of ALDH1L2 is possible or not is unclear at present, but a preliminary analysis of this gene did not indicate such a splice variant.
5. Conclusion
BLAST and BLAT analyses of several vertebrate genome databases were undertaken using amino acid sequences reported for human ALDH1L1 (cytosolic) and ALDH1L2 (mitochondrial) enzymes for interrogation of vertebrate genomes. Evidence is presented for ALDH1L1 and ALDH1L2 genes in all vertebrate genomes examined, with the exception of opossum, platypus and chicken genomes, for which only ALDH1L2 sequences were observed. This may be due to an incomplete annotation of ALDH1L1 sequences or to an alternative mechanism (such as differential splicing of ALDH1L2) in generating a cytosolic form of ALDH1L in these species. Predicted amino acid sequences for vertebrate ALDH1L-like subunits showed a high degree of similarity with the corresponding human enzymes. Phylogenetic analyses supported a hypothesis concerning the molecular evolution of vertebrate ALDH1L-like genes: vertebrate ALDH1L1 and ALDH1L2 genes were generated within a common ancestral genome (for vertebrates) by a duplication of the gene encoding cytosolic ALDH1L1, prior to the appearance of osseous fish, more than 500 million years ago [47].
Acknowledgments
This study was supported by the National Institutes of Health grant DK054388 (SAK) and a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral MD/PhD Fellows F30DK083215 (KCS). KCS was supported by USPHS NIH grant R13-AA019612 to present this work at the 15th International Meeting on Enzymology and Molecular Biology of Carbonyl Metabolism in Lexington, KY USA.
Abbreviations
- ALDH
aldehyde dehydrogenase
- FDH
10-formyltetrahydrofolate dehydrogenase
- mtFDH
mitochondrial FDH
- THF
tetrahydrofolate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wagner C. Biochemical role of folate in cellular metabolism. In: Bailey LB, editor. Folate in Health and Disease. Marcel Dekker, Inc; New York: 1995. pp. 23–42. [Google Scholar]
- 2.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 3.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 4.Krupenko SA. FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism. Chem Biol Interact. 2009;178(1–3):84–93. doi: 10.1016/j.cbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1(2):171–176. [PubMed] [Google Scholar]
- 6.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. Faseb J. 1991;5(12):2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 7.Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem. 2007;282(24):17623–17631. doi: 10.1074/jbc.M702526200. [DOI] [PubMed] [Google Scholar]
- 8.Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One. 2009;4(6):e5839. doi: 10.1371/journal.pone.0005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titus SA, Moran RG. Retrovirally mediated complementation of the glyB phenotype. Cloning of a human gene encoding the carrier for entry of folates into mitochondria. J Biol Chem. 2000;275(47):36811–36817. doi: 10.1074/jbc.M005163200. [DOI] [PubMed] [Google Scholar]
- 10.Garrow TA, Brenner AA, Whitehead VM, Chen XN, Duncan RG, Korenberg JR, Shane B. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J Biol Chem. 1993;268(16):11910–11916. [PubMed] [Google Scholar]
- 11.Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem. 1997;272(3):1842–1848. doi: 10.1074/jbc.272.3.1842. [DOI] [PubMed] [Google Scholar]
- 12.Hum DW, Bell AW, Rozen R, MacKenzie RE. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988;263(31):15946–15950. [PubMed] [Google Scholar]
- 13.Christensen KE, Patel H, Kuzmanov U, Mejia NR, MacKenzie RE. Disruption of the mthfd1 gene reveals a monofunctional 10-formyltetrahydrofolate synthetase in mammalian mitochondria. J Biol Chem. 2005;280(9):7597–7602. doi: 10.1074/jbc.M409380200. [DOI] [PubMed] [Google Scholar]
- 14.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285(7):4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook RJ, Lloyd RS, Wagner C. Isolation and characterization of cDNA clones for rat liver 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1991;266(8):4965–4973. [PubMed] [Google Scholar]
- 16.Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 285(30):23056–23063. doi: 10.1074/jbc.M110.128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrow TA, Admon A, Shane B. Expression cloning of a human cDNA encoding folylpoly(gamma-glutamate) synthetase and determination of its primary structure. Proc Natl Acad Sci U S A. 1992;89(19):9151–9155. doi: 10.1073/pnas.89.19.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freemantle SJ, Moran RG. Transcription of the human folylpoly-gamma-glutamate synthetase gene. J Biol Chem. 1997;272(40):25373–25379. doi: 10.1074/jbc.272.40.25373. [DOI] [PubMed] [Google Scholar]
- 19.Jolivet J, Dayan A, Beauchemin M, Chahla D, Mamo A, Bertrand R. Biochemical and molecular studies of human methenyltetrahydrofolate synthetase. Stem Cells. 1996;14(1):33–40. doi: 10.1002/stem.140033. [DOI] [PubMed] [Google Scholar]
- 20.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 21.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci U S A. 107(29):12872–12876. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R. Human methylenetetrahydrofolate reductase: isolation of cDNA mapping and mutation identification. Nat Genet. 1994;7(4):551. [PubMed] [Google Scholar]
- 23.Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7(2):195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig ML, Matthews RG. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 25.Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2009;284(34):22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murley LL, Mejia NR, MacKenzie RE. The nucleotide sequence of porcine formiminotransferase cyclodeaminase. Expression and purification from Escherichia coli. J Biol Chem. 1993;268(30):22820–22824. [PubMed] [Google Scholar]
- 27.Hennig D, Scales SJ, Moreau A, Murley LL, De Mey J, Kreis TE. A formiminotransferase cyclodeaminase isoform is localized to the Golgi complex and can mediate interaction of trans-Golgi network-derived vesicles with microtubules. J Biol Chem. 1998;273(31):19602–19611. doi: 10.1074/jbc.273.31.19602. [DOI] [PubMed] [Google Scholar]
- 28.Mao Y, Vyas NK, Vyas MN, Chen DH, Ludtke SJ, Chiu W, Quiocho FA. Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer. EMBO J. 2004;23(15):2963–2971. doi: 10.1038/sj.emboj.7600327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84(7):246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli LL, Watanabe K. Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J Biol Chem. 1998;273(24):15085–15090. doi: 10.1074/jbc.273.24.15085. [DOI] [PubMed] [Google Scholar]
- 31.Binzak BA, Vockley JG, Jenkins RB, Vockley J. Structure and analysis of the human dimethylglycine dehydrogenase gene. Mol Genet Metab. 2000;69(3):181–187. doi: 10.1006/mgme.2000.2980. [DOI] [PubMed] [Google Scholar]
- 32.Binzak BA, Wevers RA, Moolenaar SH, Lee YM, Hwu WL, Poggi-Bach J, Engelke UF, Hoard HM, Vockley JG, Vockley J. Cloning of dimethylglycine dehydrogenase and a new human inborn error of metabolism, dimethylglycine dehydrogenase deficiency. Am J Hum Genet. 2001;68(4):839–847. doi: 10.1086/319520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eschenbrenner M, Jorns MS. Cloning and mapping of the cDNA for human sarcosine dehydrogenase, a flavoenzyme defective in patients with sarcosinemia. Genomics. 1999;59(3):300–308. doi: 10.1006/geno.1999.5886. [DOI] [PubMed] [Google Scholar]
- 34.Chumanevich AA, Krupenko SA, Davies C. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain. J Biol Chem. 2004;279(14):14355–14364. doi: 10.1074/jbc.M313934200. [DOI] [PubMed] [Google Scholar]
- 35.Tsybovsky Y, Donato H, Krupenko NI, Davies C, Krupenko SA. Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases. Biochemistry. 2007;46(11):2917–2929. doi: 10.1021/bi0619573. [DOI] [PubMed] [Google Scholar]
- 36.Donato H, Krupenko NI, Tsybovsky Y, Krupenko SA. 10-formyltetrahydrofolate dehydrogenase requires a 4′-phosphopantetheine prosthetic group for catalysis. J Biol Chem. 2007;282(47):34159–34166. doi: 10.1074/jbc.M707627200. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 40.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–99. [Google Scholar]
- 41.Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Chambridge: 1983. [Google Scholar]
- 42.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10(5):569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, Guyer M, Peck AM, Derge JG, Lipman D, Collins FS, Jang W, Sherry S, Feolo M, Misquitta L, Lee E, Rotmistrovsky K, Greenhut SF, Schaefer CF, Buetow K, Bonner TI, Haussler D, Kent J, Kiekhaus M, Furey T, Brent M, Prange C, Schreiber K, Shapiro N, Bhat NK, Hopkins RF, Hsie F, Driscoll T, Soares MB, Casavant TL, Scheetz TE, Brown-stein MJ, Usdin TB, Toshiyuki S, Carninci P, Piao Y, Dudekula DB, Ko MS, Kawakami K, Suzuki Y, Sugano S, Gruber CE, Smith MR, Simmons B, Moore T, Waterman R, Johnson SL, Ruan Y, Wei CL, Mathavan S, Gunaratne PH, Wu J, Garcia AM, Hulyk SW, Fuh E, Yuan Y, Sneed A, Kowis C, Hodgson A, Muzny DM, McPherson J, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madari A, Young AC, Wetherby KD, Granite SJ, Kwong PN, Brinkley CP, Pearson RL, Bouffard GG, Blakesly RW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Griffith M, Griffith OL, Krzywinski MI, Liao N, Morin R, Palmquist D, Petrescu AS, Skalska U, Smailus DE, Stott JM, Schnerch A, Schein JE, Jones SJ, Holt RA, Baross A, Marra MA, Clifton S, Makowski KA, Bosak S, Malek J. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14(10B):2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krupenko SA, Wagner C, Cook RJ. Expression, purification, and properties of the aldehyde dehydrogenase homologous carboxyl-terminal domain of rat 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1997;272(15):10266–10272. doi: 10.1074/jbc.272.15.10266. [DOI] [PubMed] [Google Scholar]
- 47.Donoghue PC, Benton MJ. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol Evol. 2007;22(8):424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Krupenko SA, Wagner C, Cook RJ. Domain structure of rat 10-formyltetrahydrofolate dehydrogenase. Resolution of the amino-terminal domain as 10-formyltetrahydrofolate hydrolase. J Biol Chem. 1997;272(15):10273–10278. doi: 10.1074/jbc.272.15.10273. [DOI] [PubMed] [Google Scholar]
- 49.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4(11):865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 50.Moore AD, Bjorklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem Sci. 2008;33(9):444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. Domain rearrangements in protein evolution. J Mol Biol. 2005;353(4):911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 52.Hawkins AR, Lamb HK. The molecular biology of multidomain proteins. Selected examples. Eur J Biochem. 1995;232(1):7–18. doi: 10.1111/j.1432-1033.1995.tb20775.x. [DOI] [PubMed] [Google Scholar]
- 53.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 54.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14(2):208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Spivey HO, Ovadi J. Substrate channeling. Methods. 1999;19(2):306–321. doi: 10.1006/meth.1999.0858. [DOI] [PubMed] [Google Scholar]
- 56.Bhaumik P, Koski MK, Glumoff T, Hiltunen JK, Wierenga RK. Structural biology of the thioester-dependent degradation and synthesis of fatty acids. Curr Opin Struct Biol. 2005;15(6):621–628. doi: 10.1016/j.sbi.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 57.Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem. 2004;279(32):33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 58.Ivanetich KM, Santi DV. Bifunctional thymidylate synthase-dihydrofolate reductase in protozoa. FASEB J. 1990;4(6):1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 59.Champion KM, Cook RJ, Tollaksen SL, Giometti CS. Identification of a heritable deficiency of the folate-dependent enzyme 10-formyltetrahydrofolate dehydrogenase in mice. Proc Natl Acad Sci U S A. 1994;91(24):11338–11342. doi: 10.1073/pnas.91.24.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giometti CS, Tollaksen SL, Grahn D. Altered protein expression detected in the F1 offspring of male mice exposed to fission neutrons. Mutat Res. 1994;320(1–2):75–85. doi: 10.1016/0165-1218(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 61.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13(5):227–236. [PubMed] [Google Scholar]
- 62.Oleinik NV, Krupenko NI, Priest DG, Krupenko SA. Cancer cells activate p53 in response to 10-formyltetrahydrofolate dehydrogenase expression. Biochem J. 2005;391(Pt 3):503–511. doi: 10.1042/BJ20050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anguera MC, Field MS, Perry C, Ghandour H, Chiang EP, Selhub J, Shane B, Stover PJ. Regulation of Folate-mediated One-carbon Metabolism by 10-Formyltetrahydrofolate Dehydrogenase. J Biol Chem. 2006;281(27):18335–18342. doi: 10.1074/jbc.M510623200. [DOI] [PubMed] [Google Scholar]
- 64.Tephly TR. The toxicity of methanol. Life Sci. 1991;48(11):1031–1041. doi: 10.1016/0024-3205(91)90504-5. [DOI] [PubMed] [Google Scholar]
- 65.Casteels M, Sniekers M, Fraccascia P, Mannaerts GP, Van Veldhoven PP. The role of 2-hydroxyacyl-CoA lyase, a thiamin pyrophosphate-dependent enzyme, in the peroxisomal metabolism of 3-methyl-branched fatty acids and 2-hydroxy straight-chain fatty acids. Biochem Soc Trans. 2007;35(Pt 5):876–880. doi: 10.1042/BST0350876. [DOI] [PubMed] [Google Scholar]
- 66.Hang YD, Woodams EE. Influence of apple cultivar and juice pasteurization on hard cider and eau-de-vie methanol content. Bioresour Technol. 101(4):1396–1398. doi: 10.1016/j.biortech.2009.09.069. [DOI] [PubMed] [Google Scholar]
- 67.Tephly TR. Comments on the purported generation of formaldehyde and adduct formation from the sweetener aspartame. Life Sci. 1999;65(13):PL157–160. doi: 10.1016/s0024-3205(99)00382-3. [DOI] [PubMed] [Google Scholar]
- 68.Cook RJ, Champion KM, Giometti CS. Methanol toxicity and formate oxidation in NEUT2 mice. Arch Biochem Biophys. 2001;393(2):192–198. doi: 10.1006/abbi.2001.2485. [DOI] [PubMed] [Google Scholar]
- 69.Popov VO, Lamzin VS. NAD(+)-dependent formate dehydrogenase. Biochem J. 1994;301(Pt 3):625–643. doi: 10.1042/bj3010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christensen KE, MacKenzie RE. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. Bioessays. 2006;28(6):595–605. doi: 10.1002/bies.20420. [DOI] [PubMed] [Google Scholar]