Summary
Escherichia coli DksA is an RNA polymerase (RNAP)-binding protein required for the regulation of a number of promoters, including those for the biosynthesis of rRNA, many ribosomal proteins, components of the flagellum, and several amino acids. Previous work demonstrated that DksA protein levels do not vary greatly in different growth conditions. We show here that DksA is a stable protein whose levels are kept constant by a negative feedback loop in which transcription from the dksA promoter is inhibited by DksA protein in conjunction with its cofactor ppGpp. We map the primary dksA promoter by primer extension, show that its activity increases in a strain lacking DksA, that the DksA protein accumulates when expressed from an exogenous promoter, that inhibition of transcription by DksA is direct since it occurs with purified components in vitro, and that inhibition correlates with effects of DksA on the lifetime of the dksA promoter complex. This work provides a mechanistic basis for the maintenance of constant cellular levels of DksA in E. coli and supports the model that transcription regulation by ppGpp/DksA derives from fluctuations in the concentrations of the small molecule cofactor rather than of DksA itself.
Introduction
E. coli DksA is a 151 amino acid zinc-containing protein that binds to RNA polymerase (RNAP) and is essential for the proper regulation of the rrn P1 promoters and many other promoters in vivo and in vitro (for reviews, Paul et al., 2004b; Magnusson et al., 2005; Haugen et al., 2008; Srivatsan and Wang, 2008; Dalebroux et al., 2010). By binding in the secondary channel of RNAP, DksA modifies the promoter complex, altering the kinetics of transcription initiation (Paul et al., 2004a; Perederina et al., 2004; Rutherford et al., 2009; Lennon et al., 2009). DksA also alleviates problems caused by collisions between the DNA replication and transcription machineries and facilitates DNA repair (Meddows et al., 2005; Trautinger et al., 2005; Tehranchi et al., 2010).
We have suggested that the concentrations of two cofactors that work in conjunction with DksA, namely ppGpp (used here to refer to both guanosine-3',5'-(bis)pyrophosphate and its pentaphosphate precursor) and the initial NTP in the transcript (iNTP) fluctuate in response to changes in nutritional conditions, resulting in altered transcriptional output (Murray et al., 2003). For example, when cells are starved for amino acids, ppGpp levels increase dramatically, leading to the dramatic decrease in rRNA promoter activity characteristic of the stress response known as stringent control (Potyrkus and Cashel, 2008). Similarly, during carbon source downshifts in exponential growth, increases in ppGpp concentrations lead to inhibition of rRNA promoter activity, and in early stationary phase, increases in ppGpp concentrations coupled with decreases in iNTP concentrations combine to decrease rRNA promoter activity. As cells progress through stationary phase, ppGpp concentrations return to basal levels and iNTP concentrations remain low, keeping rRNA promoter activity low (Murray et al., 2003). In contrast, during outgrowth from stationary phase, iNTP concentrations increase rapidly, increasing rRNA transcription, and during carbon source upshifts, ppGpp concentrations decrease, increasing rRNA promoter activity (Murray et al., 2003). Mutants lacking dksA fail to exert responses to each of these changes in ppGpp and/or iNTP concentrations (Paul et al., 2004a).
ppGpp/DksA inhibits a variety of promoters (Paul et al., 2004a; Mallik et al., 2006; Lemke et al., 2009; Lemke et al., 2011) and directly activates other promoters (Paul et al., 2005; Nakanishi et al., 2006; Sharma and Payne, 2006; Costanzo et al., 2008). Expression profiling has suggested that the effects of ppGpp/DksA are truly global in scale, resulting in alterations of the transcriptional profiles of hundreds of transcripts (J.J.L. and R.L.G., unpublished data; Magnusson et al., 2007; Durfee et al., 2008; Traxler et al., 2008), some directly, some indirectly. It has been reported that DksA and ppGpp may even have opposite effects in certain cases (Aberg et al., 2009).
Our previous studies suggested that DksA protein levels do not fluctuate with changes in growth rate or growth phase. DksA concentrations do not increase in stationary phase to account for the decrease in rRNA transcription at that growth stage, and DksA levels do not decrease during outgrowth to account for the increase in rRNA transcription at that time (Paul et al., 2004a; Rutherford et al., 2007). Therefore, we proposed that (at least in E. coli) fluctuations in ppGpp and/or iNTPs levels, and not changes in DksA levels, serve as the regulatory signals generated by the translational state of the cell (Paul et al., 2004a; Rutherford et al., 2007). However, it was also reported that DksA is a target of degradation by ClpXP (Flynn et al., 2003). This observation suggested that DksA levels might be modulated under some conditions by a mechanism that operates at the level of protein stability. Furthermore, it has been reported recently that DksA levels do vary in Chlamydia and Pseudomonas (see Discussion and Perron et al., 2005; Mukhopadhyay et al., 2006; Blaby-Haas et al., 2010).
In order to investigate the mechanism(s) responsible for DksA action further, we examined its synthesis and stability in E. coli. We demonstrate here that DksA is long-lived, making it unlikely that changes in its stability could modulate rapid transcriptional responses. Instead, by inhibiting transcription from its own promoter, DksA creates a negative feedback loop that keeps its levels constant. Thus, our data support the model that in E. coli DksA sensitizes RNAP to changes in the levels of ppGpp and NTPs, whose fluctuations account for the rapid responses of DksA-regulated promoters to changes in nutritional conditions.
Results
DksA is a stable protein
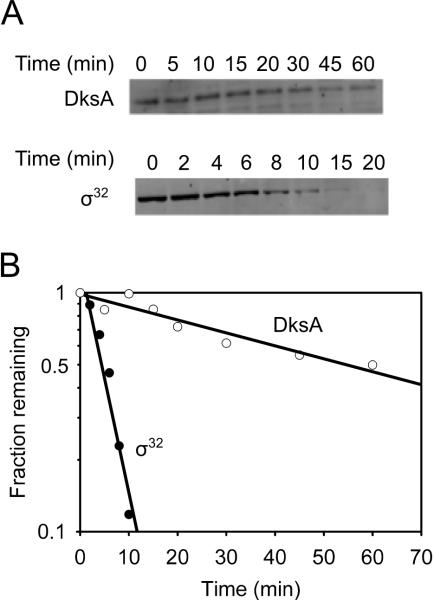
Ribosomal RNA promoters are inhibited very rapidly following starvation for amino acids (Murray et al., 2003). In theory, rapid stabilization of an unstable inhibitor of rRNA transcription such as DksA could lead to a rapid increase in the concentration of the inhibitor and a rapid decrease in rRNA transcription. To test if DksA is an unstable protein, we measured its lifetime. Samples from a culture were withdrawn at fixed time intervals after addition of a translation inhibitor, and the amount of DksA remaining at each time was determined using Western blots. DksA had a half-life of 44 ± 9 min (Fig. 1). As a control that our methods were capable of measuring short-lived proteins, we found that σ32 had a lifetime of 6 ± 1 min following a heat shock (Fig. 1), consistent with the lifetime of this protein reported previously (Grossman et al., 1987). We conclude that DksA is too long-lived under these conditions for a reduction in its turnover to lead to increased DksA levels and rapid inhibition of rRNA transcription (Murray et al., 2003).
Figure 1. DksA is a stable protein.
(A) Representative Western blots used to determine the stabilities of DksA and σ32 (as a control) (see Experimental Procedures). (B) Quantitation of western blot shown in (A). Assays were performed 3 times. The mean half-lives (with standard deviations) were 44 min ± 9 for DksA and 6 min ± 1 for σ32.
We showed previously that DksA concentrations remained constant throughout growth and declined slightly (two-fold) only after many hours in stationary phase (Rutherford et al., 2007). These data suggested that increased ppGpp and reduced NTP concentrations, rather than changes in the concentration of DksA, account for the reduction in rRNA transcription observed in stationary phase (Murray et al., 2003). Consistent with the observed high levels of DksA in all phases of growth, we found that DksA is also stable in stationary phase (18 hours after culture inoculation; data not shown).
Identification of the dksA promoter
DksA is stable and yet it does not accumulate during stationary phase when it would not be diluted out by cell division. Therefore, we reasoned there must be a mechanism regulating its synthesis, perhaps inhibition of transcription initiation.
Previous S1 nuclease mapping studies (Kang and Craig, 1990) had suggested that there might be two dksA transcription start sites in the intergenic region between sfsA and dksA. As a first step in examining control of synthesis of DksA, we constructed dksA promoter-lacZ fusions that contained either the full intergenic region between dksA and the gene upstream from dksA, sfsA (construct 1), or each of the putative promoters individually (constructs 2 and 3) (Fig. 2A; sequence endpoints of promoter fragments are in Table 1). Construct 2 accounted for most of the transcription in log phase (Fig. 2B), and we found little activity associated with the fragment correlating with the upstream region, construct 3 (Kang and Craig, 1990). Consistent with the model that the synthesis of DksA is inhibited in stationary phase, the activity of construct 2 was much lower in stationary than in log phase (Fig. 2C).
Figure 2. Identification of the dksA transcription start site.
(A) Schematic diagram of the sfsA-dksA region. Arrow indicates position of the dksA transcription start site determined below. DNA fragments 1-3 used to make promoter-lacZ fusions (construct 1, RLG10100, construct 2, RLG10101; construct 3, RLG10102). Sequence endpoints are indicated; +1 is the major transcription start site. (B) Log phase and (C) stationary phase ß-galactosidase activities from promoter-lacZ fusions using fragments 1-3 illustrated in (A). Error bars represent the standard deviations from 6 independent cultures. (D) Primer extension analysis to determine the transcription start site for dksA. The lane labeled “RNA” indicates that the product derived from extension of a primer annealed to the RNA transcript. Fragments used for promoter-lacZ fusions in (B) and (C) are aligned with the DNA sequences at left. (E) DNA sequence of the promoter region. The putative -35 element and extended -10 element are underlined. The transcription start site is indicated by an arrow.
Table 1. Strains and plasmids used in this study.
Promoter end-points are numbered relative to the transcription start site. All promoter-lacZ fusions are on phage lambda prophage on the bacterial chromosome.
| Strain | Relevent genotype | Promoter-lacZ fusion | Promoter endpoints | Source |
|---|---|---|---|---|
| VH1000 | RLG3499=MG1655 pyrE+ lacZ- lacl- | Gaal et al., 1997 | ||
| RLG4996 | VH1000 | rrnB P1 | -46/+1 | Paul et al., 2004a |
| RLG6348 | VH1000 dksA::tet | rrnB P1 | -46/+1 | Paul et al., 2004a |
| RLG4998 | VH1000 | lacUV5 | -59/+36 | Barker et al., 2001 |
| RLG8950 | VH1000 dksA::tet | lacUV5 | -59/+36 | Lemke et al., 2009 |
| RLG10100 | VH1000 | PdksA (construct 1) | -180/+37 | This work |
| RLG10101 | VH1000 | PdksA (construct 2) | -100/+37 | This work |
| RLG10102 | VH1000 | PdksA (construct 3) | -180/100 | This work |
| RLG10103 | RLG10101 dksA::tet | PdksA | -100/+37 | This work |
| RLG10104 | RLG10103 pRLG6333 | PdksA | -100/+37 | This work |
| RLG10109 | VH1000 pRLG10106 | This work | ||
| RLG10112 | VH1000 pRLG10108 | This work |
| Plasmid | Description | Promoter endpoints | Source |
|---|---|---|---|
| pRLG770 | Transcription vector | Ross et al., 1990 | |
| pMSB1 | Cloning vector for lambda recombination | Rao et al., 1994 | |
| pRLG2222 | pRLG770 containing PlacUV5 | -59/+36 | Ross et al., 1990 |
| pRLG3422 | pRLG770 containing PlacUV5 | -46/+1 | Gaal et al., 1997 |
| pRLG6332 | Complementation vector derived from pINIIIAI | Masui et al., 1984 | |
| pRLG6333 | pRLG6332 encoding DksA | Paul et al., 2004a | |
| pRLG6798 | pRLG770 containing rrnB P1 | -66/+50 | Haugen et al., 2006 |
| pRLG8110 | pRLG770 containing Pfis | -166/+83 | Mallik et al., 2006 |
| pRLG10105 | pRLG770 containing PdksA | -100/+37 | This work |
| pRLG10106 | pRLG770 containing PdksA promoter and dksA gene = PdksA-dksA | This work |
We mapped a major transcription start site within the region covered by construct 2 by primer extension from exponential phase RNA, corresponding to a C residue 54 bp upstream of the dksA translation start site (Fig. 2A, 2D). This dksA transcription start site is separated from a likely -10 element (TATTTA) by 7 bp, a likely extended -10 element (TG) is separated from the -10 element by 1 bp, and a likely -35 element (TTGCTA) is separated from the -10 element by 17 bp (Fig. 2E). This transcription start site corresponds to one of the products identified in the earlier report (Kang and Craig, 1990) and is annotated as a promoter in EcoCyc (Keseler et al., 2009). Consistent with the hypothesis that the dksA promoter is inhibited in stationary phase, this start site signal was much weaker or not detected with RNA from stationary phase cells (data not shown). No products were detected in the region corresponding to construct 3, using the same primer used for detecting the transcription start site within the region covered by construct 2 (Fig. 2D) or using primers that hybridized within the region covered by construct 3 (data not shown). Since the β-galactosidase activity derived from the region covered by construct 3 is at least 100-fold above background, we suspect that there may be multiple sequences in this region that serve as promoters but whose individual activities are still too weak to be detected by primer extension. Additional information will be required to explain why construct 1 has a higher activity in stationary phase than the sum of constructs 2 and 3.
Regulation of DksA levels is promoter-specific
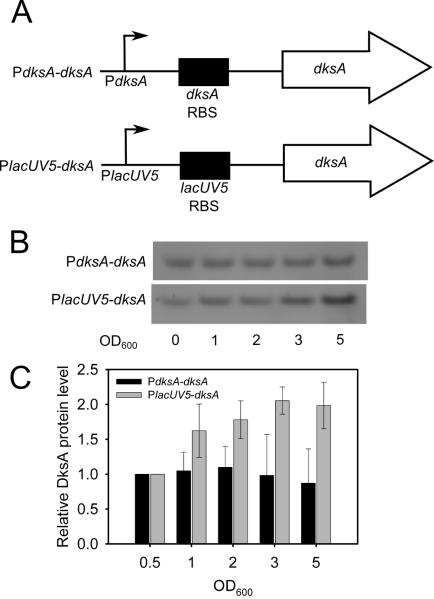
To determine if inhibition of the dksA promoter prevents accumulation of DksA in stationary phase, we used Western blots to compare the amount of DksA made from multicopy plasmid constructs in which the dksA gene was either expressed from its natural promoter (PdksA-dksA) in a ΔdksA mutant strain or from the lacUV5 promoter (PlacUV5-dksA) (Fig. 3A). In the PlacUV5 construct, the fusion junction was at the transcription start site, i.e. the mRNA derived entirely from the dksA gene (see Experimental Procedures).
Figure 3. DksA expressed from an exogenous promoter accumulates in stationary phase.
(A) Schematic diagrams of promoter constructs. Arrows represent the transcription start site, and RBS represents the ribosome binding site. (B) Representative western blots from ΔdksA cells transformed either with the plasmid in which DksA was expressed from its native promoter (pRLG10106) or from the lacUV5 promoter (pRLG10108) at 37°C in LB. Samples were collected at the indicated optical densities. The cells reached early stationary phase by OD600 = 1. (C) Quantitation of DksA protein levels from three independent experiments. The amount of DksA at each cell density was normalized to that at OD600 = 0.5.
DksA protein levels from the plasmids described above were measured from log phase and stationary phase cultures in rich medium (Fig. 3B and 3C). Relative to total protein and normalized to the amount of DksA present in log phase, DksA levels remained constant from log and stationary phase cells when the protein was expressed from the dksA promoter (black bars). However, when DksA was expressed from the lacUV5 promoter (grey bars), it accumulated by OD600 ~5.0 (stationary phase) to > 2-fold the amount at OD600 ~0.5 (log phase). We conclude that regulation of the dksA promoter prevents accumulation of DksA in stationary phase.
DksA inhibits transcription from its own promoter in vivo
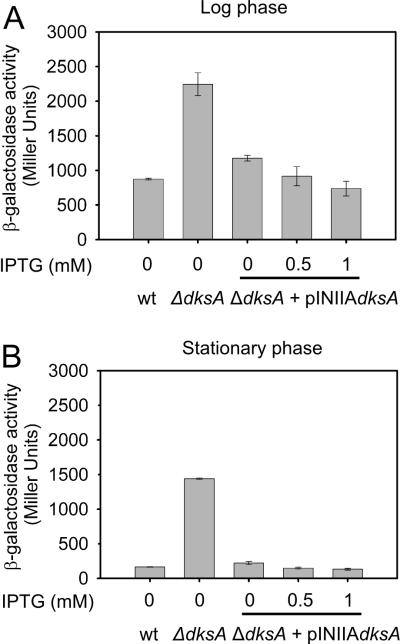
Since DksA is a transcription regulator, we next examined whether it autoregulates its own promoter by comparing the activities of a dksA promoter – lacZ fusion in strains wild-type or mutant for dksA. The dksA promoter was much more active in log phase in wild-type cells (leftmost bar in Fig. 4A) than in stationary phase (leftmost bar in Fig. 4B), as in Fig. 2. Most important, dksA promoter activity was 2 to 3-fold higher in a ΔdksA mutant than in a wild-type strain in log phase (OD600 ~0.4) (second bar in Fig. 4A) and ~15-fold higher in stationary phase (OD600 ~5) (second bar in Fig. 4B), consistent with the model that DksA negatively autoregulates its own synthesis. Furthermore, a plasmid producing close to wild-type levels of DksA from an uninduced lpp-lac promoter (Rutherford et al., 2007; see Experimental Procedures) reduced PdksA promoter activity to almost its native level (middle bar in Figs. 4A and 4B), and overexpression of DksA by induction with 0.5 or 1 mM IPTG (a condition in which DksA is overexpressed ~10-fold compared to wild-type cells; Rutherford et al., 2007), reduced dksA promoter activity more (to its native level; rightmost two bars in Figs. 4A and 4B). Taken together, these data suggest that DksA inhibits transcription from its own promoter.
Figure 4. The dksA promoter is negatively regulated by DksA in vivo.
ß-galactosidase activities were measured from a dksA promoter-lacZ fusion in wild-type (RLG10101) or ΔdksA (RLG10103) mutant strains in the presence or absence of a plasmid encoding DksA expressed from an IPTG-inducible promoter (pRLG6333). The lacZ fusion contains dksA promoter sequences from -100 to +37 with respect to the transcription start site (construct 2 in Fig. 2). Induction was performed with 0.5 or 1 mM IPTG, as indicated. Error bars represent the standard deviations from 6 independent cultures.
The dksA promoter is subject to stringent and growth rate-dependent control
The results described in the previous sections suggested that the dksA promoter would display regulatory properties similar to other promoters that are controlled by DksA (see Introduction). Therefore, we examined whether PdksA activity would decrease rapidly during amino acid starvation (i.e. would display a stringent response) and would increase in proportion to the growth rate (i.e. would be growth rate-dependent).
For the stringent control experiments, promoter activity was monitored by primer extension after the addition of the serine analog serine hydroxamate (SHX) to starve the cell for this amino acid and induce ppGpp synthesis. Although the response of PdksA to amino acid starvation appeared a bit slower than that of rrnB P1, transcription from the dksA and the rrnB P1 promoters both decreased ~5-fold by 15 min after addition of SHX (Fig. 5A, 5B). dksA transcript levels remained almost constant after SHX addition in a ΔdksA mutant strain (Figs. 5A, 5B). In contrast, the lacUV5 promoter fused to the same mRNA was unaffected by SHX treatment in the wild-type or ΔdksA mutant strains (Fig. 5C). We conclude that PdksA is a member of the stringent control regulon.
Figure 5. The dksA promoter is subject to stringent and growth rate-dependent control.
A,B,C: Primer extension was performed on RNA isolated from wild-type (wt) or ΔdksA strains containing promoter-lacZ fusions (see Table 1). (A) PdksA (wt, RLG10101; ΔdksA; RLG10103), (B) rrnB P1 (wt, RLG4996; ΔdksA, RLG6348), (C) lacUV5 (wt, RLG4998; ΔdksA, RLG8950). RNA was isolated at varying times after addition of serine hydroxamate and processed using the same primer hybridizing to lacZ for each promoter construct (see Experimental Procedures). Error bars shown are from 3 independent experiments. D,E,F: Growth rate-dependence of PdksA and rrnB P1 promoter-lacZ fusions in strains wild-type or mutant for dksA. (D) PdksA (wt, RLG10101; ΔdksA; RLG10103) (E) rrnB P1 (wt, RLG4996; ΔdksA, RLG6348). (F) Promoter activities normalized to 1 at 0.4 doublings/hr to facilitate comparison of the slopes determined in (D) and (E), as described in Experimental Procedures. The media used to vary the growth rates were (◆,◇) LB; (■,□) MOPS minimal medium supplemented with 0.4% glycerol, 0.4% casamino acids, 40 μg/ml tryptophan, and 10 μg/ml thiamine; (●,○) MOPS with 0.4% glycerol, all 20 amino acids (80 μg/ml each except 40 μg/ml for tryptophan and tyrosine), and 10 μg/ml thiamine; (▽) MOPS with 0.4% glycerol and 10 mg/ml thiamine.
The activities of the PdksA and rrnB P1 promoters were also compared at different steady-state growth rates using promoter-lacZ fusions as reporters of promoter activity (Fig. 5D-5F). When plotted against the doubling time without normalization (Fig. 5D, 5E) or normalized to the same activity at a low growth rate to facilitate comparison of the slopes (Fig. 5F), PdksA activity increased as the nutritional quality of the medium improved (i.e. as the growth rate increased), characteristic of promoters responding to ppGpp/DksA. This property was dependent on the presence of a wild-type dksA gene (Fig. 5; Paul et al., 2004a). As with some other ppGpp/DksA-responsive promoters (e.g. Pfis; Mallik et al., 2006), the slope was not quite as positive as that for rrnB P1. Taken together, the data suggest that the dksA promoter is a member of the regulon controlled by ppGpp/DksA.
DksA and ppGpp directly inhibit transcription in vitro and decrease the half-life of PdksA-RNA polymerase complexes
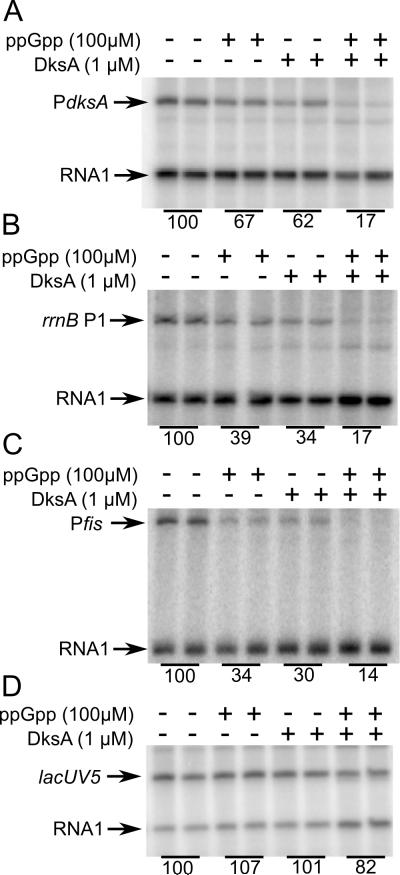
Although the results described above strongly suggested that the dksA promoter is autoregulated by ppGpp/DksA, they did not address whether these effects were direct. Therefore, we performed in vitro transcription experiments on PdksA with ppGpp and/or DksA (Fig. 6A). The rrnB P1 (Fig. 6B) and fis (Fig. 6C) promoters, both of which are ppGpp/DksA-regulated, were used as positive controls, and the lacUV5 promoter (Fig. 6D) was used as a negative control. DksA or ppGpp alone inhibited transcription from PdksA < 2-fold, and the two together inhibited PdksA ~6-fold (Fig. 6A). ppGpp or DksA alone inhibited both rrnB P1 and the fis promoter ~3-fold and together 7 to 8-fold (Fig. 6B and 6C). ppGpp and DksA individually or together had little or no effect on lacUV5 (Fig 6D). These results demonstrate that feedback regulation of the dksA promoter is direct.
Figure 6. DksA and ppGpp inhibit transcription of the dksA promoter directly.
Single round in vitro transcription from plasmid templates (see Table 1 for sequence endpoints of promoters) in the presence or absence of 1 µM DksA and/or 100 µM ppGpp. (A) PdksA (pRLG10105 ), (B) rrnB P1 (pRLG6798), (C) Pfis (pRLG8110), and (D) PlacUV5 (pRLG2222). The plasmid-derived RNA 1 transcript served as a loading control. Average transcription from duplicate reactions, relative to that from reactions without either ppGpp or DksA, is indicated below the lanes in the gel image. Representative experiments are shown, but 3 independent assays were performed with very similar results (variation was ~5% between experiments).
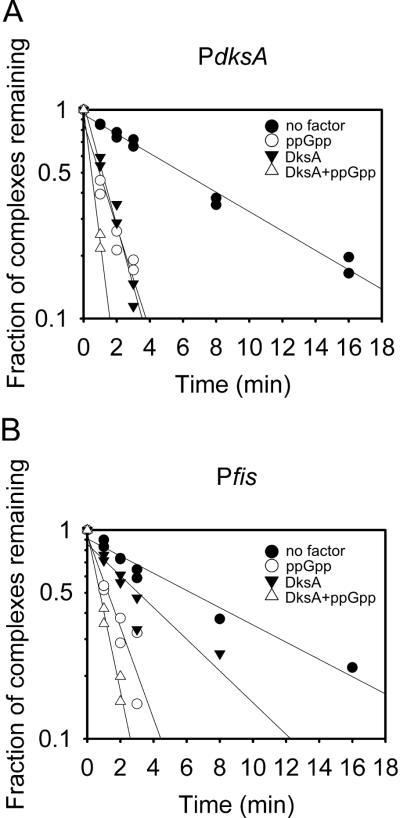
DksA and/or ppGpp inhibit transcription by reducing the rate of open complex formation. Measurements of dissociation rates of preformed promoter complexes have shown that ppGpp/DksA reduces the lifetimes of complexes formed by all promoters, but we have suggested that they inhibit transcription only from the subset of promoters that form intrinsically short-lived open complexes (Barker et al., 2001; Paul et al., 2004a; Mallik et al., 2006). As a first step in addressing the mechanism of inhibition of PdksA, promoter complexes were formed, challenged with the competitor heparin, and the fraction of complexes remaining at different times was measured by transcription (see Experimental Procedures and Fig. 7). The stability of PdksA-RNAP complexes was measured in the presence and absence of ppGpp, DksA, or the two together. Pfis, a previously described ppGpp/DksA regulated promoter (Mallik et al., 2006), was used for comparison.
Figure 7. DksA decreases the stability of the dksA promoter complex.
(A) RNAP-promoter complex half-lives were determined using a transcription based assay in the presence or absence of 1 μM DksA and/or 100 μM ppGpp. Individual reactions contained 1 nM supercoiled PdksA (pRLG10105) or Pfis (pRLG8110), and half-lives were measured as described in Experimental Procedures. The absolute half-lives varied slightly from experiment to experiment, but the ratio of the half-lives ± ppGpp/DksA were the same. Data from two independent experiments are plotted together after normalization of the two datasets at zero time. Half-lives of PdksA complex: ~6 min in the absence of DksA and ppGpp, ~2 min with ppGpp or DksA alone, and < 1 min with both DksA and ppGpp. Half-lives of Pfis complex: ~6 min in the absence of DksA and ppGpp, ~3 min with DksA alone, ~2 min with ppGpp alone, and < 1 min with both DksA and ppGpp.
The Pfis and PdksA complexes had similar intrinsic stabilities (half-life ~6 min), and ppGpp and DksA together decreased the half-lives of both to 1 min or less (Fig. 7A and 7B). The effect of DksA by itself was significantly greater on the PdksA complex than on the Pfis complex, but the relevance of this observation to inhibition of transcription is uncertain given that transcription from the two promoters was inhibited similarly when both ppGpp and DksA were present (Fig. 6).
Discussion
Biological significance of DksA autoregulation
We show here that DksA is a stable protein whose levels are kept constant by a negative feedback loop in which transcription from the dksA promoter is inhibited directly by DksA in conjunction with its cofactor ppGpp. We propose that this regulation fine-tunes dksA transcription initiation with DksA concentration, maintaining DksA at a constant level in growing cells and preventing DksA from accumulating during stationary phase when it is not diluted out by cell division. DksA levels do not go down in stationary phase because DksA is stable, and DksA does not accumulate in stationary phase because dksA transcription is inhibited by its own protein product. Unlike DksA, ppGpp and NTPs turn over rapidly, and their levels can change without new protein synthesis. Utilization of ppGpp and NTP fluctuations as signals of translational status affords the cell a rapid, promoter-specific mechanism for altering the activity of the transcription apparatus.
One could imagine that E. coli might also have evolved a mechanism to vary the concentrations of DksA itself for regulatory purposes. Although we have not detected such variation, we have not exhaustively examined the range of nutritional environments in which E. coli can grow, nor have we ruled out the possibility that additional mechanisms, working at different steps in dksA gene expression, could contribute to the regulation of DksA levels or activity. For example, we noticed that DksA protein levels increase slightly in the presence of glucose in the culture medium and that this increase is dependent on the dksA ribosome binding site region (unpublished data). In this regard, we note that the RNA binding protein CsrA binds to the dksA leader region (A. Edwards and T. Romeo, personal communication). Further studies will be needed to address whether CsrA plays a role in DksA expression.
Although we have not identified a condition where DksA levels fluctuate dramatically in E. coli, this does occur in other bacteria. For example, in Chlamydia pneumoniae, DksA protein levels increase during the transition from the intracellular, replicating reticulate form to the extracellular, infectious elementary form of the bacterium (Mukhopadhyay et al., 2006). It has also been reported that changes in the concentration of DksA (or DksA homologs) in Pseudomonas aeruginosa are used for regulatory purposes (Perron et al., 2005; Blaby-Haas et al., 2010).
In E. coli, DksA also plays an important role in preventing replication blocks under certain conditions (Tehranchi et al., 2010). Variation in the concentration of DksA could be detrimental for this function. It is possible that feedback regulation of the DksA promoter evolved primarily to keep DksA concentrations constant in growing cells so that the replication/repair roles of DksA are not compromised. Keeping a constant cellular level of a transcription factor requires just as elaborate a regulatory mechanism as one in which the levels of the factor change dramatically with environmental conditions.
As described in the Introduction, inhibition of rRNA transcription is a signature of the bacterial stringent response. It may seem counterintuitive that dksA expression is inhibited during a stringent response when the protein is needed for the response of the transcriptional apparatus to ppGpp. However, because DksA is stable, it does not get depleted on the time scale of the rapid responses of promoters to changes in the nutritional conditions. In natural starvation conditions, ppGpp concentrations return to basal levels when charged tRNA concentrations have recovered. Because the stringent response is transient, continued DksA synthesis is unnecessary in the short-term. Likewise, although DksA promoter activity increases with steady-state growth rate, DksA synthesis rates do not increase as rapidly with growth rate as rRNA synthesis rates. Apparently, the growth rate-dependent increase in PdksA activity is only sufficient to keep intracellular DksA protein levels constant and to compensate for depletion of DksA by cell division.
A previous study identified DksA as a substrate for degradation by the intracellular protease ClpXP (Flynn et al., 2003). The half-life of DksA reported in that study (~11 min) was much shorter than that reported here (44 min) in the same growth conditions (Fig. 1) or in pulse-chase experiments we have performed in different growth phases or conditions (data not shown). We have not been able to determine the basis for the difference in DksA half-life reported by the two laboratories. However, we note that a half-life of 11 min would probably still not be short enough to allow protein stabilization to serve a regulatory function in responses as rapid as those described for rRNA transcription (Murray et al., 2003).
Mechanism of inhibition of the dksA promoter by ppGpp/DksA
At rrnB P1, DksA binds to closed complexes, inhibits open complex formation, and thereby decreases transcription (Rutherford et al., 2009). However, because the kinetic properties of different promoters vary, the effects of ppGpp/DksA on different promoters vary as well: on some promoters ppGpp/DksA inhibits transcription, on some it has no effect, and on others it stimulates transcription (Paul et al., 2005).
ppGpp/DksA decreases the stability of pre-formed promoter-RNAP complexes at all promoters tested to date. However, the decay rate reported in our stability assay is a composite of multiple steps in the mechanism, with the complex at one step in equilibrium with those at the previous and following steps. The measured composite decay rate can include different steps at different promoters, because the promoter complex that is occupied by RNAP at equilibrium differs among promoters. Although there is a perfect inverse correlation between promoter complex lifetime and inhibition of transcription by ppGpp/DksA in vivo and in vitro for the rrnB P1 promoter and its variants (Haugen et al., 2006), the correspondence between intrinsic half-life and the potential for ppGpp/DksA to inhibit transcription is not always so clear for other promoters.
The PdksA complex is longer-lived than rRNA promoter complexes, but its lifetime is similar to that of several other promoters regulated by ppGpp/DksA (Pfis, PflhDC, some ribosomal protein promoters) (Mallik et al., 2006; Lemke et al., 2009; Lemke et al., 2011). Thus, we suggest that the rate of formation of some intermediate on the pathway to open complex formation at PdksA is reduced by ppGpp/DksA, just as at rRNA promoters, even though the measured intrinsic dissociation rate is slower.
We have identified promoter sequences that contribute to the instability of the rrnB P1 open complex and to inhibition of transcription by ppGpp/DksA. These include a cytosine residue on the non-template strand two bp downstream from the -10 hexamer that is detrimental to interactions with sigma subunit region 1.2, the absence of an extended -10 element needed for interactions with sigma region 3.0, and a 16 bp spacer between the -10 and -35 elements (Haugen et al., 2006). PdksA does not share any of these sequence features (Fig. 2E) even though it is regulated by ppGpp/DksA.
Understanding the properties of the RNAP-promoter interaction that sensitize it to regulation by DksA and/or ppGpp remains a long-term goal of our laboratory. Although we were motivated originally to investigate the dksA promoter in order to understand how DksA levels remain constant, PdksA could also provide important clues to understand the promoter sequence features that contribute to regulation by ppGpp/DksA.
Experimental Procedures
Strains,plasmid construction, and proteins
Strain and plasmid names are provided in Table 1. Construction of promoter–lacZ fusions carried on phage λ and isolation of λ lysogens was performed as described (Simons et al., 1987; Rao et al., 1994). Plasmids used for in vitro transcription were constructed by insertion of the promoter fragment of interest into the EcoRI and HindIII sites of plasmid pRLG770 (Ross et al., 1990) and were verified by sequencing. The end-points of the promoter fragments used for these constructions are provided in Table 1. Construction of ΔdksA strains was performed by transduction of the dksA::tet insertion-deletion from strain RLG8124 with P1vir (Rutherford et al., 2009).
pINdksA (pRLG6333 in Table 1) is a plasmid in which the lpp-lac promoter is fused to the dksA gene. The amount of DksA produced from this plasmid is ~50% of that in wild-type cells and complements a ΔdksA mutant for inhibition of transcription from an rRNA promoter and for rescue of amino acid auxotrophy (Paul et al. 2004a; Paul et al. 2005; Rutherford et al. 2007).
pdksA-dksA (pRLG10106 in Table 1) contains the dksA promoter, leader, and coding regions. It was constructed by amplification of the dksA gene by PCR from promoter position -100 to the dksA translation stop codon and insertion into the EcoRI and HindIII sites of pRLG770.
placUV5-dksA (pRLG10108 in Table 1) contains dksA under the control of the lacUV5 promoter and was constructed in two steps. First, the pRLG770 based plasmid pRLG10113, which contains a lacUV5 promoter fragment extending from -46 to +1 followed by an XbaI site, was constructed from pRLG3422 (Gaal et al., 1997) using the primer 5’-ACGTTCTAGATCCACACATTATACGAGCCGGAAGCATAAA-3’ (the XbaI site is underlined). Then, the dksA gene from promoter position +2 to the dksA translation stop codon was inserted into the XbaI and HindIII sites of pRLG10113 to form placUV5-dksA. All constructs were verified by sequencing.
RNA polymerase and DksA were purified as described (Lemke et al. 2009).
Western Blot Assay
Cultures were inoculated from fresh colonies on plates to an OD600 ~0.02 in LB, and samples were removed at the culture densities indicated in Fig. 3. Cultures were incubated on ice for 30 min, lysed by sonication, centrifuged, and total protein concentration in the soluble fraction was quantified by Bradford assay. Lysates were separated on 10 or 12% polyacrylamide gels (Invitrogen), and transferred to a PVDF membrane using a semi-dry transfer apparatus. Western blots were performed by standard procedures using a pre-cleared rabbit polyclonal anti-DksA serum (a gift from D. Downs, UW-Madison) or a monoclonal anti-σ32 antibody (a gift from R. Burgess, UW-Madison) and an HRP-conjugated secondary antibody (Santa Cruz Biotechnologies) and the ECL+ reagent (GE).
Protein half-life assay
To measure DksA stability, cells were grown in LB at 30°C to OD600 ~0.1. An aliquot was removed before the addition of chloramphenicol (34 μg/ml final concentration) as a zero time point. To measure σ32 stability, cultures containing RLG3499 were grown at 30°C to OD600 ~0.3, incubated at 42°C for 5 min, and shifted back to 30°C. Cell samples were collected at the indicated times, lysed and precipitated by addition of TCA to 50%, and the precipitates were centrifuged and then suspended in 2% SDS, 20 mM sodium phosphate buffer pH 7.5, 10 mM EDTA, 0.1 M PMSF. Western blots were performed as above.
β-galactosidase assays
Cells were grown in MOPS minimal medium supplemented with 0.4% glycerol, 0.4% casamino acids, 40 μg/ml tryptophan, and 10 μg/ml thiamine at 37°C from OD600 ~0.02 to OD600 ~0.3 for exponential phase measurements and OD600 ~5.0 for stationary phase. At the indicated OD600, 1 ml culture samples were removed to tubes on ice containing 4 ml of Z buffer (0.06 M Na2HPO4.7 H2O, 0.04 M NaH2PO4.H2O, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β–mercaptoethanol) for at least 10 min and lysed by sonication. β-galactosidase activity was determined as described previously (Barker et al., 2001). Background activity in fusions lacking promoters is very low in this system (~1-2 Miller units; Simons et al., 1987; Rao et al., 1994).
RNA extraction, primer extension, and stringent control measurements
RNA transcripts were quantified from lysogenized promoter-lacZ fusions in wild-type or ΔdksA cells with or without amino acid starvation (Ross and Gourse, 2009; Lemke et al., 2009). Briefly, cultures were grown at 37°C in MOPS minimal medium supplemented with 0.4% glucose, 0.4% casamino acids, 40 μg/ml tryptophan, and 10 μg/ml thiamine to OD600 ~0.3. For Fig. 5, half the culture was removed to a prewarmed flask containing enough serine hydroxamate (SHX) to achieve a final concentration of 1 mg/ml. At the indicated times, samples were lysed with boiling SDS solution, added to hot phenol, RNA was extracted, and reverse transcription was performed using a primer that paired to the RNA 39-59 nt downstream from the lacZ AUG (Lemke et al., 2009). To map the dksA transcription start site, the product of the primer extension reaction was run on an 11% polyacrylamide urea gel next to a sequencing ladder generated by the fmol cycle sequencing kit (Promega) and plasmid pJK537 (which contains the sfsA-dksA intergenic region; Kang and Craig, 1990). The primer (5’-TTCTCCTTAACACGCACTATCGATCCCCATG-3’) hybridized to the RNA 17-47 bp downstream from the dksA transcription start site.
Growth rate-dependent control measurements
Different growth rates were obtained by growing lysogens containing promoter-lacZ fusions (Table 1) in different media (see Fig. 5 legend). Cultures were inoculated from colonies on plates and incubated with shaking at 30°C for at least three generations before sampling. Aliquots were removed at OD600 ~0.3.
The growth rates at 30°C were: LB (wild type ~1.7 doublings/hour, ΔdksA ~1.3 doublings/hour); MOPS-glycerol-casamino acids (wild type ~1.2 doublings/hour, ΔdksA ~0.92 doublings/hour); MOPS-glycerol- 20 amino acids (wild type ~0.74 doublings/hour, ΔdksA ~0.73 doublings/hour); MOPS-glycerol (wild type ~0.49 doublings/hour; ΔdksA mutants cannot grow in media without amino acids; Paul et al., 2005).
For visual comparison of growth rate-dependences of promoters with different activities (Fig. 5F), a slope representing the activities at different growth rates was determined by linear regression, and the resulting equation of the line was used to calculate the promoter activity at a growth rate of 0.4 doublings/hour. The activities of each promoter were then normalized to 1 at 0.4 doublings/hour, and the normalized slopes for the different promoters were then plotted on the same scale (Dickson et al. 1989).
In vitro transcription assays
Single round in vitro transcription assays were performed essentially as described (Paul et al., 2004a) using Eσ70 RNAP (10-14 nM) and a supercoiled plasmid template (1 nM). The reactions were incubated at 30°C for 10 min and contained 40 mM Tris-HCl (pH 8), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 μg/μl BSA, 500 μM ATP, 200 μM CTP and GTP, 10 μM UTP, ~1 μCi [α-32P]-UTP, and DksA/ppGpp at the concentrations indicated. 10 μg/ml heparin was added with the NTPs as a competitor to prevent reinitiation. Samples were electrophoresed on a 7 M urea-6% polyacrylamide gel and quantified by phosphorimaging.
RNAP-promoter complex half-life assays
RNAP-promoter half-life assays were performed as described (Barker et al., 2001) using transcription as a reporter of the fraction of complexes remaining at the indicated times. Supercoiled plasmid DNA (1 nM) was preincubated with 1 μM DksA and/or 100 μM ppGpp for 10 min as in the section above. Heparin (10 μg/μl, Sigma) was used as a competitor. Aliquots (10 μl) were removed at various times after competitor addition to a tube containing NTPs at the concentrations provided above. As described above, transcription was allowed to proceed for 10 min at 30°C, and the transcripts were electrophoresed and analyzed by phosphorimaging.
Acknowledgments
We thank Wilma Ross and other members of the lab for comments on the manuscript. We also thank Adrianne Edwards, Tony Romeo, and Tania Baker for discussion. This work was supported by a grant from the National Institutes of Health to R.L.G. (R37 GM37048). P.C. was supported in part by a Genetics Training Grant from the N.I.H.
References
- Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas CE, Furman R, Rodionov DA, Artisomovich IA, de Crécy-Lagard V. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol. 2010;79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RR, Gaal T, deBoer HA, deHaseth PL, Gourse RL. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J Bacteriol. 1989;171:4862–4870. doi: 10.1128/jb.171.9.4862-4870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- Grossman AD, Straus DB, Walter WA, Gross CA. Sigma 32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–70. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci USA. 2011 Mar 14; doi: 10.1073/pnas.1019383108. [Epub ahead of print]PMID: 21402902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Gaal T, Ross W, Gourse RL. Escherichia coli DksA binds to free RNA polymerase with higher affinity than to RNA polymerase in an open complex. J Bacteriol. 2009;191:5854–5858. doi: 10.1128/JB.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol. 2006;188:5775–5782. doi: 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y, Mizuno T, Inouye M. Novel high-level expression cloning vehicles: 104-fold amplification of Escherichia coli minor protein. Biotechnology. 1984;2:81–85. [Google Scholar]
- Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. RecN protein and transcription factor DksA combine to promoter faithful recombinational repair of DNA double-strand breaks. Mol Microbiol. 2005;57:97–110. doi: 10.1111/j.1365-2958.2005.04677.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Good D, Miller RD, Graham JE, Matthews SA, Timms P, Summersgill JT. Identification of Chlamydia pneumoniae proteins in the transition from reticulate to elementary body formation. Mol Cell Proteomics. 2006;5:2311–2318. doi: 10.1074/mcp.M600214-MCP200. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schenider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA control gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004a;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004b;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. PNAS. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovich I, Vassylyev DG. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Perron K, Comte R, van Delden C. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol Microbiol. 2005;56:1087–1102. doi: 10.1111/j.1365-2958.2005.04597.x. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, et al. Factor independent activation of rrnB P1: an ‘extended’ promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- Ross W, Thompson JF, Newlands JT, Gourse RL. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gourse RL. Analysis of RNA polymerase-promoter complex formation. Methods. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:26–48. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol. 2006;62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Tehranchi AK, Blankenschein MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]