Abstract
Both peripheral neuropathy and distal myopathy are well-established inherited neuromuscular disorders characterized by progressive weakness and atrophy of the distal limb muscles. A complex phenotype of peripheral neuropathy, myopathy, hoarseness and hearing loss was diagnosed in a large autosomal dominant Korean family. A high density SNP-based linkage study mapped the underlying gene to a region on chromosome 19q13.3. The maximum multipoint LOD score was 3.794. Sequencing of 34 positional candidate genes in the segregating haplotype revealed a novel c.2822G>T (p.Arg941Leu) mutation in the gene MYH14, which encodes the nonmuscle myosin heavy chain 14. Clinically we observed a sequential pattern of the onset of muscle weakness starting from the anterior to the posterior leg muscle compartments followed by involvement of intrinsic hand and proximal muscles. The hearing loss and hoarseness followed the onset of distal muscle weakness. Histopathologic and electrodiagnostic studies revealed both chronic neuropathic and myopathic features in the affected patients. While mutations in MYH14 have been shown to cause nonsyndromic autosomal dominant hearing loss (DFNA4), the peripheral neuropathy, myopathy, and hoarseness have not been associated with MYH14. Therefore, we suggest that the identified mutation in MYH14 significantly expands the phenotypic spectrum of this gene.
Keywords: hearing loss, hoarseness, genomewide scan, MYH14, myopathy, neuropathy
INTRODUCTION
Hereditary motor and sensory neuropathy, which is also called Charcot-Marie-Tooth disease (CMT), is a genetically and clinically heterogeneous disorders, and exhibits clinical symptoms of distal muscle weakness and atrophy, pes caus, and impaired sensation [Reilly and Shy, 2009]. Autosomal dominant CMT is classically divided into two types; the demyelinating form (CMT1) and the axonal form (CMT2) [Pareyson and Marchesi, 2009]. CMT1 has severely reduced median motor nerve conduction velocities (NCVs) (< 38 m/sec), whereas CMT2 has slightly reduced or normal NCVs with decreased amplitudes [Harding and Thomas, 1980]. To date, more than 50 causative genes or loci have been reported to be associated with CMT at the Inherited Peripheral Neuropathies Mutation Database (IPNMD) (http://www.molgen.ua.ac.be/CMTMutations/Mutations/).
Distal myopathies are also clinically and genetically heterogeneous inherited disorders with more than 10 genes and chromosomal loci identified to date [Udd, 2009]. A major symptom of distal myopathies is weakness of skeletal muscles in the lower legs and hands occasionally concurrent with cardiomyopathy and vocal dysfunction [Malicdan and Nonaka, 2008]. Autosomal dominant distal myopathies include tibial muscular dystrophy (TMD; MIM# 600334) caused by mutations in TTN (MIM# 188849) [Hackman et al., 2002], Laing distal myopathy due to changes in MYH7 (MIM# 160760) [Meredith et al., 2004], distal caveolinopathy associated with CAV3 (MIM# 6012530) mutations [Minetti et al., 1998], and myofibrillar myopathies (MFM) caused by mutations in CRYAB (MIM# 123590), MYOT (MIM# 6041030), and LDB3/ZASP (MIM# 605906) [Vicart et al., 1998; Selcen and Engel, 2004, 2005]. MATR3 (MIM# 164015) have recently been reported as the underlying causes of dominant distal myopathy [Senderek et al., 2009]. Desminopathy and other forms of myofibrillar myopathy are caused by mutations in the DES gene (MIM# 125660) with both autosomal dominant and recessive inheritance [Goldfarb et al., 2004; Arias et al., 2006].
Several patients have been reported to have an unusual incidental combination of two neuromuscular diseases of CMT and muscle diseases, such as myotonic dystrophy, Becker muscular dystrophy (MIM# 300376) and facioscapulohumeral muscular dystrophy [Bütefisch et al., 1998; Bergmann et al., 2000; Hodapp et al., 2006; Kim et al., 2010]. Recently, a Dutch CMT neuropathy family that also showed complex phenotypes of myotonic dystrophy, encephalopathic attacks and hearing loss revealed an atypical complex mutations at the DM1 locus (MIM# 160900) [Spaans et al., 2009; Braida et al., 2010]. However, there has not been reported a mutation which causes autosomal dominant peripheral neuropathy, distal myopathy, hoarseness and hearing loss.
Although many causative genes have been reported to be associated with CMT or distal myopathies, lots of patients are still waiting to uncover their genetic causes. In the present study, we identified a large autosomal dominant family with a complex phenotype of peripheral neuropathy, distal myopathy, hoarseness, and hearing loss. Genomewide linkage analysis mapped on the underlying gene to chromosome 19q13.3 and sequencing of candidate genes revealed a c.2822G>T (p.Arg941Leu; NM_001077186.1) mutation in the nonmuscle myosin heavy chain 14 (MYH14) gene.
MATERIALS AND METHODS
Subjects
This study included a total of 33 members (15 affecteds and 18 unaffecteds) of a large Korean autosomal dominant family with a complex phenotype of peripheral neuropathy, distal myopathy, hoarseness, and hearing loss (family ID: FC317, Fig. 1), and 283 healthy controls, who had no clinical features and family history of neuromuscular disorders and hearing loss. Total DNA was extracted from peripheral blood samples using the QIAamp Blood DNA mini kit (Qiagen, Hilden, Germany). Informed consent was obtained from all participants and from parents of patients younger than 18 years of age according to the protocol approved by the Institutional Review Board for Ewha Womans University, Mokdong Hospital.
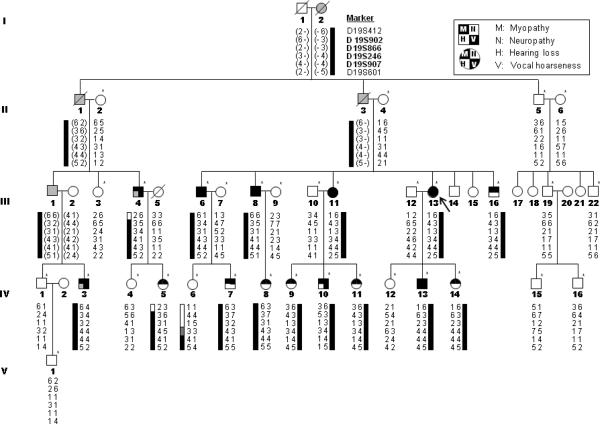
Figure 1.
Pedigree of family FC317. Haplotypes (indicated below each familial member) were determined using genotypes of microsatellite markers at chromosome 19q13.3 (genotypes in parentheses were inferred). Asterisks (*) indicate individuals whose DNA was used for linkage analysis. Black color indicates confirmed phenotypes by clinical exam, and grey indicates presumptive phenotypes based on history talking. Arrow (↖) indicates the proband.
Genomewide SNP Linkage Scan
A genomewide SNP linkage scan was performed on 28 members of family FC317 applying the Infinium II Human Linkage-12 Panel (Illumina, San Diego, CA). The chip included 6,090 SNP markers that are uniformly distributed on every chromosome with an average gap of 441 kbp and 0.58 cM. Genotyping data were scanned on the Illumina BeadStation 500G array scanner and analyzed with the Merlin-1.1.2 software using an autosomal dominant parametric model.
Chromosomal Fine Mapping
Fine mapping of the chromosome 19q13 region was performed by genotyping 29 fluorescent-labeled microsatellites. PCR products were resolved on the automated genetic analyzer ABI3100, and data were analyzed using the Genotype program (Applied Biosystems, Foster city, CA). By applying the Mlink software, two-point LOD scores were obtained under an autosomal dominant model.
Determination of Copy Number Variation
Copy number variations (CNV) in the chromosome 19q13 region were determined by using a custom designed high-density comparative genomic hybridization (CGH) 135K microarray (Roche-NimbleGen, Madison, WI). The array covered a region on chromosome 19 between 36606560-60238000 bp (UCSC hg18, NCBI Build 36.1). The mean probe size and spacing length of the array were 60-mer and 215 bp, respectively. The CGH data were analyzed with NimbleScan (ver. 2.4) and SignalMap (ver. 1.9) softwares. The gain and loss threshold used in this study were log2 ratio >0.3 and <−0.3, respectively.
Mutational Screening
Sequencing analysis of all coding exons and flanking intronic sequences was performed on 34 candidate genes in the linkage region at 19q13.3. In addition, mutational screening was performed on several previously reported genes with autosomal dominant CMT or distal myopathy. Particularly, tri-nucleotide extension was examined in the DMPK (MIM# 605377) in DM1 locus and CNBP1 (ZNF9; MIM# 116955) in DM2 locus. Entire mitochondrial DNA (mtDNA) was also amplified and sequenced using the mitoSEQr resequencing system (Applied Biosystems). PCR primer sequences and conditions are available upon request. PCR products were sequenced on the automatic genetic analyzer ABI3100 using the BigDye terminator cycle sequencing kit (Applied Biosystems). Sequence variations were confirmed by analyzing both DNA strands. Nucleotides were counted by cDNA numbering with +1 corresponding to the A of the ATG initiation codon in GenBank RefSeq NM_001077186.1, according to the journal guidelines (www.hgvs.org/mutnomen)..
Karyotyping and MYH14 Expression in Muscle
Karyotyping was performed using cultured metaphase white blood cells obtained from proband (III-13) and her sibling (III-16). Metaphase chromosomes were visualized with Gimsa staining. MYH14 expression in biopsied gastrocnemius muscle from a patient (III-16) was determined by quantitative real-time PCR using the QuantiTect Primer Assay (QT00080248, Qiagen).
Clinical Assessments
Clinical information was obtained through history taking, physical examinations, clinical observations, and electrophysiological investigations. Clinical observations included detailed neurological exams, including assessments of muscle weakness, sensory impairment, hearing loss, hoarseness, and reflexes. Electrocardiogram (ECG), echocardiography, and blood chemistry including creatine kinase (CK), lactic acid and pyruvic acid were done. The laryngeal study was performed in 4 patients (III-6, 11, 13, and IV-10) using a flexible fiberoptic laryngoscope. To determine hearing loss, pure-tone audiometry with air and bone conduction was performed in 17 individuals (11 affecteds and 6 unaffecteds) after otoscopic examination using an AC40 audiometry (Interacoustic, Denmark).
Electrophysiological Studies
Electrophysiological studies were carried out on 12 affected individuals (6 males and 6 females). Motor NCVs of the median, ulnar, peroneal, and tibial nerves were determined. Amplitudes of compound muscle action potential (CMAP) were measured from positive to negative peak values. Sensory NCVs were obtained from the median, ulnar, and sural nerves by an orthodromic scoring. Amplitudes of sensory nerve action potential (SNAP) were measured from positive peaks to negative peaks. An electromyography (EMG) was performed in the first dorsal interosseous, biceps brachii, tibialis anterior, medial gastrocnemius, and vastus lateralis muscles.
MRI studies of Lower Limbs
Fourteen individuals (12 affecteds and 2 unaffecteds) were studied with MRI of the lower limbs using a 1.5-T system (Siemens Vision, Siemens, Germany). Lower leg imaging was carried out in axial and coronal planes applying the following protocols: T1-weighted spin-echo (SE) (TR/TE 570–650/14–20, 512 matrixes), T2-weighted SE (TR/TE 2800–4000/96–99, 512 matrixes), and fat-suppressed T2-weighted SE (TR/TE 3090–4900/85–99, 512 matrixes). Muscles were graded on a five-point scale, as follows: 0 = no fat signal in muscle, 1 = some fatty streaks, 2 = fat occupying a minor part of muscle, 3 = similar amount of fat and muscle tissue, and 4 = fat occupying the greater part of muscle.
Histopathologic Studies
Muscle biopsies of left vastus lateralis and right lateral gastrocnemius were performed in patients III-6 and III-16, respectively. Serial frozen sections were stained with hematoxylin and eosin (H-E), NADH-tetrazolium reductase (NADH-TR), succinate dehydrogenase (SDH), modified Gomori trichrome (mGT), periodic acid-Schiff (PAS), Oil-red-O, and ATPase reaction with different pH preincubation. Immunostaining of myosin heavy chain (fast) (NCL-MHCf, monoclonal, 1:20) and myosin heavy chain (slow) (NCL-MHCs, monoclonal, 1:40: Vision Biosystems, Newcastle, UK), dystrophin, sarcoglycan, dysferlin, and titin was done. For the electron microscopic observation specimen were fixed in 2% glutaraldehyde in 25 mM cacodylate buffer at pH 7.4, and processed for semithin and ultrathin studies.
RESULTS
Clinical Manifestations
The clinical features of 15 affected members of the present family are shown in Table 1. Muscle weakness and atrophy started and predominated in distal portions of the legs and were noted to a lesser extent distally in the upper limbs. Distal muscle weakness of the lower limbs varied from asymptomatic to severe, and the progressive leg muscle atrophy was associated with disease duration (Supp. Fig. S1). Mean age at onset of distal weakness was 10.6±2.4 yrs (range 5–14 yrs) and disease duration was 17.5±15.3 yrs (range 1–41 yrs). All 15 affected patients showed distal leg muscle weakness, and three patients (III-4, 6 and 8) showed mild proximal thigh muscle weakness. The frequency of foot deformities was high (67%), and the decreased knee jerk reflexes were found in the majority of the patients. Serum CK levels were normal to slightly elevated, which was consistent with the other autosomal dominant distal myopathies [Saperstein et al., 2001]. ECG or echocardiography was normal.
Table 1.
Clinical phenotypes of 15 affected individuals
| Patient | III-4 | III-6 | III-8 | III-11 | III-13 | III-16 | IV-3 | IV-5 | IV-7 | IV-8 | IV-9 | IV-10 | IV-11 | IV-13 | IV-14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex/current age (yrs) | M/52 | M/52 | M/48 | F/45 | F/41 | M/33 | M/31 | F/18 | M/15 | F/11 | F/18 | M/16 | F/15 | M/15 | F/11 |
| Age of onset (yrs)a | 12 | 11 | 10 | 12 | 13 | 9 | 10 | 12 | 10 | 9 | 13 | 12 | 14 | 5 | 7 |
| Disease duration (yrs) | 40 | 41 | 38 | 33 | 28 | 24 | 21 | 6 | 5 | 2 | 5 | 4 | 1 | 10 | 4 |
| Muscle weakness | D>P | D>P | D>P | Distal | Distal | Distal | Distal | Distal | Distal | Distal | Distal | Distal | Distal | Distal | Distal |
| Distal muscle atrophy | Severe (L>U) | Severe (L>U) | Severe (L>U) | Severe (L>U) | Moderate (L>U) | Moderate (L>U) | Severe (L>U) | Mild (L>U) | Mild (L>U) | Absent | Mild (L>U) | Mild (L=U) | Absent | Mild (L>U) | Mild (L=U) |
| Sensory loss | − | − | ± | − | − | − | − | − | − | − | − | − | − | − | − |
| Hearing loss | NA | + | + | + | + | − | NA | NA | − | − | − | − | − | + | NA |
| Hoarseness | + | + | + | + | + | − | + | − | − | − | − | + | − | + | − |
| Cardiac involvement | NA | − | − | − | − | − | NA | NA | − | − | − | − | − | − | − |
| Foot deformity | + | + | + | + | + | + | + | + | − | − | − | + | − | + | − |
| Knee jerk reflex | Areflex | Areflex | Areflex | Areflex | Areflex | Areflex | Areflex | Normal | Normal | Normal | Normal | Hyporeflex | Normal | Hyporeflex | Normal |
| Additional symptom | − | Seizure | Tremor | − | − | Tremor | Tremor | Arthritis | − | − | − | Seizure | − | − | − |
| Creatine kinase (IU/L)b | NA | WNL | 2.5 fold | WNL | WNL | 1.4 fold | NA | NA | WNL | WNL | WNL | 1.3 fold | WNL | 2.1 fold | NA |
| Lactic acid (mg/dl)c | NA | 13.0 | NA | 19.0 | 9.0 | 11.0 | NA | NA | NA | NA | 10.0 | 13.0 | 7.0 | 12.0 | NA |
| Pyruvic acid (mg/dl)d | NA | 0.5 | NA | 1.0 | 0.7 | 0.7 | NA | NA | NA | NA | 0.7 | 1.1 | 0.6 | 0.7 | NA |
| MRI of lower limbe | |||||||||||||||
| Leg | NA | 3.9±0.3 | 3.6±0.5 | 3.4±0.8 | 3.5±0.6 | 2.5±0.9 | NA | NA | 0.2±0.4 | 0.1±0.3 | 0.3±0.5 | 0.2±0.4 | 0.1±0.3 | 0.8±1.0 | 0.3±0.5 |
| Thigh | NA | 1.4±0.7 | 1.1±0.6 | 0.4±0.5 | 0.3±0.4 | 0 | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Muscle biopsyf | NA | Groupingg | NA | NA | NA | Groupingg | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Abbreviations: D = distal muscle; L = lower limb; NA = not available; P = proximal muscle; U = upper limb; WNL = within normal limit; +/− = positive or negative finding; ± = equivocal finding.
Age at onset of distal muscle muscle weakness.
Creatine kinase values are listed in relations to the upper limits of normal for our laboratory.
Reference range: each 4.5–14.4 mg/dl
Reference range: each 0.3–0.9 mg/dl.
Degree of fatty infiltration was graded as five-point scale (mean±S.D.).
Muscle biopsy was done at vastus lateralis (III-6) and at gastrocnemius muscles (III-16).
Grouping of muscle fiber types and frequent paracrystalline inclusions.
A hoarse voice was present in 8 of 15 affected individuals (53%), albeit some affecteds had a hypophonic voice only. Six patients (III-4, 6, 8, 11, 13 and IV-3) experienced hoarseness when they were in their twenties, and in individuals IV-10 and IV-13, a hoarse voice was observed at ages 15 and 10, respectively. However, difficulty with swallowing, aspiration, dyspnea or ocular movement was not found, and flexible laryngoscope did not show paresis of vocal cord. Audiological studies showed bilateral sensorineural hearing loss in 5 of 11 affected patients (45%) (Supp. Table S1). Hearing loss was present in individual III-6 from about age 25 and in individuals III-8, III-11 and III-13 when they were in their thirties. High-frequency loss was observed in individual IV-13 at age 12 during a general physical examination. Unaffected family members noticed no symptoms of hearing loss.
Mapping of a New CMT Locus
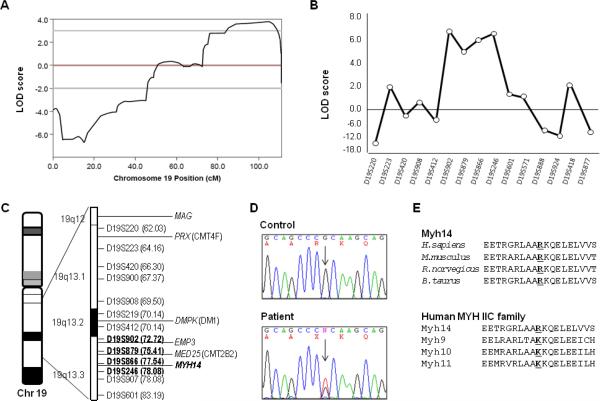
Genomewide SNP linkage analysis revealed a maximum multipoint LOD score of 3.79 at SNP marker rs1058511 under an autosomal dominant inheritance model (0.0001, 0.1, 0.9, 0.99). A haplotype co-segregated among affecteds that spans a region from approximately 75–110 Mbp (rs2041975 to rs1051500) on chromosome 19q13.2–3 (NCBI Build 36.1) (Fig. 2A). No additional chromosomal region showed LOD scores higher than 2 under an autosomal dominant model.
Figure 2.
Linkage analysis of family FC317 and identification of a missense mutation in MYH14 gene. (A) Two-point LOD scores of chromosome 19. (B) Chromosomal fine mapping of the 19q13.2-3 region. Genotyping of 29 microsatellites revealed a ~13Mb linkage region flanked by markers D19S902 and D19S246. (C) Diagram of chromosome 19q13.2-3. Markers that co-segregate with the phenotype are indicated in bold. (D) Sequencing chromatograms of the c.2822G>T (Arg941Leu) mutation in the MYH14 gene. The variant co-segregated with the affected family members. (E) High conservation of the Arg94 residue is illustrated by alignment of the amino acid sequences of MYH14 from different species and the MYH IIC family of proteins (ClustalX program, ver. 1.83) (H. sapiens MYH14-NP_001070654.1, MYH9-NP_002464.1, MYH10-NP_005955.1, MYH11-NP_002465.1; M. musculus Myh14-NP_082297.1, R. norvegicus Myh14- NP_001094160.1, B. Taurus Myh14- XM_882711.4).
Fine mapping of the chromosomal linkage region revealed a two-point maximum LOD score of 6.360 at D19S246 under an autosomal dominant model (Theta: 0.00). Haplotype and segregation analysis narrowed the chromosomal region at 19q13.3 to approximately 13 cM (D19S412 to D19S601) (Fig. 1 and 2B). The DMPK gene is close to this region, but could be excluded based on haplotype analysis (Fig. 2C). The linkage study also excluded main autosomal dominant CMT loci, such as 1p36.2 (MFN2; MIM# 608507), 1q23.3 (MPZ; MIM# 159440), 7p15 (GARS; MIM# 600287), 7q11.23 (HSPB1; MIM# 602195), 8p21.2 (NEFL; MIM# 162280) 8q21.1 (GDAP1; MIM# 606598), 9q34.13 (ALS4; MIM# 602433), 10q21.2 (EGR2; MIM# 129010), 12q24 (HSPB8; MIM# 608014), 16p13.13 (LITAF; MIM# 603795), 17p12 (PMP22; MIM# 601097) and 19p13.2 (DNM2; MIM# 602378).
Identification of a Missense Mutation in MYH14
Sequencing analysis was performed on all the coding exons of 34 candidate genes in the 19q13.3 linkage region (Fig. 2C). Particularly, 41 primer pairs were designed to amplify all exons and the promoter region of the MYH14 gene. Because CMT4F (PRX; MIM# 605725) and CMT2B2 (MED25; MIM# 610197) loci are located near or within the disequilibrium region [Berghoff et al., 2004; Kabzinska et al., 2006; Leal et al., 2009], we carefully examined both genes, but did not identify significant nucleotide changes. Similarly, no causative mutation was found in EMP3 (MIM# 602335), which is located within the linkage region and shows highly conserved homology with PMP22 myelin gene (CMT1A). Although the DMPK gene at the DM1 locus was excluded by the haplotype analysis, the careful examination of DMPK revealed neither abnormal CTG repeats nor a causative mutation in the coding regions. We also excluded chromosomal duplication and deletion events in the 19q13 region by applying a custom-designed high density CGH array to 3 individuals of the family (2 affecteds and 1 unaffected) (Supp. Fig. S2A). The karyotyping analysis in each affected male and female individual revealed no chromosomal abnormality (Supp. Fig. S2B).
In the screen of 34 candidate genes, the only functionally significant variant identified was a missense change in MYH14, which encodes the nonmuscle myosin heavy chain 14 (MYH14) protein [Leal et al., 2003]. The mutation was a G-to-T transversion at position 2282, c.2282G>T, which leads to substitution of an arginine residue with leucine, p.Arg941Leu (Fig. 2D). The c.2282G>T mutation has not been previously reported in the dbSNP database (http://www.ncbi.nlm.nih.gov/snp/). This mutation completely co-segregated with all affected members in the pedigree. Sequencing analysis confirmed the absence of this variant in unaffected members of the pedigree and in 566 ethnically matched healthy control chromosomes. The mutation is located in the tail domain of MYH14 and it is highly conserved in other species (Fig. 2E). The amino acid corresponding to the mutation site in other human MYHIIC protein family members is lysine, which is a basic residue and thus similar to arginine. Additional polymorphic variants identified in MYH14 are listed in Supp. Table S2.
Although MYH14 expression in skeletal muscle, cochlea, brain and peripheral nerves has been reported [Leal et al., 2003; Donaudy et al., 2004; Golomb et al., 2004], we confirmed MYH14 expression in biopsied gastrocnemius muscle with quantitative real-time PCR. The expression level in gastrocnemius muscle from an affected member (III-16) was slightly decreased compared to a normal male (Supp. Fig. S3).
Electrophysiological Findings
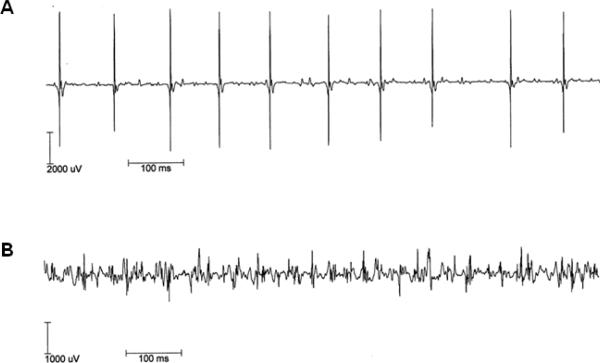
The electrophysiological findings of 29 nerves in 12 affected members (6 males and 6 females) are shown in Table 2. Nerve conduction studies demonstrated mildly reduced or normal median, ulnar and sural sensory NCVs, and reduced median and ulnar CMAPs were always associated with normal NCVs. Noteworthy, severely reduced CMAPs were observed in bilateral peroneal nerves. Needle EMG was performed in 12 patients, and they showed fibrillation potentials, but myotonic discharges were not found. Two affecteds (IV-8 and -11) did not show distal muscle atrophy, but they showed fibrillation potentials in the tibialis anterior muscles. In the same affected individuals, we observed both a large amplitude with long duration motor unit action potential (MUAP), which was usually seen in neuropathy (Fig. 3A), and also a small amplitude with short duration polyphasic MUAP seen in myopathy (Fig. 3B). These findings suggested that the affected patients have evidence of both neuropathy and myopathy.
Table 2.
Nerve conduction velocities in 12 affected family members
| Patient | Age (yrs) | R/L | Median motor | Ulnar motor | Peroneal motor | Tibial motor | Median sensory | Ulnar sensory | Sural sensory | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | |||
| III-6 | 50 | R | 4.2 | 56.4 | 7.9 | 58.3 | A | A | 0.6 | 41.1 | 37.3 | 42.3 | 22.6 | 38.7 | 31.4 | 38.5 |
| 50 | L | 6.3 | 57.4 | 6.4 | 70.0 | A | A | 0.6 | 49.3 | 24.2 | 44.1 | 22.3 | 39.1 | 20.4 | 38.7 | |
| 52 | R | 4.8 | 56.5 | 5.9 | 56.4 | A | A | 0.7 | 45.6 | 33.5 | 44.1 | 24.9 | 40.4 | 22.2 | 39.3 | |
| 52 | L | 6.0 | 52.8 | 6.6 | 54.3 | A | A | 0.5 | 44.3 | 36.7 | 45.5 | 24.6 | 39.2 | 30.8 | 39.5 | |
| III-8 | 48 | R | 2.9 | 53.5 | 6.1 | 54.1 | A | A | 12.5 | 36.1 | 39.8 | 35.7 | 31.7 | 34.2 | 23.4 | 30.3 |
| 48 | L | 5.4 | 59.8 | 5.1 | 55.3 | A | A | 7.7 | 37.5 | 32.2 | 38.3 | 18.6 | 34.5 | 28.2 | 28.1 | |
| III-11 | 45 | L | 7.9 | 58.8 | 5.2 | 69.0 | 1.5 | 49.3 | 7.3 | 47.7 | 41.0 | 46.9 | 32.0 | 41.7 | 30.3 | 42.3 |
| III-13 | 40 | R | 10.3 | 56.1 | 9.9 | 56.4 | A | A | 16.5 | 45.6 | 47.8 | 42.3 | 23.6 | 41.7 | 24.4 | 39.5 |
| 40 | L | 6.9 | 59.5 | 7.7 | 63.9 | A | A | 14.7 | 43.8 | 59.1 | 42.9 | 28.7 | 39.1 | 24.0 | 34.1 | |
| 41 | R | 11.0 | 53.5 | 11.0 | 55.7 | A | A | 17.0 | 46.8 | 57.1 | 41.1 | 26.2 | 40.3 | 24.1 | 35.7 | |
| 41 | L | 7.3 | 53.7 | 7.3 | 57.5 | A | A | 16.8 | 43.3 | 49.0 | 41.7 | 22.6 | 39.1 | 22.3 | 36.1 | |
| III-16 | 33 | R | 7.1 | 59.0 | 7.0 | 64.7 | A | A | 10.6 | 48.1 | 43.7 | 48.4 | 26.0 | 40.3 | 27.1 | 41.2 |
| 33 | L | 8.2 | 57.3 | 4.8 | 64.7 | A | A | 7.5 | 48.6 | 57.8 | 46.9 | 33.9 | 43.8 | 29.4 | 42.2 | |
| IV-7 | 14 | R | 6.2 | 53.4 | 18.1 | 54.8 | 4.6 | 43.9 | 15.1 | 42.2 | 22.3 | 40.1 | 14.6 | 43.9 | 10.2 | 33.1 |
| 14 | L | 7.3 | 54.5 | 16.2 | 60.5 | 5.9 | 37.7 | 12.1 | 38.6 | 22.0 | 43.4 | 15.9 | 37.7 | 12.9 | 34.8 | |
| 15 | R | 7.0 | 52.2 | 17.8 | 52.4 | 3.7 | 41.5 | 13.2 | 44.7 | 26.1 | 41.7 | 8.4 | 37.9 | 11.4 | 35.3 | |
| 15 | L | 8.4 | 55.3 | 17.8 | 53.7 | 5.1 | 37.5 | 14.1 | 38.4 | 19.2 | 40.5 | 13.4 | 38.6 | 11.9 | 38.2 | |
| IV-8 | 11 | R | 6.5 | 57.2 | 6.3 | 54.7 | 0.3 | 44.2 | 14.7 | 49.3 | 31.8 | 41.7 | 34.6 | 38.8 | 27.2 | 40.5 |
| 11 | L | 8.1 | 56.1 | 4.5 | 58.1 | 0.2 | 46.9 | 10.8 | 48.0 | 45.1 | 41.7 | 32.4 | 41.7 | 28.6 | 41.7 | |
| IV-9 | 18 | R | 15.4 | 63.2 | 10.0 | 60.8 | 3.4 | 47.8 | 8.2 | 47.3 | 55.0 | 40.4 | 35.3 | 39.5 | 29.4 | 36.6 |
| 18 | L | 11.5 | 58.3 | 14.0 | 63.6 | 4.6 | 45.1 | 8.0 | 47.9 | 53.7 | 43.0 | 26.1 | 40.3 | 31.6 | 33.4 | |
| IV-10 | 16 | R | 12.3 | 60.8 | 11.6 | 66.2 | 0.2 | 47.6 | 7.0 | 48.6 | 27.0 | 42.3 | 15.7 | 43.1 | 27.6 | 45.5 |
| 16 | L | 14.7 | 64.7 | 10.6 | 67.5 | 0.6 | 48.6 | 7.1 | 47.2 | 41.2 | 43.5 | 32.4 | 41.7 | 30.5 | 47.8 | |
| IV-11 | 15 | R | 10.6 | 56.3 | 14.5 | 62.1 | 2.9 | 49.2 | 14.6 | 47.8 | 30.9 | 40.4 | 19.7 | 39.5 | 18.1 | 35.5 |
| 15 | L | 10.5 | 64.3 | 13.9 | 62.9 | 3.3 | 45.7 | 12.7 | 45.7 | 30.0 | 41.7 | 20.7 | 39.5 | 21.5 | 37.2 | |
| IV-13 | 15 | R | 7.8 | 53.0 | 5.4 | 53.7 | A | A | 14.7 | 45.1 | 31.9 | 37.8 | 18.0 | 33.1 | 24.1 | 38.5 |
| 15 | L | 5.2 | 55.3 | 4.1 | 57.9 | A | A | 10.5 | 46.2 | 35.0 | 36.9 | 17.9 | 35.7 | 26.7 | 38.2 | |
| IV-14 | 11 | R | 9.1 | 52.9 | 11.4 | 56.1 | 2.1 | 42.9 | 12.5 | 43.8 | 31.5 | 40.4 | 22.4 | 39.4 | 14.3 | 36.0 |
| 11 | L | 10.6 | 56.1 | 6.9 | 57.3 | 1.6 | 42.8 | 15.0 | 43.4 | 31.5 | 41.7 | 20.4 | 40.3 | 17.2 | 36.8 | |
Boldface represents abnormal values. A = absent response; Age = age at examination; Amp = amplitude (motor: by mV; sensory: by μV). CV = conduction velocity (m/s); R/L = right/left. Normal CVs: motor median ≥ 50.5, ulnar ≥ 51.1, peroneal ≥ 41.2, tibial ≥ 41.1, sensory median ≥ 39.3, ulnar ≥ 37.5, and sural ≥ 32.1. Normal amplitudes: motor median ≥ 6; ulnar ≥ 8, peroneal ≥ 6, tibial ≥ 6, sensory median ≥ 8, ulnar ≥ 7.9, and sural ≥ 6.0.
Figure 3.
Motor unit action potential (MUAP). Recordings of the interference patterns in patient III-8 were made from the right vastus lateralis (A), and the first dorsal interosseous (B). The patient has evidence, clinically and electrophysiologically, of both neuropathy and myopathy. His electrophisiological studies revealed both a large amplitude with long duration (neuropathic, A) and a small amplitude with short duration polyphasic (myopathic) motor units (B).
Sequential Fatty Infiltration in Distal Muscle
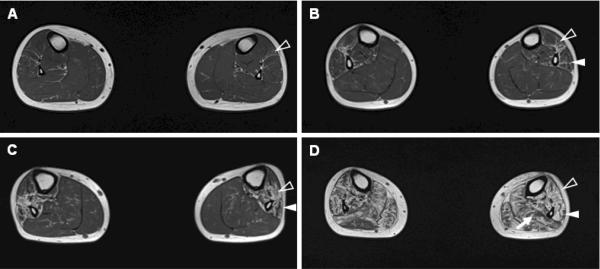
All examined affected individuals showed abnormal fatty infiltrations on MRI (Table 1). Eight of 12 patients showed fatty replacements of muscle tissue only in their legs, and all of them had anterior compartment involvement. Four patients (III-6, 8, 11, and 13) showed both leg and thigh muscle involvement; in all these patients, the posterior compartment of the thighs was involved, and in two, the anterior and medial compartments were involved. We could observe a sequential pattern of onset of muscle involvement associated with disease duration. In early stage of the disease, fatty infiltrations were present in the anterior and lateral compartments of the legs (Fig. 4A–B), and in later stages, posterior compartment leg muscles were also affected, and calf muscle atrophy was noticed (Fig. 4C–D).
Figure 4.
A sequential pattern of muscle involvement associated with disease duration (DD) in the T1-weighted axial MRI. (A) A 16-yr-old male patient (IV-10, DD=4 yrs) showed mild streaky fatty infiltrations of the anterior compartment muscles of the legs. (B) A 15-yr-old male patient (IV-13, DD=5 yrs) displayed more fatty infiltrations, including the anterior and the lateral compartments. (C) A 33-yr-old male (III-16, DD=24 yrs) showed prominent involvement of the anterior and lateral compartments of leg muscles with mild involvement of the posterior compartment. (D) A 41-yr-old female patient (III-13, DD=28 yrs) showed severe fatty involvement of all muscles compartments including a calf muscle atrophy (anterior compartment: blank arrowheads, lateral compartment: arrowheads, posterior compartment: arrow).
Histopathologic Findings
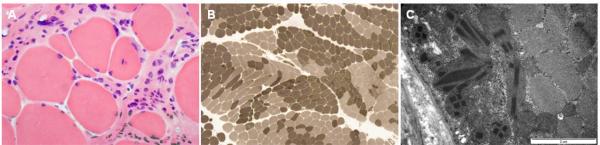
The histopathologic features of muscle biopsies were similar in two patients (III-6 and III-16). Myofibers showed moderate to marked variation of fiber size and shape (Fig. 5A). Immunostaining with dystrophin, sarcoglycan, dysferin, and titin did not show abnormal findings, and no inflammatory infiltration was present. The grouping of the histochemical muscle fiber types were observed by ATPase with pH 9.4 preincubation and immunostaining with myosin heavy chain (fast), myosin heavy chain (slow), and myosin IIa (Fig. 5B). NADH-TR and SDH staining showed multifocal subsarcolemmal accumulation of mitochondria, although mGT staining did not show ragged-red fibers or rimmed vacuoles. Notably, the electron micrographs frequently revealed subsarcolemmal accumulation of enlarged mitochondria with variable sized rectangular or elongated rhomboidal paracrystalline inclusions (Fig. 5C).
Figure 5.
Histopathologic findings of lateral gastrocnemius muscle in patient III-16. (A) A frozen section showing marked variation of fiber size and shape with many small rounded fibers, and degenerating fibers. Endomysial fibrosis was prominent on the background of degenerating myofibers without inflammatory cell infiltration by H&E stain. (B) Grouping of histochemical fiber types. ATPase reaction with different pH preincubation and immunostaining with myosin heavy chain (fast), myosin heavy chain (slow), and myosin IIa showed grouping of histochemical fiber types. (C) Subsarcolemmal accumulation of abnormal mitochondria. Electron micrographs revealed subsarcolemmal accumulation of frequent abnormal mitochondria including variable sized rectangular or elongated rhomboidal paracrystalline inclusions. Magnification: × 200 (A), × 40 (B), and × 30,000 (C).
DISCUSSION
We have mapped a new chromosomal locus for autosomal dominant peripheral neuropathy, myopathy, hoarseness, and hearing loss to 19q13.3. Sequencing analysis of 34 candidate genes revealed a novel c.2822G>T (Arg941Leu) mutation in the gene MYH14, that encodes the nonmuscle myosin heavy chain 14. MYH14 consists of 41 exons (40 coding), which comprise 108 kbp of genomic sequence. It encodes 1995 amino acids and four conserved functional domains: the amino-terminal myosin domain, the myosin head, two IQ domains and the myosin tail. Northern blot analyses have revealed strong expression of the 7 kbp transcript in the skeletal muscle, small intestine, colon and cochlea, but it is also expressed in a wide range of tissue including brain and peripheral nerves [Leal et al., 2003; Donaudy et al., 2004; Golomb et al., 2004]. We also confirmed MYH14 expression in the gastrocnemius muscle.
Our data support that the p.Arg941Leu mutation in the MYH14 is responsible for the complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss in family FC317 due to the following reasons: (1) co-segregation of the mutation with affected members in the pedigree, (2) no detection of the same mutation in 566 ethnicity matched control chromosomes, (3) high conservation of amino acids at the mutation site among different species, (4) previous involvement of MYH14 in non-syndromic hearing loss, and (5) absence of alternative causative mutations in known CMT or distal myopathy genes. Moreover, CNV examination revealed no significant duplication or deletion in the linkage region. Karyotyping also showed no chromosomal abnormality. Recently complex mutations in DMPK were reported as the underlying cause of a sensorimotor neuropathy family with myotonic dystrophy, encephalopathic attacks and hearing loss [Spaans et al., 2009; Braida et al., 2010]. However, we excluded mutations in DMPK, which falls near 19q13.3 and also sequenced additional candidate genes, including PRX (CMT4F), MED25 (CMT2B2), MAG, and EMP3. The complex phenotype of Dutch family is in some extent similar to the present Korean family, however, any myotonic discharges were not found.
Electrodiagnostic and histopathologic studies showed both chronic neuropathic and myopathic features in the affected patients. Needle EMG showed a small amplitude short duration myopathic MUAPs, and also revealed neuropathic MUAPs in the same affected individuals (Fig. 3). Histopathologic findings revealed marked variation of fiber size with many small round or angulated fibers, degenerating fibers, and endomyseal fibrosis, which were usually observed in myopathy, but also showed grouping of histochemical muscle fiber types, which was one of well known features noted in neurogenic changes of skeletal muscle (Fig. 5). Additionally, variable sized rectangular or rhomboidal paracrystalline inclusions were frequently found in electron micrographs. Because the histopathologic abnormalities were reminiscent of cases with mitochondrial diseases, we completely sequenced the mtDNA from two biopsied patients (III-6 and III-13), but found no causative mutation (Supp. Table S2). The histological study suggested that the pathogenic mechanism of the disease may relate with abnormal translocation and dysfunction of mitochondria.
Myosins are a superfamily with a domain which interacts with actin in order to produce movement under hydrolysis of ATP. The three known nonmuscle myosins are encoded by the MYH9 (MIM# 160775), MYH10 (MIM# 160776), and MYH14 genes located on chromosomes 22q11.2, 17p13.3, and 19q13.3, respectively (Simons et al. 1991; Leal et al. 2003). Myosin heavy chain genes underlie several forms of hereditary hearing loss. The hearing loss loci, DFNB2 (MIM# 600060) and DFNA17 (MIM# 600652), are associated with MYH7A (MIM# 160760) [Astuto et al., 2002] and MYH9 mutations, respectively [Lalwani et al., 2000]. The nonsyndromic autosomal dominant form of hearing impairment, DFNA4 (MIM# 600652), is caused by mutations in MYH14 [Mirghomizadeh et al., 2002; Donaudy et al., 2004]. In contrast, the studied Korean family exhibited a syndromic phenotype, dominated by peripheral neuropathy, myopathy, hoarseness, and hearing loss. Therefore, we suggest that this study significantly extents the known phenotype associated with MYH14 mutations. Future molecular studies of similar patients should consider MYH14 as the underlying cause.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients and their family members who participated in this study for their cooperation. This study was supported by the Mid-career Researcher Program through NRF grant funded by the MEST (R01-2008-000-20604-0 and KRF-2008-313-C00750), the Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs (A090500), Republic of Korea, and the National Institutes of Health, USA (R01NS052767 to SZ).
Grant sponsor: NRF (R01-2008-000-20604-0 and KRF-2008-313-C00750), the Healthcare Technology R&D Project (A090500) in Korea, and NIH (R01NS052767) in USA.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
REFERENCES
- Arias M, Pardo J, Blanco-Arias P, Sobrido MJ, Arias S, Dapena D, Carracedo A, Goldfarb LG, Navarro C. Distinct phenotypic features and gender-specific disease manifestations in a Spanish family with desmin L370P mutation. Neuromuscul Disord. 2006;16:498–503. doi: 10.1016/j.nmd.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Astuto LM, Kelley PM, Askew JW, Weston MD, Smith RJ, Alswaid AF, Al-Rakaf M, Kimberling WJ. Searching for evidence of DFNB2. Am J Med Genet. 2002;109:291–297. doi: 10.1002/ajmg.10384. [DOI] [PubMed] [Google Scholar]
- Berghoff C, Berghoff M, Leal A, Morera B, Barrantes R, Reis A, Neundörfer B, Rautenstrauss B, Del Valle G, Heuss D. Clinical and electrophysiological characteristics of autosomal recessive axonal Charcot-Marie-Tooth disease (ARCMT2B) that maps to chromosome 19q13.3. Neuromuscul Disord. 2004;5:301–306. doi: 10.1016/j.nmd.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Bergmann C, Senderek J, Hermanns B, Jauch A, Janssen B, Schröder JM, Karch D. Becker muscular dystrophy combined with X-linked Charcot-Marie-Tooth neuropathy. Muscle Nerve. 2000;23:818–823. doi: 10.1002/(sici)1097-4598(200005)23:5<818::aid-mus23>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Braida C, Stefanatos RK, Adam B, Mahajan N, Smeets HJ, Niel F, Goizet C, Arveiler B, Koenig M, Lagier-Tourenne C, et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum Mol Genet. 2010;19:1399–1412. doi: 10.1093/hmg/ddq015. [DOI] [PubMed] [Google Scholar]
- Bütefisch CM, Lang DF, Gutmann L. The devastating combination of Charcot-Marie-Tooth disease and facioscapulohumeral muscular dystrophy. Muscle Nerve. 1998;21:788–791. doi: 10.1002/(sici)1097-4598(199806)21:6<788::aid-mus11>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Desmin myopathy. Brain. 2004;127:723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- Hackman P, Vihola A, Haravuori H, Marchand S, Sarparanta J, De Seze J, Labeit S, Witt C, Peltonen L, Richard I, Udd B. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980;103:259–280. doi: 10.1093/brain/103.2.259. [DOI] [PubMed] [Google Scholar]
- Hodapp JA, Carter GT, Lipe HP, Michelson SJ, Kraft GH, Bird TD. Double trouble in hereditary neuropathy: Concomitant mutations in the PMP-22 gene and another gene produce novel phenotypes. Arch Neurol. 2006;63:112–117. doi: 10.1001/archneur.63.1.112. [DOI] [PubMed] [Google Scholar]
- Kabzinska D, Drac H, Sherman DL, Kostera-Pruszczyk A, Brophy PJ, Kochanski A, Hausmanowa-Petrusewicz I. Charcot-Marie-Tooth type 4F disease caused by S399fsx410 mutation in the PRX gene. Neurology. 2006;66:745–747. doi: 10.1212/01.wnl.0000201269.46071.35. [DOI] [PubMed] [Google Scholar]
- Kim HS, Chung KW, Kang SH, Choi SK, Cho SY, Koo H, Kim SB, Choi BO. Myotonic dystrophy type I combined with X-linked dominant Charcot-Marie-Tooth neuropathy. Neurogenetics. 2010;11:425–433. doi: 10.1007/s10048-010-0246-5. [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN. Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet. 2000;67:1121–1128. doi: 10.1016/s0002-9297(07)62942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal A, Endele S, Stengel C, Huehne K, Loetterle J, Barrantes R, Winterpacht A, Rautenstrauss B. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/s0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- Leal A, Huehne K, Bauer F, Sticht H, Berger P, Suter U, Morera B, Del Valle G, Lupski JR, Ekici A, et al. Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models. Neurogenetics. 2009;10:275–287. doi: 10.1007/s10048-009-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicdan MC, Nonaka I. Distal myopathies a review: highlights on distal myopathies with rimmed vacuoles. Neurol India. 2008;56:314–324. doi: 10.4103/0028-3886.43450. [DOI] [PubMed] [Google Scholar]
- Meredith C, Herrmann R, Parry C, Liyanage K, Dye DE, Durling HJ, Duff RM, Beckman K, de Visser M, van der Graaff MM, et al. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1) Am J Hum Genet. 2004;75:703–708. doi: 10.1086/424760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati MA, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- Mirghomizadeh F, Bardtke B, Devoto M, Pfister M, Oeken J, Konig E, Vitale E, Riccio A, De Rienzo A, Zenner HP, et al. Second family with hearing impairment linked to 19q13 and refined DFNA4 localization. Eur J Hum Genet. 2002;10:95–99. doi: 10.1038/sj.ejhg.5200769. [DOI] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009;8:654–667. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- Reilly MM, Shy ME. Diagnosis and new treatments in genetic neuropathies. J Neurol Neurosurg Psychiatry. 2009;80:1304–1314. doi: 10.1136/jnnp.2008.158295. [DOI] [PubMed] [Google Scholar]
- Saperstein DS, Amato AA, Barohn RJ. Clinical and genetic aspects of distal myopathies. Muscle Nerve. 2001;24:1440–1450. doi: 10.1002/mus.1167. [DOI] [PubMed] [Google Scholar]
- Selcen D, Engel AG. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- Selcen D, Engel AG. Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann Neurol. 2005;57:269–276. doi: 10.1002/ana.20376. [DOI] [PubMed] [Google Scholar]
- Senderek J, Garvey SM, Krieger M, Guergueltcheva V, Urtizberea A, Roos A, Elbracht M, Stendel C, Tournev I, Mihailova V, et al. Autosomal dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am J Hum Genet. 2009;84:511–518. doi: 10.1016/j.ajhg.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L. Human non-muscle myosin heavy chains are encoded by two genes located on different chromosomes. Circulation Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- Spaans F, Faber CG, Smeets HJ, Hofman PA, Braida C, Monckton DG, de Die-Smulders CE. Encephalopathic attacks in a family co-segregating myotonic dystrophy type 1, an intermediate Charcot-Marie-Tooth neuropathy and early hearing loss. J Neurol Neurosurg Psychiatry. 2009;80:1029–1035. doi: 10.1136/jnnp.2008.170126. [DOI] [PubMed] [Google Scholar]
- Udd B. 165th ENMC International Workshop: Distal myopathies III. Neuromusc Disord. 2009;19:429–438. doi: 10.1016/j.nmd.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prévost MC, Faure A, Chateau D, Chapon F, Tomé F, Dupret JM, et al. A missense mutation in the alpha B-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.