Abstract
A new caged xanthone (1), a new prenylxanthone (2), seven known xanthones, and a known sterol glucoside were isolated from the stems of Cratoxylum cochinchinense, collected in Vietnam. Compounds 1 and 2 were determined structurally by analysis of their spectroscopic data. In addition, five new (10 and 16–19) and eight known prenylated xanthone derivatives were synthesized from the known compounds, α-mangostin (3) and cochinchinone A (6). Several of these substances were found to be cytotoxic towards HT-29 human colon cancer cells, with the most potent being 3,6-di-O-acetyl-α-mangostin (8, ED50, 1.0 μM), which was tested further in an in vivo hollow fiber assay, but found to be inactive at the highest dose used (20 mg/kg; ip). Of the substances evaluated in a NF-κB p65 inhibition assay, 1,3,7-trihydroxy-2,4-diisoprenylxanthone (5) exhibited the most potent activity (IC50, 2.9 μM). In a mitochondrial transmembrane potential (MTP) assay, two new compounds, 1 (IC50, 3.3 μM) and 10 (IC50, 1.4 μM), and two known compounds, 3 (α-mangostin, IC50, 0.2 μM) and 11 (3,6-di-O-methyl-α-mangostin, IC50, 0.9 μM), were active. A preliminary analogue development study showed that 3,6-di-acetylation and 6-benzoylation both slightly increased the cytotoxicity of α-mangostin (3), whereas methylation reduced such activity. In contrast, neither acetylation, benzoylation, nor methylation enhanced the cytotoxicity of cochinchinone A (6).
Cratoxylum (Clusiaceae) is a small genus distributed in Southeast Asia, with some of its species used medicinally.1 Previous phytochemical investigations have demonstrated that xanthones2,3 are the most characteristic biologically active components of this genus.4–7Cratoxylum cochinchinense (Lour.) Bl. is a tropical plant used in folk medicine to treat a number of diseases, including cough, diarrhea, fever, and ulcers.8 Detailed phytochemical studies of C. cochinchinense have resulted in the isolation of several xanthones,3,9–12 including some caged xanthones10,13,14 and α-mangostin.9,10,14 The latter compound has shown cytotoxicity towards the NCI-H187 human lung and HT-29 human colon cancer cell lines,3,10 and acts by inducing caspase-3 dependent apoptosis.4–6
As a part of a research program on the discovery of new natural product anticancer agents from diverse organisms,15 it was found that a crude methanol extract of the stems of C. cochinchinense, collected in Vietnam, exhibited cytotoxicity towards HT-29 human colon cancer cells. Using column chromatography, a new caged xanthone (1) and a new prenylxanthone (2), together with seven known xanthone compounds and a known sterol glucoside were isolated from this species. Two of the known compounds were identified as α-mangostin (3) and cochinchinone A (6), which showed ED50 values of 4.1 and 16.1 μM, respectively, when tested for cytotoxicity against the HT-29 human colon cancer cell line. Several semi-synthetic derivatives of the lead compounds 3 and 6 were prepared in an attempt to obtain more potent bioactive analogues. All isolated and semi-synthetic compounds were evaluated for their cytotoxicity towards HT-29 cells in vitro, with the most active compound obtained (3,6-di-O-acetyl-α-mangostin, 8) being tested in an in vivo hollow fiber assay. Compounds obtained in this investigation were assessed for their NF-κB inhibitory and mitochondrial transmembrane potential (MTP) activities, where the quantities isolated permitted.
Results and Discussion
A chloroform-soluble extract of the methanol extract of the dried and ground stems of C. cochinchinense was separated by column chromatography over silica gel and yielded 15 pooled fractions. Combined fractions 3 and 4 were separated by silica gel column chromatography to afford cochinchinone A (6)13 and 3-geranyloxy-1,7-dihydroxyxanthone (cochinchinone G).11,14 The most active fractions 5 and 6 were separated by silica gel column chromatography to furnish the new compounds, cochinchinoxanthone (1) and cochinensoxanthone (2), along with the known compounds, α-mangostin (3),16 γ-mangostin (4),17 1,3,7-trihydroxy-2,4-diisoprenylxanthone (5),18 euxanthone,19 1,7-dihydroxy-4-methoxyxanthone,20 and β-sitosterol 3-O-β-D-glucopyranoside.21 Analogue development of α-mangostin (3) and cochinchinone A (6), which were both obtained in large amounts, yielded five new derivatives, 10 and 16–19. All these new compounds were determined structurally by comparison of their spectroscopic data with those of their parent compounds, 3 and 6. Also obtained were eight known analogues, 3-O-acetyl-α-mangostin (7),22 3,6-di-O-acetyl-α-mangostin (8),23 3,6,7-tri-O-acetyl-α-mangostin (9),24 3,6-di-O-methyl-α-mangostin (11),25 6-O-benzoyl-α-mangostin (12),26 18-O-formyl-3-isomangostin hydrate (13),27 3-isomangostin hydrate (14),17, 27 and 1-isomangostin hydrate (15).17,27 These known semi-synthetic compounds were identified by comparison of their spectroscopic data with those of the parent compounds and from literature values.
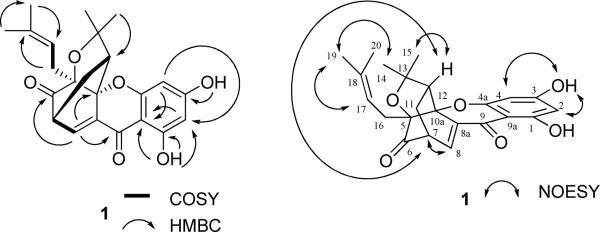
Compound 1 was isolated as an amorphous light yellow powder, with a molecular formula of C23H24O6, as determined by HRESIMS (m/z 419.1468 [M + Na]+, calcd for 419.1471). Both the UV (λmax 226, 327, and 346 nm) and IR [νmax 1744 (unconjugated carbonyl group), 1644 (hydrogen-bonded α,β-conjugated carbonyl group) cm−1] spectra showed absorption bands characteristic of a caged xanthone.28 This preliminary structural assignment was supported from the four three-proton methyl singlets at δ 1.08, 1.28, 1.37, and 1.67, a one-proton multiplet at δ 3.51 (m), and a one-proton triplet at δ 4.42 (J = 7.0 Hz) in the 1H NMR spectrum (Table 1). In turn, a signal at δ 203.1 for a carbonyl group and three signals at δ 84.0, 84.7, and 90.3 for three quaternary carbons, observed in the 13C NMR spectrum of 1 (Table 2), were typical signals for a caged xanthone.29–32 The NMR spectra of 1 indicated the presence of two prenyl groups, as supported by its elemental formula. In addition, the 1H NMR spectrum also revealed a hydrogen-bonded hydroxy group at C-1 (δ 12.45) and a second hydroxy group at C-3 (δ 6.26), which were confirmed by HMBC correlations between these hydroxy groups and C-1 and C-3, respectively (Figure 1). Furthermore, the HMBC correlations between H-11/C-6, C-8 and H-16/C-5, C-10a suggested that two prenyl groups are connected to C-5 and C-7, respectively. Therefore, 1 was determined as a 1,3-dihydroxy-5,7-diprenyl caged xanthone (Figure 1).
Table 1.
1H NMR Spectroscopic Data of Compounds 1, 2, 17, and 18
| position | 1a | 2a | 17b | 18b |
|---|---|---|---|---|
| 2 | 6.01 d (1.5) | |||
| 4 | 6.06 d (1.5) | |||
| 5 | 7.23 m | 7.38 d (9.0) | 7.40 d (9.0) | |
| 6 | 7.23 m | 7.29 dd (3.0, 9.0) | 7.29 dd (3.0, 9.0) | |
| 7 | 3.51m | |||
| 8 | 7.42 d (8.4) | 8.35 d (2.4) | 7.61 d (3.0) | 7.62 d (3.0) |
| 11 | 2.35 m | 2.78 dd (5.1, 17.1) | 3.46 d (7.0) | 3.41 d (6.8) |
| 1.31 m | 2.95 dd (5.1, 17.1) | |||
| 12 | 2.45 m | 3.91 t (5.1) | 5.27 m | 5.26 t (6.8) |
| 13 | ||||
| 14 | 1.28 s | 1.45 s | 1.74 s | 1.68 s |
| 15 | 1.67 s | 1.57 s | 1.83 s | 1.79 s |
| 16 | 2.61 d (7.0) | 3.61 d (7.2) | 3.57 d (8.0) | 3.53 d (6.7) |
| 17 | 4.42 t (7.0) | 5.28 m | 5.27 m | 5.21 t (6.7) |
| 19 | 1.37 s | 1.86 s | 1.86 s | 1.86 s |
| 20 | 1.08 s | 2.10 m | 2.09 m | 2.09 m |
| 21 | 2.10 m | 2.08 m | 2.09 m | |
| 22 | 5.03 brs | 5.03 t (8.0) | 5.00 t (6.8) | |
| 24 | 1.65 s | 1.62 s | 1.56 s | |
| 25 | 1.56 s | 1.56 s | 1.51 s | |
| OH-1 | 12.45 s | 13.21s | 12.97 s | |
| OH-3 | 6.26 s | |||
| OH-7 | 6.44 s | |||
| OMe-3 | 3.80 s | |||
| OMe-7 | 3.89 s | 3.90 s |
Data (δ) measured in CDCl3 at 300 MHz.
Data (δ) measured in CDCl3 at 400 MHz. s = singlet, brs = broad singlet, d = doublet, t = triplet, m = multiplet, dd = double doublet. J values presented in Hz and omitted if the signals overlapped as multiplets.
Table 2.
. 13C NMR Spectroscopic Data of Compounds 1, 2, 17, and 18
| position | 1a | 2a | 17b | 18b |
|---|---|---|---|---|
| 1 | 165.5 C | 155.1 C | 158.4 C | 158.7 C |
| 2 | 97.1 CH | 106.4 C | 109.1 C | 116.9 C |
| 3 | 165.4 C | 159.0 C | 160.9 C | 163.6 C |
| 4 | 95.4 CH | 105.0 C | 105.1 C | 113.0 C |
| 4a | 161.3 C | 153.5 C | 153.0 C | 153.0 C |
| 5 | 84.0 C | 117.9 CH | 119.0 CH | 119.2 CH |
| 6 | 203.1 C | 123.5 CH | 124.7 CH | 124.1 CH |
| 7 | 47.0 CH | 153.5 C | 155.9 C | 155.9 C |
| 8 | 133.8 CH | 111.8 CH | 105.1 CH | 105.9 CH |
| 8a | 135.6 C | 122.9 C | 120.6 C | 120.6 C |
| 9 | 179.4 C | 176.7 C | 181.0 C | 181.6 C |
| 9a | 101.3 C | 103.0 C | 103.3 C | 105.0 C |
| 10a | 90.3 C | 149.2 C | 150.8 C | 151.1 C |
| 11 | 25.4 CH2 | 26.2 CH2 | 21.6 CH2 | 22.7 CH2 |
| 12 | 49.0 CH | 68.7 CH | 121.6 CH | 125.2 CH |
| 13 | 84.7 C | 78.3 C | 135.1 C | 131.4 C |
| 14 | 29.2 CH3 | 24.5 CH3 | 25.9 CH3 | 25.6 CH3 |
| 15 | 30.5 CH3 | 22.0 CH3 | 18.0 CH3 | 17.9 CH3 |
| 16 | 29.2 CH2 | 22.2 CH2 | 21.8 CH2 | 22.5 CH2 |
| 17 | 118.3 CH | 121.3 CH | 121.6 CH | 122.8 CH |
| 18 | 134.1 C | 140.3 C | 137.9 C | 135.3 C |
| 19 | 25.8 CH3 | 16.3 CH3 | 16.3 CH3 | 16.3 CH3 |
| 20 | 17.1 CH3 | 39.7 CH2 | 39.7 CH2 | 39.7 CH2 |
| 21 | 26.2 CH2 | 26.4 CH2 | 26.5 CH2 | |
| 22 | 123.4CH | 123.9 CH | 122.6 CH | |
| 23 | 132.3 C | 131.9 C | 131.9 C | |
| 24 | 25.7 CH3 | 25.7 CH3 | 25.7 CH3 | |
| 25 | 17.7 CH3 | 17.7 CH3 | 17.7 CH3 | |
| OMe-3 | 61.9 CH3 | |||
| OMe-7 | 56.0 CH3 | 56.0 CH3 |
Data (δ) measured in CDCl3 at 75.5 MHz.
Data (δ) measured in CDCl3 at 100.6 MHz.
Figure 1.
COSY, selected NOESY, and key HMBC correlations of 1.
An isomer of 1 has been reported in previous studies.13,14,29,31 This was derived chemically from bractatin treated with formic acid,29 and later isolated from the roots of C. cochinchinense,13 representing at the time the first discovery of caged xanthones from a plant genus other than Garcinia. To further investigate this compound, Li et al. developed a method to synthesize the scaffold of this compound.31 However, the finalized structural information and detailed spectroscopic data concerning this compound have not been reported.
Several caged xanthones have now been isolated from C. cochinchinense, but their absolute configuration was not determined.10,13,14,29,31 Some caged xanthones isolated from C. cochinchinense contain the same structural skeleton but show opposite (positive or negative) specific rotation values.13 Comparison of the literature data of this earlier caged xanthone29 with those of 1 revealed that both compounds exhibit similar NMR spectra, except for differences between the methyl group signals, which appeared at δ 1.32 (H-20), 1.42 (H-24, equal to H-14 of 1), 1.44 (H-19), and 1.70 (H-25, equal to H-15 of 1) in the substance described in the literature,29 but were displayed at δ 1.08 (H-20), 1.28 (H-14), 1.37 (H-19), and 1.67 (H-15) in the 1H NMR spectrum of compound 1. These differences suggested 1 to be an isomer of the literature xanthone, exhibiting one or more different configurations at C-5, -7, -10a, and/or -12.
In a previous collaborative study on the determination of the configuration of caged xanthones using electronic circular dichroism (ECD), the absolute configuration of (−)-morellic acid was determined as 5R, 7S, 10aS, and 27S, respectively.32 The CD spectrum of (−)-morellic acid showed a positive and a negative Cotton effect at 293.5 and at 360.5 nm.32 However, the CD spectrum of compound 1 exhibited a negative and a positive Cotton effect at 292 nm at 353 nm. Based on a comparison of the CD spectrum of 1 with that of (−)-morellic acid, the absolute configuration of C-5 and C-7 of 1 may be proposed as 5S and 7R. NOESY correlations between H-7/H-8, -12, H-12/H-8, -15, H-14, -17/H-19 (Figure 1) indicated the R configuration for C-10a and C-12. Accordingly, the absolute configuration of compound 1 could be defined as 5S, 7R, 10aR, and 12R. Therefore, 1 was determined as (1S,3aR,5R,12aR)-3,3a,4,5-tetrahydro-8,10-dihydroxy-3,3-dimethyl-1-(3-methyl-2-buten-1-yl)-1,5-methano-1H,7H-furo[3,4-d]xanthene-7,13-dione.33 This compound has been assigned the trivial name, cochinchinoxanthone.
Compound 2 was isolated and purified as an amorphous yellow powder and showed a purple color under UV light at 365 nm. The HRESIMS exhibited a sodiated molecular ion peak at m/z 487.2131 (calcd 487.2097), consistent with a molecular formula of C28H32O6. The UV (λmax 242, 308 nm) and IR (νmax 3344, 1632, 1616, 1587, 1457 cm−1) spectra showed the absorption characteristics of a prenylxanthone.34 Analysis of the 1H and 13C NMR data gave evidence of 2 being a C-1, -2, -3, -4, and -7 penta-substituted xanthone. The 1H NMR spectrum (Table 1) showed three aromatic protons at δ 7.23 × 2 and 8.35 (Table 1). The 13C NMR and DEPT spectra contained signals consistent with the presence of a xanthone unit substituted by a prenyl group at C-2 and a geranyl group at C-4. These assignments were supported by HMBC correlations between H-15/C-12 and H-16/C-3, -4a (Figure 2). Comparison of the 1H and 13C NMR data of 2 with those of compound 613 indicated the same partial structures due to their xanthone unit and the geranyl group at C-4, but a difference in regard to the prenyl group at C-2 of these compounds. The signals at δ 121.6 (CH) and 135.0 (C) observed for 6 were replaced in 2 by those at δ 68.7 (CH) and 78.3 (C) (Table 2), due to a 3-hydroxy-2,2-dimethyldihydropyran ring,34 which was confirmed by the mass spectrum of 2 that showed the molecule to contain an extra oxygen atom when compared with 6. The HMBC correlation between H-15/C-12, together with the NOESY correlations between H-15/H-11, H-12 indicated that this hydroxy group is linked to C-12 (Figure 2). In the 1H NMR spectrum (Table 1), the lack of a signal at ca. δ 12.8 due to a hydrogen-bonded hydroxy group in 6 suggested that the 3-hydroxy-2,2-dimethyldihydropyran ring is located at C-1 and C-2 rather than at C-2 and C-3. This determination was substantiated by the signals evident for C-12 at δ 69.0/70.4/68.8 (CH) and for C-13 at δ 78.5/79.6/78.2 (C) of some 3-hydroxy-2,2-dimethyldihydropyranoxanthones.9,35,36
Figure 2.
COSY, selected NOESY, and key HMBC correlations of 2.
The absolute configuration at C-3 of a 3-hydroxy-2,2-dimethyldihydropyranoxanthone has not been determined. It has been reported that several natural products containing one stereocenter in a 3(R)-hydroxy-2,2-dimethyldihydropyran residue show negative specific rotation values, while their S isomers exhibit positive specific rotation values.37,38 For example, a model compound containing a 3(R)-hydroxy-2,2-dimethylchroman system showed a specific rotation value of −102 in pyridine (−11 in CHCl3), while its isomer with a 3(S)-hydroxy-2,2-dimethylchroman system exhibited a specific rotation value of +102 in pyridine.37 Another model compound, (2R)-2-hydroxy-1,2-dihydroacronycine, exhibited a specific rotation value of −14.9 in CHCl3, but its isomer, (2S)-2-hydroxy-1,2-dihydroacronycine, gave a specific rotation value of +15.2 in CHCl3.38 The absolute configuration at C-12 of 2 was postulated as being S because it displays a positive specific rotation value of +11 in CH2Cl2 and is consistent with those of 3(S)-hydroxy-2,2-dimethyldihydropyran derivatives.37,38 Therefore, 2 was proposed as 3,4-dihydro-3(S),5,10-trihydroxy-2,2-dimethyl-6-[3,7-dimethylocta-2(E),6-dien-1-yl]-2H,12H-pyrano[2,3-a]xanthen-12-one,39 and it has been assigned the trivial name, cochinensoxanthone.
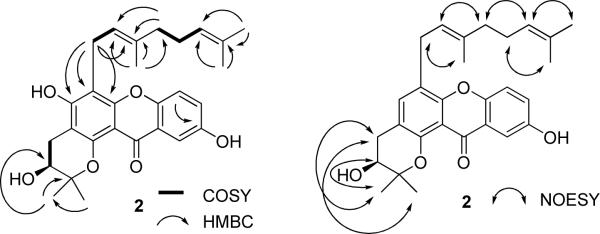
The most potent cytotoxic compound (α-mangostin, 3) obtained in a relatively large quantity from C. cochinchinense, was modified chemically by a series of synthetic methods, including acetylation, methylation, benzoylation, and cyclization (Schemes S1–S4, Supporting Information). All the semi-synthetic derivatives obtained were determined structurally by comparison of their spectroscopic data with those of the parent compound (Tables 1–4 and Tables S1–S6, Supporting Information,). The new compound 10, obtained by methyl iodide methylation of 3, gave a sodiated molecular ion peak at m/z 475.2052 (C27H32O6Na), indicating the presence of three additional methyl groups when compared with 3. A signal at δ 13.45 in the 1H NMR spectrum suggested a hydrogen-bonded hydroxy group at C-1, and a signal at δ 106.0 in the 13C NMR spectrum was consistent with a methyl group being linked to C-4, because this signal appeared at 93.3 in the 13C NMR spectrum of 3. Thus, 10 was determined as 4-methyl-3,6-di-O-methyl-α-mangostin.
Table 4.
13C NMR Spectroscopic Data of Compounds 10, 16, and 19
| position | 10a | 16b | 19a |
|---|---|---|---|
| 1 | 158.6 C | 158.5 C | 158.7 C |
| 2 | 116.6 C | 116.9C | 109.7 C |
| 3 | 162.8 C | 154.3 C | 161.5 C |
| 4 | 106.0 C | 112.8 C | 105.6 C |
| 4a | 158.3 C | 153.6 C | 153.7 C |
| 5 | 98.3 CH | 119.0 CH | 118.3 CH |
| 6 | 152.3 C | 129.5 CH | 129.1 CH |
| 7 | 144.0 C | 146.6 C | 146.9 C |
| 8 | 137.3 C | 118.0 CH | 119.0 CH |
| 8a | 111.9 C | 120.8 C | 121.3 C |
| 9 | 182.8 C | 181.4 C | 180.7 C |
| 9a | 107.6 C | 107.0 C | 103.5 C |
| 10a | 155.7 C | 152.5 C | 153.1 C |
| 11 | 22.4 CH2 | 22.9 CH2 | 21.8 CH2 |
| 12 | 122.8 CH | 121.3 CH | 121.6 CH |
| 13 | 131.9 C | 132.3 C | 135.5 C |
| 14 | 25.8 CH3 | 25.7 CH3 | 25.9 CH3 |
| 15 | 18.2 CH3 | 17.9 CH3 | 17.9 CH3 |
| 16 | 26.2 CH2 | 23.1 CH2 | 22.1 CH2 |
| 17 | 123.1 CH | 121.3 CH | 121.7 CH |
| 18 | 131.6 C | 135.8 C | 138.1 C |
| 19 | 25.9 CH3 | 16.3 CH3 | 16.5 CH3 |
| 20 | 17.9 CH3 | 39.6 CH2 | 39.9 CH2 |
| 21 | 26.5 CH2 | 26.6 CH2 | |
| 22 | 124.0 CH | 124.1 CH | |
| 23 | 131.5 C | 132.1 C | |
| 24 | 25.6 CH3 | 26.1 CH3 | |
| 25 | 17.7 CH3 | 18.2 CH3 | |
| OMe-3 | 61.0 CH3 | ||
| OMe-6 | 56.1 CH3 | ||
| OMe-7 | 61.1 CH3 | ||
| Me-4 | 8.5 CH3 | ||
| OAc-3 | 168.3 C | ||
| 20.6 CH3 | |||
| OCOH-6 | |||
| OAc-7 | 169.2 C | ||
| 21.0 CH3 | |||
| 1' | 165.4 C | ||
| 2' | 129.3 C | ||
| 3' | 130.5 CH | ||
| 4' | 128.9 CH | ||
| 5' | 134.1 CH | ||
| 6' | 128.9 CH | ||
| 7' | 130.5 CH |
Data (δ) measured in CDCl3 at 100.6 MHz.
Data (δ) measured in CDCl3 at 75.5 MHz.
A second compound (cochinchinone A, 6) isolated in a reasonable quantity from C. cochinchinense, was derivatized chemically using the same methods as described for compound 3. The identities of these derivatives were determined by comparison of their spectroscopic data with those of the parent xanthone (Tables 1–4, and Tables S1 and S2, Supporting Information). Compound 16 gave a sodiated molecular ion peak at m/z 555.2416, corresponding to a molecular formula of C32H36O7, indicating the presence of two additional acetyl groups when compared to 6. A signal at δ 12.80 in the 1H NMR spectrum was assigned to a hydrogen-bonded hydroxy group at C-1, consistent with the structure of 16 being 3,7-di-O-acetylcochinchinone A. Likewise, the pseudomolecular ion peak at m/z 485.2355 (C29H34O5Na) indicated that 17 contains an additional methyl group when compared with 6. In the 13C NMR spectrum of 17, a signal at δ 155.9 indicated a methoxy group linked to C-7, because the signal for this carbon appeared at δ 152.3 in the 13C NMR spectrum of 6. Therefore, 17 was assigned as 7-O-methylcochinchinone A. The HRESIMS at m/z 499.2484 (C30H36O5Na) showed that 18 contains an additional methyl group when compared with 17. A signal at δ 12.97 due to a hydrogen-bonded hydroxy group in the 1H NMR spectrum was consistent with the structure of 18 being 3,7-di-O-methylcochinchinone A. Compound 19, also prepared from cochinchinone A (6), gave a sodiated ion peak at m/z 575.2432 (C35H36O6Na), consistent with the presence of a benzoyl group. A signal at δ 158.7 in the 13C NMR spectrum indicated a benzoyl group at C-7, because this carbon signal appeared at δ 152.3 in the 13C NMR spectrum of 6. Therefore, 19 is 7-O-benzoylcochinchinone A.
All isolated and semi-synthesized compounds were tested in terms of their cytotoxicity against the HT-29 human colon cancer cell line, using paclitaxel as positive control (Table 5). Compounds 1, 3, 4, 7–9, and 12–14 were found to be cytotoxic, of which 8 and 12 were the most potent active substances, with ED50 values of 1.0 and 1.9 μM, respectively. Further, compound 8 (3,6-di-O-acetyl-α-mangostin) was tested in an in vivo hollow fiber assay, but was found to be inactive at the highest dose used (20 mg/kg; ip).
Table 5.
Biological Activity of Compounds Isolated and Derivatized from the Stems of C. cochinchinense and their Analogues
| compound | cytotoxicitya | NF-κB p65 inhibitionb | MTPc |
|---|---|---|---|
| 1 | 5.8 | >10 | 3.3 |
| 2 | >10 | NTd | NTd |
| 3 | 4.1 | >10 | 0.2 |
| 4 | 4.0 | NTd | >10 |
| 5 | >10 | 2.9 | NTd |
| 6 | >10 | >10 | >10 |
| 7 | 8.8 | >10 | NTd |
| 8 | 1.0 | >10 | 1.8 |
| 9 | 6.0 | >10 | NTd |
| 10 | >10 | >10 | 0.9 |
| 11 | >10 | >10 | 1.4 |
| 12 | 1.9 | >10 | >10 |
| 13 | 4.4 | NTd | >10 |
| 14 | 4.4 | NTd | >10 |
| 15 | >10 | >10 | >10 |
| 16 | >10 | >10 | NTd |
| 17 | >10 | >10 | >10 |
| 18 | >10 | >10 | NTd |
| 19 | >10 | >10 | >10 |
Data presented as ED50 values (μM) towards HT-29 cells, with paclitaxel used as a positive control (ED50, 0.10 nM).
Data presented as IC50 values (μM), with rocaglamide used as positive control (IC50, 0.075 μM).
Mitochondrial transmembrane potential, data presented as IC50 values (μM), with staurosporine used as positive control (IC50, 2.0 nM).
Not tested.
Comparison of the cytotoxicity of compound 1 with those of the caged xanthones obtained in a previous study32 indicated that the prenyl groups at C-2 and C-4 and a carboxyl group at C-18 are not necessary for the mediation of cytotoxicity of caged xanthones. Chemical modification of compound 3 (α-mangostin, ED50, 4.1 μM) demonstrated that both 3,6-di-acetylation (e. g., 8, ED50, 1.0 μM) and 6-benzoylation (e. g., 12, ED50, 1.9 μM) improved the resultant cytotoxicity. Also, cyclization at C-2 and C-3 (e. g., 13, ED50, 4.4 μM and 14, ED50, 4.4 μM) retained the initial cytotoxic potency of 3, but cyclization at C-1 and C-2 (e. g., 15, ED50, > 20 μM) and 3,6-dimethylation (e. g., 10, ED50, >20 μM and 11, ED50, > 20 μM) greatly decreased such activity. In turn, neither acetylation, methylation, nor benzoylation enhanced the cytotoxicity of cochinchinone A (6, ED50, 16.1 μM).
Some of the compounds with sufficient quantities were tested in a NF-κB p65 inhibition assay, using rocaglamide as positive control (Table 5). Compound 5 (1,3,7-trihydroxy-2,4-diisoprenylxanthone) exhibited the most potent activity (IC50 value of 2.9 μM) of the substances tested in this assay. In a mitochondrial transmembrane potential (MTP) assay (Table 5), the two new compounds, 1 (IC50, 3.3 μM) and 10 (IC50, 1.4 μM), were active. The two known compounds, 3 (α-mangostin) and 11 (3,6-di-O-methyl-α-mangostin), exhibited promising activity with IC50 values of 0.2 μM and 0.9 μM, respectively.
Experimental Section
General Experimental Procedures
Specific rotation values were measured with a Perkin-Elmer model 343 polarimeter. UV spectra were recorded on a Hitachi U2910 UV spectrophotometer. CD measurements were performed using a JASCO J-810 spectropolarimeter. IR spectra were recorded on a Nicolet 6700 FT-IR spectrometer. 1H and 13C, DEPT, HMQC, HMBC, NOESY, and COSY NMR spectra were recorded at room temperature on a Bruker Avance DPX-300 or DRX-400 NMR spectrometer, with TMS as internal standard. ESIMS and HRESIMS were recorded on a LCT-TOF mass spectrometer. Column chromatography was conducted using silica gel (70–230 mesh, Merck, Darmstadt, Germany). Analytical and preparative thin-layer chromatography (TLC) were performed on precoated silica gel 60 F254 plates (Sorbent Technologies, Atlanta, GA). Sephadex LH-20 was purchased from Amersham Biosciences, Uppsala, Sweden. For visualization of TLC plates, H2SO4 reagent was used. Fluorescence was tested under a Spectroline (model ENF-260C) UV light source. HPLC was performed on a Hitachi (Hitachi High Technologies America) HPLC instrument with a model Prep pump, an Elite LaChrom L-2200 autosampler, and an Elite LaChrom L-2400 UV detector. Chromatograms were recorded using EZChrom Elite software (Hitachi). Analytical HPLC was carried out on a YMC (PH12S05-1546 WT) column (150 mm × 4.6 mm i.d., 5 μm) used together with a YMC pre-column, having an injector with a 500 μL loop. Semi-preparative HPLC was carried out on a YMC (PH12S05-1520 WT) column (150 mm × 20 mm i.d., 5 μm), again used together with a YMC pre-column. Solvents A and B of the mobile phase were MeOH and H2O, respectively. A linear gradient was applied from a 50:50 A/B mixture to a 80:20 A/B mixture, over 30 min at room temperature, at a flow rate of 1 mL/min for the analytical column or 5 mL/min for the semi-preparative column, and the detection wavelength was at 250 nm. All other procedures were carried out using anhydrous solvents purchased from commercial sources and employed without further purification. All reagents for chemical synthesis were purchased from Sigma except where indicated. Reactions were monitored by TLC using precoated silica gel plates.
Plant Material
A sample of the stems of Cratoxylum cochinchinense was collected at Hon Ba Nature Reserve, Dien Khanh District, Khanh Hoa Province, Vietnam, in November, 2004. The voucher herbarium specimen (Soejarto et al. 13598), was identified by D.D.S. as C. cochinchinense (Lour.) Bl. (Clusiaceae), and was deposited at the John G. Searle Herbarium of the Field Museum of Natural History, Chicago, under accession number FM 2257409.
Extraction and Isolation
The ground stems of C. cochinchinens (450 g) were extracted with MeOH (3 L × 5) at room temperature. The solvent was evaporated in vacuo. The resultant dried MeOH extract (33 g, 7.3%) was suspended in 10% H2O in MeOH (600 mL) and partitioned with n-hexane (300 mL × 3) to yield a hexane-soluble residue (6.0 g, 1.3%). The aqueous-MeOH layer was then partitioned with CHCl3 (500, 400, 300, 200 mL) to afford a chloroform-soluble extract (3.3 g, 0.7%), which was washed with a 1% aqueous solution of NaCl, to partially remove any plant polyphenols present.
The active chloroform-soluble extract (3.0 g, ED50 < 20 μg/mL) was subjected to passage over a silica gel column (4.5 × 45 cm), eluted with gradient mixture of n-hexane–acetone (100:1→1:1; 500 mL each). Fractions were pooled by TLC analysis to give 15 combined fractions. Of these, fractions 3 and 4 (ED50 < 10 μg/mL) were combined and further chromatographed over a silica gel column (2.5 × 20 cm), eluted with a gradient mixture of nhexane–acetone (20:1→3:1, 200 mL each), to yield five combined fractions. Fractions 3-1 and 3-2 were chromatographed over silica gel using n-hexane-acetone (10:1) as solvent, and then purified by separation over a Sephadex LH-20 column (2.5 × 25 cm), eluted with CHCl3–MeOH (1:1), affording cochinchinone A (6, 250 mg). Fractions 3-3 to 3-5 were combined and separated by silica gel chromatography, eluted with n-hexane–acetone (10:1), and then purified by passage over a Sephadex LH-20 column, eluted with a mixture of CHCl3–MeOH (1:1), to afford 3-geranyloxy-1,7-dihydroxyxanthone (1.0 mg).
Fractions 5 and 6 (ED50 < 5 μg/mL) were combined and chromatographed over a silica gel column (2.5 × 20 cm), eluted with a gradient mixture of n-hexane–acetone (10:1→1:1, 300 mL each), to yield nine combined fractions. Fraction 5-1 was chromatographed over silica gel using n-hexane–acetone (5:1) as solvent, and then purified by passage over a Sephadex LH-20 column, eluted with CHCl3–MeOH (1:1), affording 2 (2.0 mg). Fraction 5-2 was purified by silica gel chromatography, eluted by n-hexane–acetone (5:1), and then purified by preparative HPLC, to afford 1,3,7-trihydroxy-2,4-diisoprenylxanthone (5) (tR 13.1 min, 3.5 mg). Fraction 5-3 was chromatographed sequentially over silica gel, eluted by n-hexane–acetone (5:1), and over Sephadex LH-20, eluted with a mixture of CHCl3–MeOH (1:1), to afford α-mangostin (3, 75 mg). Fractions 5-4 and 5-5 were combined and chromatographed over silica gel, eluted by nhexane–acetone (3:1), and then purified over a Sephadex LH-20 column, with CHCl3–MeOH (1:1) as eluent, affording 1,7-dihydroxy-4-methoxyxanthone (1 mg). Fraction 5-6 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified over a Sephadex LH-20 column, using CHCl3–MeOH (1:1) for elution, furnishing euxanthone (1.5 mg). Fraction 5-7 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then separated over Sephadex LH-20, using CHCl3–MeOH (1:1) for elution, and finally purified by semi-preparative HPLC, to yield 1 (tR 15.5 min, 5 mg). Fraction 5-8 was chromatographed over silica gel, eluted by n-hexane–acetone (1:1), and then purified over a Sephadex LH-20 column, using CHCl3–MeOH (1:1) for elution, furnishing γ-mangostin (4, 2 mg). Fraction 5-9 was chromatographed over silica gel, eluted by n-hexane–acetone (1:1), and then purified over a column containing Sephadex LH-20, using CHCl3–MeOH (1:1) for elution, affording β-sitosterol 3-O-β-D-glucopyranoside (7 mg).
Cochinchinoxanthone (1)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; [α]20D +10 (c 0.1, CH2Cl2); UV (MeOH) λmax (log ε) 226 (4.67), 327 (4.59), 346 (4.60) nm; CD (MeOH, nm) λmax (ε) 213 (−26.96), 292 (−2.41), 303.5 (−2.64), 353 (+1.97); IR (dried film) νmax 3230, 1744, 1644, 1597, 1461, 1333, 1274, 1165, 815 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 419.1468, calcd for C23H24O6Na, 419.1471.
Cochinensoxanthone (2)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; [α]20D +11 (c 0.1, CH2Cl2); UV (MeOH) λmax (log ε) 242 (4.58), 308 (4.22) nm; CD (MeOH, nm) λmax (Δε) 206.5 (+27.43), 228 (+2.63), 262.5 (+3.90), 283 (−2.83); IR (dried film) νmax 3344, 1632, 1616, 1587, 1457, 1226, 1125 cm−1;1 and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 487.2131, calcd for C28H32O6Na, 487.2097.
Acetylation of α-Mangostin (3) and Cochinchinone A (6)
To a dried 50 mL flask equipped with water condenser and magnetic stirrer, containing 12.3 mg (0.03 mmol) of α-mangostin (3), acetic anhydride (9.2 mg, 0.09 mmol) and pyridine (5 mL) were added. After the mixture was stirred at 60 °C for 1 h, the mixture was cooled to room temperature. Then, 5 mL of CHCl3 were transferred into the flask, and the solution was extracted with distilled H2O. The organic layer was washed with distilled H2O, and then evaporated at reduced pressure. The residue was separated by silica gel column chromatography, using n-hexane and acetone (5:1→1:1), to afford 3-O-acetyl-α-mangostin (7, 1.5 mg, 0.003 mmol, 11.0%) and 3,6-di-O-acetyl-α-mangostin (8, 12.0 mg, 0.024 mmol, 81.0%). Using the same procedure, reaction at 70 °C for 2 h yielded 3,6-di-O-acetyl-α-mangostin (8, 12.5 mg, 0.025 mmol, 84.3%) and 3,6,7-tri-O-acetyl-α-mangostin (9, 2.5 mg, 0.0047 mmol, 12.4%).
The same reaction as described above with 13.4 mg (0.03 mmol) of cochinchinone A (6) at 60 °C for 1 h produced 3,7-di-O-acetylcochinchinone A (16, 12 mg, 0.022 mmol, 75.2%).
3,7-Di-O-acetylcochinchinone A (16)
Amorphous white powder (n-hexane) showing a purple color under UV light at 365 nm; UV (MeOH) λmax (log ε) 235 (4.57), 260 (4.60), 286 (4.07) nm; IR (dried film) νmax 2968, 1767, 1635, 1616, 1587, 1484, 1463, 1372, 878 cm−1; 1H and 13C NMR data, see Tables 3 and 4; positive HRESIMS m/z 555.2416, calcd for C32H36O7Na, 555.2359.
Table 3.
1H NMR Spectroscopic Data of Compounds 10, 16, and 19
| position | 10a | 16b | 19b |
|---|---|---|---|
| 4 | |||
| 5 | 6.79 s | 7.46 m | 8.06 d (2.4) |
| 6 | 7.46 m | 7.58 m | |
| 8 | 7.94 brs | 7.58 m | |
| 11 | 3.39 d (6.3) | 3.28 brs | 3.47 d (7.0) |
| 12 | 5.26 m | 5.14 m | 5.25 m |
| 14 | 1.67 s | 1.67 s | 1.75 s |
| 15 | 1.79 s | 1.76 s | 1.84 s |
| 16 | 4.12 d (6.3) | 3.40 brs | 3.58 d (6.9) |
| 17 | 5.26 m | 5.03 t (6.6) | 5.25 m |
| 19 | 1.65 s | 1.83 s | 1.87 s |
| 20 | 1.83 s | 2.04 m | 2.10 m |
| 21 | 2.00 m | 2.10 m | |
| 22 | 5.14 m | 5.04 m | |
| 24 | 1.57 s | 1.66 s | |
| 25 | 1.53 s | 1.56 s | |
| OH-1 | 13.45 s | 12.80 s | 13.10 s |
| OH-3 | 6.50 s | ||
| OMe-3 | 3.78 s | ||
| OMe-6 | 3.97 s | ||
| OMe-7 | 3.77 s | ||
| Me-4 | 2.30 s | ||
| OAc | 2.34 s | ||
| OAc | 2.33 s | ||
| 3' | 8.22 d (7.5) | ||
| 4' | 7.58 m | ||
| 5' | 7.65 t (7.5) | ||
| 6' | 7.58 m | ||
| 7' | 8.22 d (7.5) |
Data (δ) measured in CDCl3 at 400 MHz.
Data (δ) measured in CDCl3 at 300 MHz. s = singlet, brs = broad singlet, d = doublet, t = triplet, m = multiplet. J values presented in Hz and omitted if the signals overlapped as multiplets.
Methylation of α-Mangostin (3) and Cochinchinone A (6)
To a dried flask (50 mL) equipped with a water condenser and a magnetic stirrer, containing 12.3 mg (0.03 mmol) of α-mangostin (3), 20.8 mg of Ag2O (0.09 mmol), 12.8 mg of MeI (0.09 mmol), and 5 mL of CH2Cl2 were added. After being stirred at 40 °C for 4 h, the mixture was cooled to room temperature, and washed with distilled H2O. The organic layer was evaporated at reduced pressure. The residue was separated by silica gel column chromatography, using n-hexane and acetone (5:1→1:1) for elution, to afford 4-methyl-3,6-di-O-methyl-α-mangostin (10, 2.0 mg, 0.0042 mmol, 14.7%) and 3,6-di-O-methyl-α-mangostin (11, 4.0 mg, 0.009 mmol, 30.4%). The same reaction performed as described above with 13.4 mg (0.03 mmol) of cochinchinone A (6) and 20.8 mg (0.09 mmol) of Ag2O gave 7-O-methylcochinchinone A (17, 2.0 mg, 0.0043 mmol, 14.4%) and 3,7-di-O-methylcochinchinone A (18, 1.5 mg, 0.0032 mmol, 10.5%).
4-Methyl-3,6-di-O-methyl-α-mangostin (10)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; UV (MeOH) λmax (log ε) 238 (4.38), 271 (4.45), 307 (4.25) nm; IR (dried film) νmax 2925, 1645, 1600, 1469, 1373, 1284, 826 cm−1; 1H and 13C NMR data, see Tables 3 and 4; positive HRESIMS m/z 475.2052, calcd for C27H32O6Na, 475.2097.
7-O-Methylcochinchinone A (17)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; UV (MeOH) λmax (log ε) 236 (4.75), 266 (4.79), 316 (4.44) nm; IR (dried film) νmax 3390, 1651, 1600, 1455, 1372, 1220, 821 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 485.2355, calcd for C29H34O5Na, 485.2304.
3,7-Di-O-methylcochinchinone A (18)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; UV (MeOH) λmax (log ε) 238 (4.41), 267 (4.53), 297 (3.96) nm; IR (dried film) νmax 2921, 1641, 1608, 1488, 1469, 1281, 821 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 499.2 [M + Na]+; positive HRESIMS m/z 499.2484, calcd for C30H36O5Na, 499.2460.
Benzoylation of α-Mangostin (3) and Cochinchinone A (6)
To a dried flask (50 mL) equipped with a water condenser and a magnetic stirrer, 12.3 mg (0.03 mmol) of α-mangostin (3), 11.0 mg of benzoic acid (0.09 mmol), 6.2 mg of DCC (0.03 mmol), and 5 mL of CH2Cl2 were added. After the mixture was stirred at 40 °C for 4 h, it was cooled to room temperature, and then washed with distilled H2O. The organic layer was evaporated under reduced pressure, and the residue separated by silica gel column chromatography, using n-hexane and acetone (5:1→1:1) as eluents, to produce 6-O-benzoyl-α-mangostin (12, 3.0 mg, 0.0058 mmol, 19.5%). The same reaction as described above was performed on 13.4 mg (0.03 mmol) of cochinchinone A (6) to afford 7-O-benzoylcochinchinone A (19, 2.0 mg, 0.0036 mmol, 12.1%).
7-O-Benzoylcochinchinone A (19)
Amorphous yellow powder (n-hexane) showing a purple color under UV light at 365 nm; UV (MeOH) λmax (log ε) 239 (4.53), 257 (4.36), 313 (4.05) nm; IR (dried film) νmax 1743, 1645, 1483, 1253, 807 cm−1; 1H and 13C NMR data, see Tables 3 and 4; positive HRESIMS m/z 575.2432, calcd for C35H36O6Na, 575.2410.
Cyclization of α-Mangostin (3)
To a dried flask (50 mL) equipped with water condenser and magnetic stirrer, containing 12.3 mg (0.03 mmol) of α-mangostin (3), 1.44 mg of formic acid (0.09 mmol), 6.18 mg of DCC (0.03 mmol), and 5 mL of CH2Cl2 were added. After the mixture was stirred at 40 °C for 4 h and cooled to room temperature, it was washed with distilled H2O. The organic layer was evaporated at reduced pressure, and the residue was separated by silica gel column chromatography, using n-hexane and acetone (5:1→1:1) as eluents, to afford 18-O-formyl-3-isomangostin hydrate (13, 1.5 mg, 0.0033 mmol, 11.0%), 3-isomangostin hydrate (14, 3.0 mg, 0.007 mmol, 23.4%), and 1-isomangostin hydrate (15, 1.5 mg, 0.0035 mmol, 11.7%).
Cytotoxicity Assay
Cytotoxicity of the samples was performed against HT-29 human colon cancer cells by a previously reported procedure.32 The cells were cultured under standard conditions and trypsinized. Then, the harvested cells were added to 96-well plates and treated by the test samples dissolved in DMSO at different concentrations, the positive control, and the negative control (DMSO). The plates were incubated at 37 °C in 5% CO2 for three days, and then the cells were fixed, incubated at room temperature for 30 min, washed with tap water, dried at room temperature overnight, and dyed using sulforhodamine B. After the dyed cells were lysed in tris-base buffer, the plates were read at 515 nm with an ELISA plate reader. Paclitaxel was used as a positive control, and the ED50 values of the test samples in serial dilutions were calculated using nonlinear regression analysis (Table Curve2Dv4; AISN Software, Inc., Mapleton, OR). Measurements were performed in triplicate and are representative of two independent experiments in which the values generally agreed within 10%.
Enzyme-Based ELISA NF-κB Inhibition Assay
A NF-κB inhibition assay was carried out using a published procedure, with an EZ-Detect Transcription Factor Assay System ELISA kit (Pierce Biotechnology, Rockford, IL).40 Thus, nuclear extracts of HeLa cells (ATCC, American Type Culture Collection) treated with the positive control and the test samples at four different concentrations were used to determine the specific binding ability of the activated p65 subunit of NF-κB to the biotinylated-consensus sequence and was measured by detecting the chemiluminescent signal in a Fluostar Optima plate reader (BMG Labtech, Inc., Durham, NC). Rocaglamide was used as a positive control,40 and measurements were performed in duplicate and are representative of two independent experiments, with the values generally agreed within 10%. The dose response curve was calculated for IC50 determinations using non-linear regression analysis (Table Curve2DV4; AISN Software Inc., Mapleton, OR).
Mitochondrial Transmembrane Potential (MTP) Assay
Changes in mitochondrial transmembrane potential were determined by a fluorescence cell-based assay, using a previously described protocol.41 In brief, HT-29 cells at a density of 6 × 104 were incubated overnight at 37 °C in a CO2 incubator on black 96-well plates. Cells were then treated by the test compounds or the positive control at four different concentrations for 2 h. Soon afterwards, cells were incubated with the lipophilic cationic dye 5,5,6,6-tetrachloro-1,1,3,3-tetraethylbenzymidazolylcarbocyanide (JC-1, Cayman Chemical Company, Ann Arbor, MI) for 30 min. After incubation, cells were washed to remove unbound staining dye. The black 96-well plates were analyzed by a Fluostar Optima fluorescence plate reader (BMG Labtech, Inc.) with excitation wavelengths of 485 nm and 560 nm, and emission wavelengths of 530 nm and 595 nm for JC-1 monomers and J-aggregates, respectively. Measurements were performed in triplicate and are representative of at least two independent experiments with values generally agreed within 10%. The dose response curve was determined for IC50 determinations using non-linear regression analysis (Table Curve2DV4; AISN Software Inc., Mapleton, OR).
In Vivo Hollow Fiber Assay
The in vivo hollow fiber assay was conducted as described previously.32,36 Three human cancer cell lines, designated HT-29 (colon adenocarcinoma), MCF-7 (breast cancer), and MDA-MB-435 (melanoma), were used in this study, and paclitaxel was used as positive control substance. A dose range of 1 – 20 mg /kg (ip) was used for both the test compounds and the positive control.
Supplementary Material
Acknowledgment
This investigation was supported by grants U01 CA52956 and P01 CA125066, funded by the National Cancer Institute, NIH, Bethesda, MD. The plant sample Cratoxylum cochinchinense (A05984) was collected under a collaborative arrangement between the University of Illinois at Chicago (USA) and the Institute of Ecology and Biological Resources of the Vietnam Academy of Science and Technology, Hanoi (Vietnam). We are grateful to Dr. David J. Hart, Department of Chemistry, The Ohio State University, for helpful advice concerning the semi-systematic names used for compounds 1 and 2. We thank Dr. Kari Green-Church, of the Mass Spectrometry and Proteomics of the Campus Chemical Instrument Center, The Ohio State University, for assistance with the MS measurements. We also thank Jack Fowble, College of Pharmacy, The Ohio State University, for access to the NMR instrumentation used in this investigation.
Footnotes
Supporting Information Available: Mass, and 1H and 13C NMR spectra of compounds 1, 2, 10, 13, and 16–19; schemes for the synthesis of analogues of α-mangostin (3) and cochinchinone A (6); 1H and 13C NMR assignments of known xanthones isolated or derivatized from C. cochinchinense; physical data of the known xanthones isolated or derivatized from C. cochinchinense; and evaluation of 3,6-di-O-acetyl-α-mangostin (8) in an in vivo hollow fiber assay. This information is available free-of-charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Iinuma M, Tosa H, Ito T, Tanaka T, Madulid DA. Phytochemistry. 1996;42:1195–1198. [Google Scholar]
- (2).Pattanaprateeb P, Ruangrungsi N, Cordell GA. Planta Med. 2005;71:181–183. doi: 10.1055/s-2005-837788. [DOI] [PubMed] [Google Scholar]
- (3).(a) Boonnak N, Karalai C, Chantrapromma S, Ponglimanont C, Fun H-K, Kanjana-Opas A, Laphookhieo S. Tetrahedron. 2006;62:8850–8859. [Google Scholar]; (b) Laphookhieo S, Maneerat W, Koysomboon S. Molecules. 2009;14:1389–1395. doi: 10.3390/molecules14041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Matsumoto K, Akao Y, Kobayashi E, Ohguchi K, Ito T, Tanaka T, Iinuma M, Nozawa Y. J. Nat. Prod. 2003;66:1124–1127. doi: 10.1021/np020546u. [DOI] [PubMed] [Google Scholar]
- (5).Nakagawa Y, Iinuma M, Naoe T, Nozawa Y, Akao Y. Bioorg. Med. Chem. 2007;15:5620–5628. doi: 10.1016/j.bmc.2007.04.071. [DOI] [PubMed] [Google Scholar]
- (6).Na Y. J. Pharm. Pharmacol. 2009;61:707–712. doi: 10.1211/jpp/61.06.0002. [DOI] [PubMed] [Google Scholar]
- (7).Duan Y-H, Dai Y, Wang G-H, Zhang X, Chen H-F, Chen J-B, Yao X-S, Zhang X-K. J. Nat. Prod. 2010;73:1283–1287. doi: 10.1021/np1001797. [DOI] [PubMed] [Google Scholar]
- (8).Laphookhieo S, Maneerat W, Buatip T, Syers JK. Can. J. Chem. 2008;86:757–760. [Google Scholar]
- (9).(a) Bennett GJ, Harrison LJ, Sia GL, Sim KY. Phytochemistry. 1993;32:1245–1251. [Google Scholar]; (b) Sia G-L, Bennett GJ, Harrison LJ, Sim KY. Phytochemistry. 1995;38:1521–1528. doi: 10.1016/0031-9422(94)00314-j. [DOI] [PubMed] [Google Scholar]; (c) Nguyen LHD, Harrison LJ. Phytochemistry. 1998;50:471–476. [Google Scholar]
- (10).Laphookhieo S, Syers JK, Kiattansakul R, Chantrapromma K. Chem. Pharm. Bull. 2006;54:745–747. doi: 10.1248/cpb.54.745. [DOI] [PubMed] [Google Scholar]
- (11).Mahabusarakam W, Rattanaburi S, Phongpaichit S, Kanjana-Opas A. Phytochemistry Lett. 2008;1:211–214. [Google Scholar]
- (12).Jin S-L, Wang N-L, Zhang X, Dai Y, Yao X-S. J. Asian Nat. Prod. Res. 2009;11:322–325. doi: 10.1080/10286020902727355. [DOI] [PubMed] [Google Scholar]
- (13).Mahabusarakam W, Nuangnaowarat W, Taylor WC. Phytochemistry. 2006;67:470–474. doi: 10.1016/j.phytochem.2005.10.008. [DOI] [PubMed] [Google Scholar]
- (14).Boonnak N, Karalai C, Chantrapromma S, Ponglimanont C, Fun H-K, Kanjana-Opas A, Chantrapromma K, Kato S. Tetrahedron. 2009;65:3003–3013. [Google Scholar]
- (15).Kinghorn AD, Carcache-Blanco EJ, Chai H-B, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sen AK, Sarkar KK, Mazumder PC, Banerji N, Uusvuori R, Hase TA. Phytochemistry. 1982;21:1747–1750. [Google Scholar]
- (17).Mahabusarakam W, Wiriyachitra P, Taylor WC. J. Nat. Prod. 1987;50:474–478. [Google Scholar]
- (18).Iinuma M, Tosa H, Tanaka T, Riswan S. Chem. Pharm. Bull. 1996;44:232–234. [Google Scholar]
- (19).Fujita T, Liu D-Y, Ueda S, Takeda Y. Phytochemistry. 1992;31:3997–4000. [Google Scholar]
- (20).Yang X-D, Xu L-Z, Yang S-L. Phytochemistry. 2001;58:1245–1249. doi: 10.1016/s0031-9422(01)00356-9. [DOI] [PubMed] [Google Scholar]
- (21).Faizi S, Ali M, Saleem R, Irfanullah, Bibi S. Magn. Reson. Chem. 2001;39:399–405. [Google Scholar]
- (22).Mahabusarkam W, Kuaha K, Wilairat P, Taylor W. Planta Med. 2006;72:912–916. doi: 10.1055/s-2006-947190. [DOI] [PubMed] [Google Scholar]
- (23).Gopalakrishnan G, Banumathi B, Suresh G. J. Nat. Prod. 1997;60:519–524. doi: 10.1021/np970165u. [DOI] [PubMed] [Google Scholar]
- (24).Sen AK, Sarkar KK, Majumder PC, Banerji N. Indian J. Chem. Sec. B: Org. Chem. Med. Chem. 1986;25B:1157–1158. [Google Scholar]
- (25).Ito C, Itoigawa M, Takakura T, Ruangrungsi N, Enjo F, Tokuda H, Nishino H, Furukawa H. J. Nat. Prod. 2003;66:200–205. doi: 10.1021/np020290s. [DOI] [PubMed] [Google Scholar]
- (26).Suphavanich K, Maitarad P, Hannongbua S, Sudta P, Suksamrarn S, Tantirungrotechai Y, Limtrakul J. Monatsh. Chem. 2009;140:273–280. [Google Scholar]
- (27).Yates P, Bhat HB. Can. J. Chem. 1970;48:680–684. [Google Scholar]
- (28).(a) Asano J, Chiba K, Tada M, Yoshii T. Phytochemistry. 1996;41:815–820. doi: 10.1016/0031-9422(95)00682-6. [DOI] [PubMed] [Google Scholar]; (b) Cao S-G, Sng VHL, Wu X-H, Sim K-Y, Tan BKH, Pereira JT, Goh S-H. Tetrahedron. 1998;54:10915–10924. [Google Scholar]
- (29).Thoison O, Fahy J, Dumontet V, Chiaroni A, Riche C, Tri MV, Sevenet T. J. Nat. Prod. 2000;63:441–446. doi: 10.1021/np9903088. [DOI] [PubMed] [Google Scholar]
- (30).Han Q-B, Wang Y-L, Yang L, Tso T-F, Qiao C-F, Song J-Z, Xu L-J, Chen S-L, Yang D-J, Xu H-X. Chem. Pharm. Bull. 2006;54:265–267. doi: 10.1248/cpb.54.265. [DOI] [PubMed] [Google Scholar]
- (31).Li N-G, Wang J-X, Liu X-R, Lin C-J, You Q-D, Guo Q-L. Tetrahedron Lett. 2007;48:6586–6589. [Google Scholar]
- (32).Ren Y, Lantvit DD, Carcache de Blanco EJ, Kardono LBS, Riswan S, Chai H, Cottrell CE, Farnsworth NR, Swanson SM, Ding Y-Q, Li X-C, Marais JPJ, Ferreira D, Kinghorn AD. Tetrahedron. 2010;66:5311–5320. doi: 10.1016/j.tet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).The semi-systematic name of 1 is as used in SciFinder™. Carbons C-5, -7, -10a, and -12 of 1 are equivalent to C-1, -5, -12a, and -3a, respectively, used in the semi-systematic name for this compound.
- (34).Chairungsrilerd N, Takeuchi K, Ohizumi Y, Nozoe S, Ohta T. Phytochemistry. 1996;43:1099–1102. [Google Scholar]
- (35).Thoison O, Cuong DD, Gramain A, Chiaroni A, Hung NV, Sevenet T. Tetrahedron. 2005;61:8529–8535. [Google Scholar]
- (36).Han A-R, Kim J-A, Lantvit DD, Kardono LBS, Riswan S, Chai H, Carcache de Blanco EJ, Farnsworth NR, Swanson SM, Kinghorn AD. J. Nat. Prod. 2009;72:2028–2031. doi: 10.1021/np900517h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lemmich J, Nielsen BE. Tetrahedron Lett. 1969:3–4. doi: 10.1016/s0040-4039(00)99764-8. [DOI] [PubMed] [Google Scholar]
- (38).Costes N, Michel S, Tillequin F, Koch M, Pierre A, Atassi G. J. Nat. Prod. 1999;62:490–492. doi: 10.1021/np980420q. [DOI] [PubMed] [Google Scholar]
- (39).The systematic name proposed for 2 is based on a numbering scheme published in Ref. 3. Carbon C-12 of 2 is equivalent to C-3 in the semi-systematic name of 2.
- (40).Ren Y, Kardono LBS, Riswan S, Chai H-B, Farnsworth NR, Soejarto DD, Carcache de Blanco EJ, Kinghorn AD. J. Nat. Prod. 2010;73:949–955. doi: 10.1021/np1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Deng Y, Balunas MJ, Kim J-A, Lantvit DD, Chin YW, Chai H-B, Sugiarso S, Kardono LBS, Fong HHS, Pezzuto JM, Swanson SM, Carcache de Blanco EJ, Kinghorn AD. J. Nat. Prod. 2009;72:1165–1169. doi: 10.1021/np9001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.