Abstract
10-Formyltetrahydrofolate dehydrogenase (FDH, ALDH1L1), an abundant cytosolic enzyme of folate metabolism, shares significant sequence similarity with enzymes of the aldehyde dehydrogenase (ALDH) family. The enzyme converts 10-formyltetrahydrofolate (10-fTHF) to tetrahydrofolate and CO2 in an NADP+-dependent manner. The mechanism of this reaction includes three consecutive steps with the final occurring in an ALDH-homologous domain. We have recently identified a mitochondrial isoform of FDH (mtFDH), which is the product of a separate gene, ALDH1L2. Its overall identity to cytosolic FDH is about 74%, and the identity between the ALDH domains rises up to 79%. In the present study, human mtFDH was expressed in E. coli, purified to homogeneity, and characterized. While the recombinant enzyme was capable of catalyzing the 10-fTHF hydrolase reaction, it did not produce detectable levels of ALDH activity. Despite the lack of typical ALDH catalysis, mtFDH was able to perform the characteristic 10-fTHF dehydrogenase reaction after reactivation by recombinant 4′-phosphopantetheinyl transferase (PPT) in the presence of coenzyme A. Using site-directed mutagenesis, it was determined that PPT modifies mtFDH specifically at Ser375. The C-terminal domain of mtFDH (residues 413-923) was also expressed in E. coli and characterized. This domain was found to exist as a tetramer and to catalyze an esterase reaction that is typical of other ALDH enzymes. Taken together, our studies suggest that ALDH1L2 has enzymatic properties similar to its cytosolic counterpart, although the inability to catalyze the ALDH reaction with short-chain aldehyde substrates remains an unresolved issue at present.
Keywords: Aldehyde dehydrogenase, ALDH1L2, folate metabolism, 10-formyltetrahydrofolate dehydrogenase, mitochondria, phosphopantetheinylation
1. Introduction
Folic acid, also known as vitamin B9, is a major coenzyme in biosynthetic reactions involving the transfer of single-carbon groups [1]. Higher animals are unable to synthesize this compound and, therefore, must obtain it from their diet [2]. Of note, folate deficiency increases the risk of neural tube defects. In 1996, in order to reduce this risk, the United States Food and Drug Administration mandated the fortification of grain products with folic acid. Since that time, a dramatic decrease in the incidence of neural tube defects (e.g. spina bifida) has been observed [3], but the far-reaching health effects of widespread folic acid supplementation are yet unclear [4]. In the cell, folic acid must be converted to its active form, tetrahydrofolate (THF), by the action of dihydrofolate reductase. THF is then derivatized by the addition of different one-carbon units. These derivatives are distinguished by the oxidation state of the carbon attached to the N5 and/or N10 atoms of THF. The various THF compounds are used in nucleotide biosynthesis, the methylation of homocysteine to regenerate methionine, amino acid conversions involving serine, glycine, and histidine, and the formylation of mitochondrial methionyl-tRNA [1]. In addition, the various forms of folate can be interconverted by the action of specialized enzymes.
Compartmentation is a basic property of eukaryotic cells, and folate metabolism has evolved to become functionally integrated into certain cellular compartments. The cytosol and mitochondria (and perhaps the nucleus) contain discrete pools of folate enzymes and THF-derivatives [5]. Current knowledge suggests that there is no free exchange of folates between the cytoplasm and mitochondria, but these compartments are connected metabolically via the transport of one-carbon donors (such as serine, glycine, and formate) which facilitate the generation of the THF derivatives required for biosynthetic reactions [6]. Several folate reactions and their corresponding enzymes are present in both cytosolic and mitochondrial compartments.
10-Formyltetrahydrofolate dehydrogenase (ALDH1L1, FDH) is an abundant cytosolic folate enzyme in the liver and kidney: it has been reported to constitute approximately 1.2% of the total cytosolic protein in rat liver [7]. While other enzymes utilize 10-formyltetrahydrofolate (10-fTHF) in biosynthetic processes, FDH is unique in being a catabolic enzyme, catalyzing the NADP+-dependent degradation of 10-fTHF to THF and CO2 [8]. Because 10-fTHF is a substrate in two steps of de novo purine biosynthesis, it has been proposed that FDH inhibits cellular proliferation by regulating intracellular purine levels [9]. Since THF and its derivatives, including 10-fTHF, can be interconverted, it is possible that FDH broadly functions to regulate folate-mediated one-carbon metabolism. In line with this hypothesis, it has been shown that ectopic expression of FDH results in depletion of both cellular 5-methyltetrahydrofolate and S-adenosylmethionine levels [10].
FDH exists as a tetramer of identical 100 kDa subunits. Each subunit is 902 amino acid residues in length and can be divided into three distinct domains. An N-terminal 10-fTHF hydrolase domain (residues 1-310) contains the folate binding site and is homologous to glycinamide ribonucleotide formyltransferase and methionyl-tRNA formyltransferase, enzymes that utilize 10-fTHF as a substrate. This domain is capable of hydrolyzing 10-fTHF to THF and formate [11]. FDH also possesses a C-terminal domain (residues 397-902) that is homologous to aldehyde dehydrogenases (ALDHs) [8]. When expressed as an independent protein, this domain is capable of oxidizing small-chain aldehydes to their corresponding carboxylic acids in an NADP+-dependent manner [12]. The intermediate domain of FDH (residues 311-396) is structurally and functionally homologous to acyl carrier proteins [13]. This domain contains the site of attachment (Ser354) for a 4′-phosphopantetheine (4-PP) prosthetic group, and this post-translational modification is necessary for the 10-fTHF dehydrogenase activity of the enzyme [13]. This moiety is attached to FDH by a broad-specificity 4′-phosphopantetheinyl transferase (PPT), an enzyme that uses coenzyme A (CoA) as the 4-PP donor molecule [14]. In the proposed catalytic mechanism of FDH, the 4-PP group acts as a swinging arm to carry the formyl group from the N-terminal hydrolase domain to the C-terminal ALDH-like domain where it is oxidized to CO2.
A mitochondrial isoform of FDH (ALDH1L2, mtFDH) has recently been identified [15]. This enzyme has 74% sequence identity to its cytosolic counterpart and is distinguished by the presence of an N-terminal mitochondrial targeting sequence of 22 amino acid residues. The mitochondrial enzyme, purified from pig liver, is capable of catalyzing the characteristic 10-fTHF hydrolase and dehydrogenase reactions [15]. In the current study, the cloning, E. coli expression, and purification of recombinant mtFDH are described, and the characterization of the catalytic activities of the enzyme, as well as its pathway of post-translational modification, are reported.
2. Materials and Methods
2.1. Cloning, Expression, and Purification of ALDH1L1 and ALDH1L2
Cloning, E. coli expression, and purification of human ALDH1L1 was carried out as previously described [14]. Briefly, an ALDH1L1/pRSET construct was transformed into a BL21-(DE3)-codon plus strain of E. coli (Invitrogen). Cells were grown at 37°C to A600 = 0.6, and expression was induced by the addition of 1 mM isopropyl 1-thio-β-D-galactopyranoside (Sigma). After induction, the culture was grown at 20 °C for 4 hours with constant shaking. The protein was purified from lysed cells using a 5-formyltetrahydrofolate affinity column followed by size exclusion chromatography. The vector for the bacterial expression of human mtFDH (ALDH1L2/pRSET) has already been reported [15]. Expression and purification of mtFDH was performed as described above for ALDH1L1.
2.2. CD-spectroscopy
Far-UV CD spectra (200–260 nm) were obtained at a resolution of 0.2 nm with a bandwidth of 2 nm using a Model 400 Circular Dichroism Spectrophotometer (AVIV Biomedical, Inc., Lakewood, NJ) at the Protein Production Core, MUSC Structural Biology Center. For each sample, three spectra were collected at 25 °C and averaged. The protein concentration was 0.5 mg/ml in these experiments. The protein spectra were corrected by subtracting the averaged spectrum of the buffer solution and presented in units of molar ellipticity. The spectra were analyzed by CD Spectra Deconvolution software, version 2.1 (® 1997 by Gerald Böhm).
2.3. Site-directed Mutagenesis
Site-directed mutagenesis was accomplished using a QuikChange kit (Stratagene). The mutagenic primers used in the generation of each construct are listed in Table 1. To generate the recombinant C-terminal domain of ALDH1L1 (Ct-FDH, encompassing residues 397-902), site-directed mutagenesis was performed using ALDH1L1/pRSET as the plasmid template. The expression vector for C-terminal ALDH1L2 (Ct-mtFDH) was generated using site-directed mutagenesis of the ALDH1L2/pRSET plasmid. Another mutant enzyme, encompassing the N-terminal and intermediate domains of mtFDH (NI-mtFDH, residues 22-429), was also produced in this manner. For each truncated construct, expression was accomplished as in Section 2.1, and the protein was purified from lysed cells using a PrepEase His-Tagged Protein Purification Kit (USB).
Table 1.
Site-Directed Mutagenesis Primers
| Construct | Primer Sequences |
|---|---|
| Ct-FDH | Forward: 5′-CATCATCATCATGGTATGGGGGACGATGAGGAGGGC-3′ Reverse: 5′-GCCCTCCTCATCGTCCCCCATACCATGATGATGATG-3′ |
| Ct-mtFDH | Forward: 5′-CATCATAAGCTGCTGAGGAAACTGAG-3′ Reverse: 5′-CACAGTTTCCTCAGCAGCTTATGATG-3′ |
| NI-mtFDH | Forward: 5′-GGTTGTAGATTAAATTTCAAAGG-3′ Reverse: 5′-CCTTTGAAATTTAATCTACAACC-3′ |
| S374A | Forward: 5′-CTGGAGCAGCCTCAATGGATGTTG-3′ Reverse: 5′-CAACATCCATTGAGGCTGCTCCAG-3′ |
| S375A | Forward: 5′-CTGGAGCAAGCGCAATGGATGTTG-3′ Reverse: 5′-CAACATCCATTGCGCTTGCTCCAG-3′ |
| S374/375A | Forward: 5′-CTGGAGCAGCCGCAATGGATGTTG-3′ Reverse: 5′-CAACATCCATTGCGGCTGCTCCAG-3′ |
Two mutant constructs, Ser374Ala (S374A) and Ser375Ala (S375A), were produced using the ALDH1L2/pRSET vector as a template. A Ser374Ala/Ser375Ala double mutant (S374/375A) was generated from the S374A vector. For each of these mutants, expression and purification was completed as for the full-length ALDH1L1 and ALDH1L2 proteins as detailed in Section 2.1. The coding regions of all plasmid constructs were confirmed by DNA sequencing at the MUSC Nucleic Acid Analysis Facility.
2.4. Assays of Enzymatic Activity
All enzyme assays were conducted at 30 °C in a 1-cm quartz cuvette using a Shimadzu 2401PC double-beam spectrophotometer. Measurements of 10-fTHF hydrolase, 10-fTHF dehydrogenase, and ALDH activities were taken according to previously published procedures [12, 16, 17]. Esterase activity was assayed according to the method of Takahashi and Weiner [18]. The substrate, p-nitrophenyl acetate (p-NPA), was dissolved in acetone to prevent spontaneous hydrolysis. The enzyme was added to a mixture containing 100 mM sodium pyrophosphate, pH 7.7, and variable amounts of substrate. Absorbance was monitored at 400 nm, and activity was calculated using a molar extinction coefficient of 16 × 103 M−1 cm−1 [19].
2.5. Activation of mtFDH by PPT
Recombinant PPT was expressed in E. coli and purified using metal affinity chromatography as we have previously described [14]. Two μg of the purified enzyme was incubated with 50 μg of E. coli-produced mtFDH in a 200 μl reaction containing 1 mM CoA and 0.5 mM MgCl2. The mixture was incubated for 10 min at room temperature and then immediately used in the 10-fTHF dehydrogenase activity assay (Section 2.4).
2.6. Fluorescent Labeling of mtFDH by PPT and Cell Lysate
The fluorescent reporter, a fluorescein maleimide derivative of coenzyme A, was synthesized as previously described [13, 20]. The reporter (10 μl of a 2 mM solution) was added to 12 μg of purified ALDH1L2 or its mutant constructs along with 6 μg of recombinant PPT (or H2O, as a control) in a solution containing 50 mM MgCl2 and 100 mM Tris-HCl, pH 7.5. This mixture was incubated at room temperature for 20 min, after which time EDTA was added to quench the reaction, the sample was immediately subjected to SDS-PAGE, and the gel was imaged under UV light followed by staining with Coomassie Blue.
Lysates from siRNA-treated A549 cells were obtained as we have previously reported [14]. The levels of PPT and actin in experimental and control lysates were assessed by Western blot using a specific polyclonal antibody. After knockdown of PPT was confirmed, the lysates were normalized for protein content, and equal volumes of lysates were added to 20 μg of NI-mtFDH in the presence of fluorescent reporter and MgCl2. Samples were then incubated, subjected to SDS-PAGE, and imaged as above.
2.7. Homology modeling
Homology models of the C-terminal domain of human mtFDH and zebrafish cytosolic FDH (zFDH) were generated with the SWISS-MODEL server using the crystal structure of the apo form of rat cytosolic Ct-FDH (PDB ID 2O2P) as the template [21]. To verify the reliability of these models, they were superimposed with the structure of each of the four monomers of template Ct-FDH using the Superpose program from the CCP4 suite [22, 23].
3. Results
3.1. Cloning, Expression and Purification of Recombinant ALDH1L2 Constructs
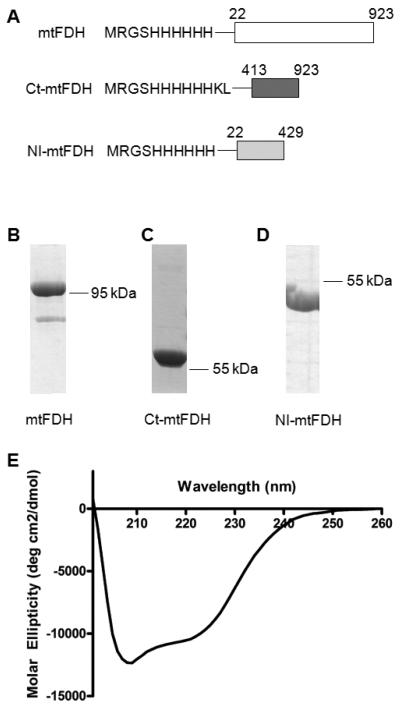
To characterize its enzymatic properties, mtFDH was expressed as a recombinant protein. Since it has been predicted that the mitochondrial leader sequence is cleaved upon translocation of the protein into mitochondria, the fragment encoding the first 21 amino acids of mtFDH was removed from the plasmid (Figure 1A). Recombinant mtFDH was expressed as a soluble protein and was purified to near-homogeneity (Figure 1B). To study the aldehyde dehydrogenase domain of mtFDH, we generated a C-terminal construct of the protein (Ct-mtFDH) encompassing residues 413-923 (Figure 1A) from the above plasmid. Ct-mtFDH was expressed as a soluble protein of ~58 kDa and was purified using metal affinity chromatography (Figure 1C). Size exclusion chromatography of the purified Ct-mtFDH demonstrated that the protein eluted at an average volume of 97.9 ml. This elution volume was similar to that of the 232 kDa standard, catalase (99.7 ml), indicating that the protein exists as a tetramer. CD spectroscopy was used to evaluate whether Ct-mtFDH was appropriately folded (Figure 1D), and our analysis revealed a similar proportion of secondary structure elements as that of the C-terminal domain of cytosolic FDH.
Figure 1.
Expression and purification steps of ALDH1L2 constructs. A, schematic depicting the construct design for full-length mtFDH, C-terminal mtFDH (Ct-mtFDH) and N-terminal Intermediate mtFDH (NI-mtFDH). B, C, D, SDS-PAGE (Coomassie staining) of purified recombinant mtFDH, Ct-mtFDH, and NI-mtFDH, respectively. E, CD spectrum of Ct-mtFDH protein.
3.2. Hydrolase Activities of ALDH1L1 and ALDH1L2
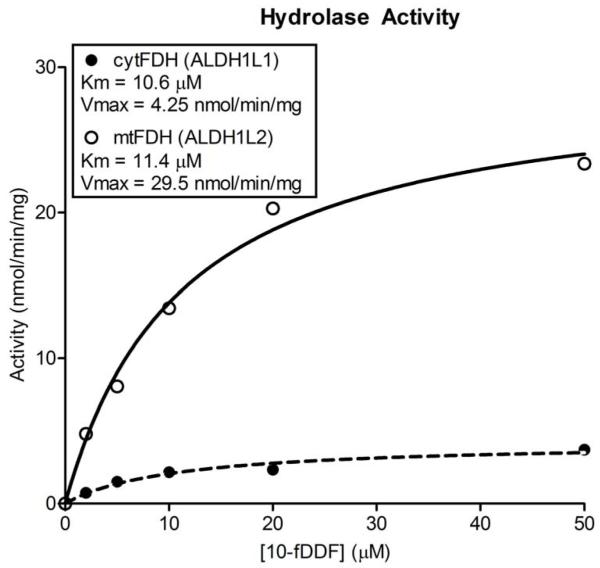
The ability of the FDH isoforms to catalyze the 10-fTHF hydrolase reaction was assessed using the substrate 10-formyldideazafolate (10-fDDF). This dideaza analogue is capable of substituting the natural substrate in the in vitro assays of FDH activity, and it has the advantage of being much more stable than 10-fTHF [16]. For mtFDH, the Km in relation to 10-fDDF was determined to be 11.4 μM with a Vmax of 29.5 nmol min−1 mg−1 (Figure 2). For comparison, recombinant cytosolic FDH expressed in E. coli was assayed in control experiments [11, 14]. The Km for the cytosolic enzyme was calculated to be 10.6 μM with a Vmax of 4.25 nmol min−1 mg−1. Thus, while both isoforms of FDH shared a similar affinity for the substrate, the mtFDH demonstrated a ~7-fold higher rate of turnover for the hydrolase reaction.
Figure 2.
Hydrolase activities of ALDH1L1 and ALDH1L2. Michaelis-Menten curves for cytFDH (closed circles) and mtFDH (open circles) hydrolase reactions.
3.3. Aldehyde Dehydrogenase and Esterase Activities of ALDH1L1 and ALDH1L2
Recombinant cytosolic FDH was shown to possess ALDH activity toward the short-chain aldehydes, propanal and acetaldehyde. Interestingly, recombinant mtFDH was unable to catalyze the oxidation of propanal or acetaldehyde using either NADP+ or NAD+ as a coenzyme. It has been previously shown that, in addition to its ability to catalyze the ALDH reaction, cytosolic FDH is capable of an esterase activity, the hydrolysis of para-nitrophenylacetate (p-NPA) [19]. We examined the recombinant mtFDH for its ability to catalyze the esterase reaction and found it to be able to do so.
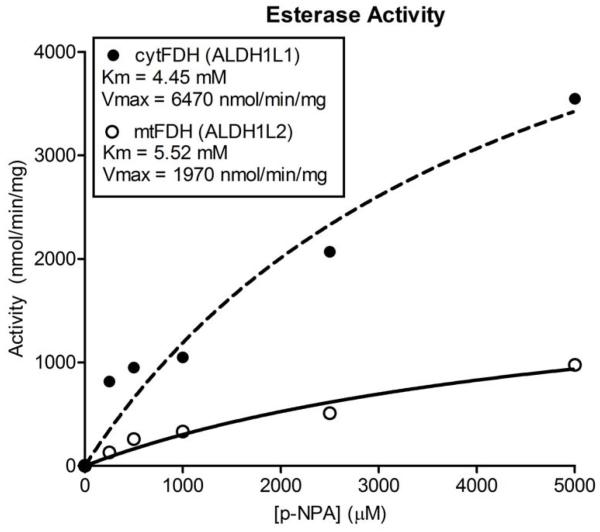
The C-terminal domain of cytosolic FDH, expressed as a recombinant protein, is a functional ALDH [12]. However, recombinant Ct-mtFDH (residues 413-923) was unable to catalyze such a reaction, similar to the full-length mitochondrial protein. Ct-mtFDH was also tested for its ability to perform the esterase reaction and was found to have a Km of 5.52 mM for the substrate, p-NPA, with a Vmax of 1970 nmol min−1 mg−1 (Figure 3). As a comparison, the C-terminal domain of cytosolic FDH was found to have a Km of 4.45 mM for this substrate and a Vmax of 6470 nmol min−1 mg−1. Thus, the C-terminal domain of mtFDH, while capable of catalyzing the esterase reaction, has more than three-fold decreased esterase activity compared to the cytosolic enzyme.
Figure 3.
Esterase activities of the C-terminal domains of ALDH1L1 and ALDH1L2. Michaelis-Menten curves for cytFDH (ALDH1L1, closed circles) and mtFDH (ALDH1L2, open circles) C-terminal constructs esterase reactions.
3.4. PPT Activates Dehydrogenase Catalysis of ALDH1L2 by Modifying Ser375
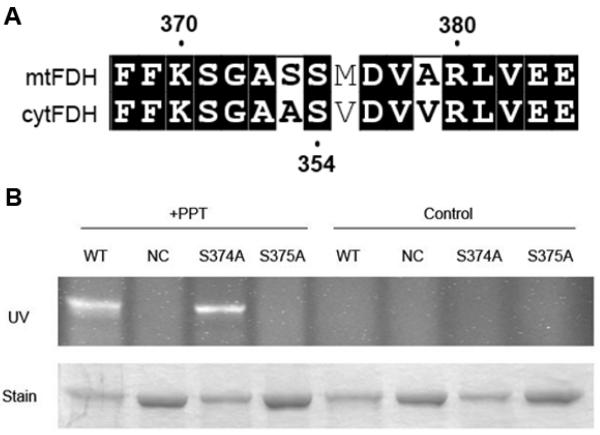
Cytosolic FDH requires a 4-PP post-translational modification at Ser354 for its 10-fTHF dehydrogenase catalytic activity [13]. Due to the fact that mtFDH also displays 10-fTHF dehydrogenase activity, and, taking into account the similarity between the cytosolic and mitochondrial isoforms, we predicted that the mitochondrial isoform also requires a post-translational modification with 4-PP for its activity. A sequence alignment of the cytosolic and mitochondrial FDH isoforms demonstrated the presence of 2 consecutive serine residues (Ser374 and Ser375) at the putative site of mtFDH phosphopantetheinylation (Figure 4A). In order to determine which of these residues is the modification site of mtFDH, we mutated each of them separately (S374A and S374A mutants) as well as both of them simultaneously (S374/S375A double mutant), and tested the corresponding proteins in an in vitro reaction using a CoA-derived fluorescent reporter and recombinant PPT. We have previously shown that ALDH1L1 can be covalently modified with the reporter when incubated with PPT in vitro [14]. In our experiments, PPT modified wild-type mtFDH and the S374A protein with the fluorescent reporter, but not the S375A or S374/S375A mutants (Figure 4B), indicating that Ser375 is the only site of 4-PP attachment.
Figure 4.
PPT modifies ALDH1L2 specifically at Ser375. A, sequence alignment of ALDH1L1 (cytFDH) and ALDH1L2 (mtFDH) depicting the residues flanking the site of 4-PP attachment (Ser354). Two adjacent serine residues (Ser374 and Ser375) are present at this location in mtFDH. B, SDS-PAGE of recombinant mtFDH mutant constructs after incubation with fluorescent reporter and either recombinant PPT (+PPT) or water (control). Upper panel, SDS-PAGE visualized under UV irradiation. Lower panel, SDS-PAGE after Coomassie staining. NC refers to the S374/375A double mutant.
Recombinant cytosolic FDH, expressed in E. coli, does not catalyze the 10-fTHF dehydrogenase reaction due to the absence of the 4-PP modification [13]. However, when incubated with PPT and CoA in vitro, this enzyme acquires dehydrogenase activity. Similar to this finding, recombinant mtFDH was found to be incapable of catalyzing the 10-fTHF dehydrogenase reaction. To test whether PPT enables this activity, recombinant mtFDH was incubated with E. coli expressed PPT, CoA, and Mg2+. The resulting mixture was tested for dehydrogenase catalysis as previously described and found to be dehydrogenase competent (Figure 5C) [14].
Figure 5.
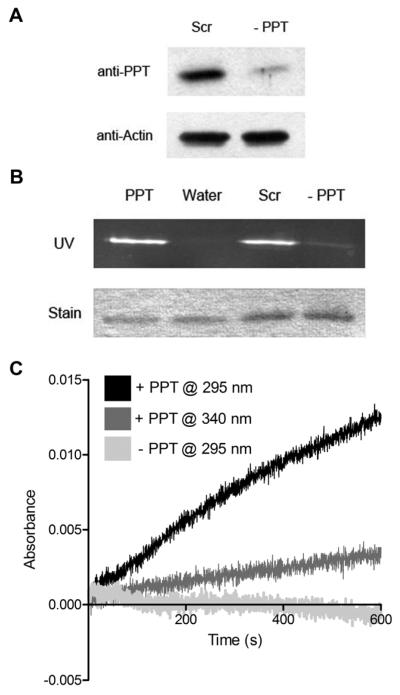
Human PPT modifies recombinant ALDH1L2 and enables its 10-fTHF dehydrogenase activity. A, knockdown of PPT in A549 cells using siRNA (−PPT) determined by immunoblot with PPT-specific antibody. B, Modification of recombinant ALDH1L2 with the fluorescent reporter by normal cell lysates (PPT) and lysates from cells treated with either scrambled (Scr) or PPT-specific siRNA (−PPT). C, Modification of the recombinant ALDH1L2 by PPT enables the 10-fTHF reaction. Appearance of the products, DDF and NADPH was followed at 295nm and 340nm, respectively. The non-modified protein reaction was monitored at 295 nm.
3.5. PPT Silencing Prevents Modification of ALDH1L2 by Mammalian Cell Lysate
It has been suggested that humans possess only a single phoshopantetheinyl transferase [24, 25], and we have recently provided experimental evidence for this hypothesis [14]. We used a method to fluorescently label recombinant FDH using mammalian cell lysates as the source of PPT [13]. We demonstrated that, after silencing PPT in A549 cells, their lysates were unable to modify cytosolic FDH in vitro, while control lysates were capable of catalyzing this modification [14]. To determine if PPT is required for the post-translational modification of mtFDH, we used a similar approach. For these experiments, we used a truncated construct of mtFDH (NI-mtFDH, residues 22-429, Figure 1D) which contained the Ser375 modification site. Successful PPT-silencing was confirmed by immunoblot using a specific polyclonal antibody (Figure 5A). In our in vitro modification reactions, recombinant NI-mtFDH was incubated with A549 cell lysates in the presence of the CoA-derived fluorescent reporter. Lysates from PPT-silenced cells were unable to modify NI-mtFDH, while lysates from cells treated with scrambled siRNA successfully labeled the protein (Figure 5B).
4. Discussion
Cytosolic FDH (ALDH1L1) is an abundant folate enzyme which is well characterized with regard to its catalytic properties and structure [11, 12, 17, 26-29]. An interesting feature of this protein is its chimeric nature. Indeed, the enzyme has three distinctive domains, one of which, the C-terminal, is a homolog of aldehyde dehydrogenases and thus defines the classification of the enzyme [8]. This complex organization enables three catalytic activities of the enzyme: the 10-fTHF hydrolase, ALDH, and 10-fTHF dehydrogenase. Numerous studies suggest that the 10-fTHF dehydrogenase reaction, believed to be the main biological function of the enzyme, is a combination of the first two reactions, the hydrolase and ALDH. While cytosolic FDH has long been described, the identification of its mitochondrial isoform is a relatively recent discovery [15]. The catalytic properties of mtFDH were previously characterized using protein purified from pig liver. A lower abundance of the endogenous enzyme compared to the cytosolic isoform, as well as an obvious inability to characterize its different domains independently, prompted us to pursue studies of recombinant mtFDH. In the present work, the catalytic properties of recombinant full-length mtFDH and its different domains, expressed in E. coli, were assessed.
Due to the high sequence similarity to cytosolic FDH, mtFDH was expected to possess the same catalytic residues and activities as the cytosolic isoform (Table 2). Indeed, mtFDH, purified from mammalian liver, was capable of catalyzing the 10-fTHF hydrolase and 10-fTHF dehydrogenase reactions [15]. With no surprise, recombinant mtFDH demonstrated strong 10-fTHF hydrolase activity. Moreover, while the Km values for this reaction were similar between the two isoforms, the Vmax for the mtFDH-catalyzed reaction was about 7-fold higher than that of cytosolic FDH. Of note, one of the products of this reaction is formate. Since it is believed that mitochondrial folate metabolism serves to generate formate for its further utilization in cytosolic folate-dependent biosynthetic reactions [6], a high hydrolase activity points toward mtFDH as a potential producer of formate from folate-bound one-carbon groups. Similar to cytosolic FDH, this reaction requires high levels of a reducing agent in the in vitro assay (2-mercaptoethanol or DTT were used in our experiments). While it is not clear at present if this reaction truly occurs in mitochondria, high concentrations of glutathione in this organelle (more than 10 mM [30]) could potentially support in vivo hydrolase activity of mtFDH.
Table 2.
Important Catalytic Residues of the Human FDH Isoforms
| ALDH1L1 | ALDH1L2 | Relevant Activities | Reference |
|---|---|---|---|
| His 106 | His 128 (predicted) | Hydrolase and DH* | [29] |
| Asp 142 | Asp 164 (predicted) | Hydrolase and DH | [17] |
| Ser 354 | Ser 375 | 4-PP attachment (DH) | [13] |
| Glu 673 | Glu 694 (predicted) | ALDH and DH | [27] |
| Cys 707 | Cys 728 (predicted) | ALDH and DH | [28] |
DH, 10-fTHF dehydrogenase activity
We have previously shown that cytosolic FDH is post-translationally modified by PPT, which adds a 4-PP prosthetic group to the enzyme at serine 354 [14]. This modification is necessary for 10-fTHF dehydrogenase catalysis [13]. Interestingly, mtFDH has two serine residues near the predicted site of 4-PP attachment (Ser374 and Ser375), and it was speculated that either of these residues could be modified. Our results, however, demonstrate that PPT modifies mtFDH exclusively at Ser375. Thus, the modification of the intermediate domain by PPT appears to be highly specific. As in the case of the cytosolic enzyme, this modification enables 10-fTHF dehydrogenase catalysis of mtFDH. In our previous studies, siRNA knockdown of PPT abrogated the ability of cell lysate to modify recombinant FDH [14]. In the present study, the same effect was seen in our experiments with mtFDH. This data, taken together with the strong antiproliferative effects of PPT knockdown seen in human cell lines [14], suggest the presence of a single PPT type enzyme in humans. Interestingly, a prior study found that PPT is a cytosolic enzyme [24]. Thus, these findings suggest that mtFDH should be modified in the cytosol prior to its translocation into mitochondria. Since PPT recognizes the ternary structure of proteins [25], mtFDH must be at least partially folded prior to modification. Overall, these observations raise important questions about the processing of mtFDH and the mechanism of its translocation. Specifically, what is the oligomeric form in which the enzyme translocates (unfolded monomer or folded tetramer) and whether mtFDH is folded into a fully functional protein in cytosol?
Surprisingly, our studies revealed that recombinant mtFDH is unable to catalyze the ALDH reaction. Since mtFDH still catalyzes the 10-fTHF dehydrogenase reaction and previous studies have implied that ALDH activity is a component of this catalysis, it would be logical to assume that the enzyme is capable of ALDH catalysis. The fact that mtFDH catalyzes the 10-fTHF dehydrogenase reaction eliminates the possibility that the lack of ALDH activity is due to incomplete or incorrect folding of the C-terminal domain. Indeed, when expressed separately, this domain forms tetramers and displays a typical CD-spectrum, both parameters indicative of a folded protein. Nevertheless, Ct-mtFDH itself does not possess ALDH activity in contrast to the corresponding domain of the cytosolic enzyme. While the lack of ALDH catalysis in mtFDH does not have a definitive explanation at present, several mechanisms for such a phenomenon could be considered. For instance, it is possible that short-chain aldehydes are not substrates for mtFDH. Another explanation would be that the 10-fTHF dehydrogenase mechanism is not a precise replica of the ALDH catalytic mechanism (i.e. active ALDH catalysis, as whole, might not be a prerequisite for 10-fTHF catalysis). Indeed, the two reactions result in different oxidation states of the carbon atom of the substrate: it is oxidized from the level of aldehyde to the level of acid in one case (ALDH) and from the level of acid to the level of carbon dioxide in the other (10-fTHF dehydrogenase reaction). ALDH catalysis includes two steps, acylation and deacylation, with a covalent intermediate [31-33]. Taking into consideration the requirement for 4-PP for the 10-fTHF dehydrogenase reaction, the difference between a typical ALDH reaction and the one catalyzed by the dehydrogenase domain of mtFDH may be in the acylation step, which in the latter enzyme probably consists of the transfer of the formyl group from the thiol of 4-PP to the thiol of the FDH catalytic cysteine. Thus, the inability to accomplish canonical acylation step could be accountable for the lack of the ALDH catalysis in mtFDH.
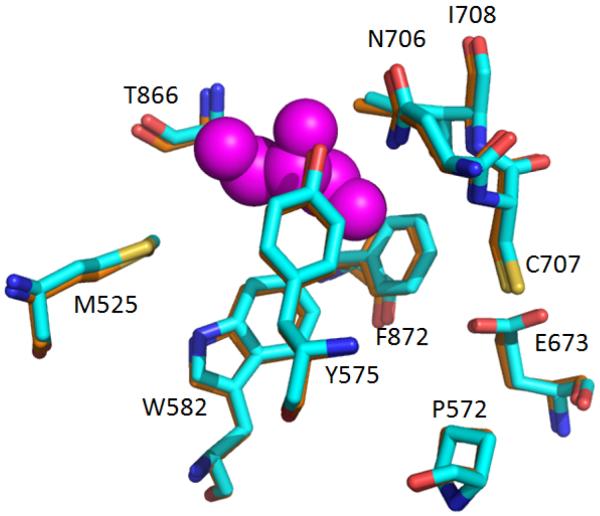
Importantly, it has recently been reported that zebrafish cytosolic FDH (zFDH) also lacks ALDH activity [34]. Of note, recombinant zFDH, similar to mtFDH, is capable of the 10-fTHF dehydrogenase catalysis (after activation with mammalian cell lysate and CoA). The C-terminal domains of human mitochondrial and zebrafish FDH are highly similar to that of the rat cytosolic enzyme (77% and 81% sequence identity, respectively). In particular, the catalytic cysteine and glutamate (Table 2), as well as the residues forming the substrate entrance tunnel, are conserved in all three proteins. The ALDH catalytic centers of the active C-terminal domain of cytosolic FDH and non-active domains of mtFDH and zFDH were analyzed for potential structural alterations using homology modeling. To confirm the utility of our models, they were examined for sterical clashes at the monomer-monomer interface, as well as for other potential hindrances due to amino acid residue replacements. These in silico experiments suggest a near identical arrangement of the active sites in the examined proteins (Figure 6). However, it can be speculated that the ALDH catalysis of the inactive enzymes may be influenced by distant residues, not participating directly in substrate binding. For example, it has been suggested that the half-of-the-site reactivity observed in some aldehyde dehydrogenases could be a result of yet to be characterized long-range interactions occurring between the catalytic site and the nucleotide binding site [35]. Alternatively, insertion of the bulky 4-PP prosthetic group in the substrate entrance tunnel may be necessary for “fine tuning” of the structures of human mitochondrial and zebrafish enzymes in order for optimal substrate binding and orientation for nucleophilic attack of the catalytic cysteine.
Figure 6.
Superposition of the active site residues in the crystal structure of rat cytosolic Ct-FDH (PDB ID 2o2p, orange) and a homology model of human mitochondrial Ct-FDH (cyan) generated with the SWISS-MODEL server [21]. Amino acid residues are numbered according to the rat cytosolic enzyme. A glycerol molecule found in the crystal structure of rat Ct-FDH is shown with magenta spheres to mark the location of the substrate entrance tunnel. Superposition with a model of zebrafish FDH gives identical results (not shown).
Aldehyde dehydrogenases also possess an esterase activity, the hydrolysis of p-NPA [36]. This activity was found in both isoforms of FDH, in contrast to ALDH activity. It appears, however, that mtFDH displays much lower esterase activity than the cytosolic enzyme. Esterase catalysis relies on the action of the same cysteine which acts as the nucleophile in ALDH catalysis [37]. Thus, the diminished esterase activity of mtFDH may reflect the apparent lack of the ALDH activity. It is hard to predict whether mtFDH or zFDH would have aldehyde dehydrogenase activity within the cellular environment. In this regard, a recent study demonstrated activation of the low-activity oriental ALDH variant by a small molecule [38]. The existence of such a compound implies a theoretical possibility for activation of mtFDH ALDH catalysis, but this activity may not have an independent physiologic function for FDH enzymes. Regardless, the main catalytic function of the FDH isoforms as 10-fTHF metabolizing enzymes appears to be competent in the absence of ALDH activity. On an evolutionary note, since the divergence between ALDH1L1 and ALDH1L2 took place at some time before the emergence of bony fish [39], it is possible that cytosolic FDH acquired the ability to catalyze the ALDH reaction at some distant point after ALDH1L1 gene duplication.
5. Conclusion
The compartmentation of folate metabolism between cytosol and mitochondria is a well-established phenomenon. One of the folate coenzymes, 10-fTHF, is present in both compartments and has separate functions in cytosol and mitochondria. In the former, it participates in de novo purine biosynthesis while in the latter it is involved in both the production of formate [40, 41] and the formylation of the translation initiator, methionyl-tRNA [42, 43].
FDH is a major enzyme metabolizing 10-fTHF in both cytosol and mitochondria. While the proposed function of the cytosolic enzyme is the overall regulation of the flux of one-carbon groups through folate pool [10], mtFDH may be involved in producing formate, a pathway shuttling one-carbon groups from mitochondria to cytosol. This enzyme might also be involved in the regulation of mitochondrial protein biosynthesis. A physiologic function for FDH-associated ALDH activity has not been experimentally established. The fact that mtFDH and cytosolic zFDH do not display a detectable level of ALDH activity implies the lack of a biological function relevant to oxidation of aldehydes for this group of enzymes.
Acknowledgments
This study was supported by the National Institutes of Health grant DK054388 (SAK) and a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral MD/PhD Fellows F30DK083215 (KCS). KCS was supported by USPHS NIH grant R13-AA019612 to present this work at the 15th International Meeting on Enzymology and Molecular Biology of Carbonyl Metabolism in Lexington, KY USA.
Abbreviations
- ALDH
aldehyde dehydrogenase
- CoA
coenzyme A
- FDH
10-formyltetrahydrofolate dehydrogenase
- mtFDH
mitochondrial 10-formyltetrahydrofolate dehydrogenase
- 10-fTHF
10-formyltetrahydrofolate
- p-NPA
para-nitrophenylacetate
- 4-PP
4′-phosphopantetheine
- PPT
4′-phosphopantetheinyl transferase
- THF
tetrahydrofolate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wagner C. Biochemical role of folate in cellular metabolism. In: Bailey LB, editor. Folate in Health and Disease. Marcel Dekker, Inc.; New York: 1995. pp. 23–42. [Google Scholar]
- [2].Herbert V. Experimental nutritional folate deficiency in man. Trans Assoc Am Physicians. 1962;75:307–320. [PubMed] [Google Scholar]
- [3].Honein MA, Paulozzi LJ, Mathews TJ, Erickson JD, Wong LY. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. JAMA. 2001;285(23):2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- [4].Lucock M, Yates Z. Folic acid fortification: a double-edged sword. Curr Opin Clin Nutr Metab Care. 2009;12(6):555–564. doi: 10.1097/MCO.0b013e32833192bc. [DOI] [PubMed] [Google Scholar]
- [5].Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- [6].Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- [7].Kisliuk RL. Folate biochemistry in relation to antifolate selectivity. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Humana Press; Totowa, New Jersey: 1999. pp. 13–36. [Google Scholar]
- [8].Krupenko SA. FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism. Chem Biol Interact. 2009;178(1-3):84–93. doi: 10.1016/j.cbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13(5):227–236. [PubMed] [Google Scholar]
- [10].Anguera MC, Field MS, Perry C, Ghandour H, Chiang EP, Selhub J, Shane B, Stover PJ. Regulation of folate-mediated one-carbon metabolism by 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 2006;281(27):18335–18342. doi: 10.1074/jbc.M510623200. [DOI] [PubMed] [Google Scholar]
- [11].Krupenko SA, Wagner C, Cook RJ. Domain structure of rat 10-formyltetrahydrofolate dehydrogenase. Resolution of the amino-terminal domain as 10-formyltetrahydrofolate hydrolase. J Biol Chem. 1997;272(15):10273–10278. doi: 10.1074/jbc.272.15.10273. [DOI] [PubMed] [Google Scholar]
- [12].Krupenko SA, Wagner C, Cook RJ. Expression, purification, and properties of the aldehyde dehydrogenase homologous carboxyl-terminal domain of rat 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1997;272(15):10266–10272. doi: 10.1074/jbc.272.15.10266. [DOI] [PubMed] [Google Scholar]
- [13].Donato H, Krupenko NI, Tsybovsky Y, Krupenko SA. 10-formyltetrahydrofolate dehydrogenase requires a 4′-phosphopantetheine prosthetic group for catalysis. J Biol Chem. 2007;282(47):34159–34166. doi: 10.1074/jbc.M707627200. [DOI] [PubMed] [Google Scholar]
- [14].Strickland KC, Hoeferlin LA, Oleinik NV, Krupenko NI, Krupenko SA. Acyl carrier protein-specific 4′-phosphopantetheinyl transferase activates 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 2010;285(3):1627–1633. doi: 10.1074/jbc.M109.080556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 2010;285(30):23056–23063. doi: 10.1074/jbc.M110.128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Krupenko SA, Wagner C, Cook RJ. Recombinant 10-formyltetrahydrofolate dehydrogenase catalyses both dehydrogenase and hydrolase reactions utilizing the synthetic substrate 10-formyl-5,8-dideazafolate. Biochem J. 1995;306(Pt 3):651–655. doi: 10.1042/bj3060651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krupenko SA, Wagner C. Aspartate 142 is involved in both hydrolase and dehydrogenase catalytic centers of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1999;274(50):35777–35784. doi: 10.1074/jbc.274.50.35777. [DOI] [PubMed] [Google Scholar]
- [18].Takahashi K, Weiner H. Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase. Biochemistry. 1981;20(10):2720–2726. doi: 10.1021/bi00513a003. [DOI] [PubMed] [Google Scholar]
- [19].Cook RJ, Wagner C. Enzymatic activities of rat liver cytosol 10-formyltetrahydrofolate dehydrogenase. Arch Biochem Biophys. 1995;321(2):336–344. doi: 10.1006/abbi.1995.1403. [DOI] [PubMed] [Google Scholar]
- [20].La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem Biol. 2004;11(2):195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- [21].Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- [23].The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- [24].Joshi AK, Zhang L, Rangan VS, Smith S. Cloning, expression, and characterization of a human 4′-phosphopantetheinyl transferase with broad substrate specificity. J Biol Chem. 2003;278(35):33142–33149. doi: 10.1074/jbc.M305459200. [DOI] [PubMed] [Google Scholar]
- [25].Bunkoczi G, Pasta S, Joshi A, Wu X, Kavanagh KL, Smith S, Oppermann U. Mechanism and substrate recognition of human holo ACP synthase. Chem Biol. 2007;14(11):1243–1253. doi: 10.1016/j.chembiol.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chumanevich AA, Krupenko SA, Davies C. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain. J Biol Chem. 2004;279(14):14355–14364. doi: 10.1074/jbc.M313934200. [DOI] [PubMed] [Google Scholar]
- [27].Tsybovsky Y, Donato H, Krupenko NI, Davies C, Krupenko SA. Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases. Biochemistry. 2007;46(11):2917–2929. doi: 10.1021/bi0619573. [DOI] [PubMed] [Google Scholar]
- [28].Krupenko SA, Wagner C, Cook RJ. Cysteine 707 is involved in the dehydrogenase activity site of rat 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1995;270(2):519–522. doi: 10.1074/jbc.270.2.519. [DOI] [PubMed] [Google Scholar]
- [29].Krupenko SA, Vlasov AP, Wagner C. On the role of conserved histidine 106 in 10-formyltetrahydrofolate dehydrogenase catalysis: connection between hydrolase and dehydrogenase mechanisms. J Biol Chem. 2001;276(26):24030–24037. doi: 10.1074/jbc.M009257200. [DOI] [PubMed] [Google Scholar]
- [30].Shen D, Dalton TP, Nebert DW, Shertzer HG. Glutathione redox state regulates mitochondrial reactive oxygen production. J Biol Chem. 2005;280(27):25305–25312. doi: 10.1074/jbc.M500095200. [DOI] [PubMed] [Google Scholar]
- [31].Feldman RI, Weiner H. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J Biol Chem. 1972;247(1):267–272. [PubMed] [Google Scholar]
- [32].Sheikh S, Ni L, Hurley TD, Weiner H. The potential roles of the conserved amino acids in human liver mitochondrial aldehyde dehydrogenase. J Biol Chem. 1997;272(30):18817–18822. doi: 10.1074/jbc.272.30.18817. [DOI] [PubMed] [Google Scholar]
- [33].Marchal S, Cobessi D, Rahuel-Clermont S, Tete-Favier F, Aubry A, Branlant G. Chemical mechanism and substrate binding sites of NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. Chem Biol Interact. 2001;130-132(1-3):15–28. doi: 10.1016/s0009-2797(00)00218-0. [DOI] [PubMed] [Google Scholar]
- [34].Chang WN, Lin HC, Fu TF. Zebrafish 10-formyltetrahydrofolate dehydrogenase is similar to its mammalian isozymes for its structural and catalytic properties. Protein Expr Purif. 2010;72(2):217–222. doi: 10.1016/j.pep.2010.04.003. [DOI] [PubMed] [Google Scholar]
- [35].Hurley TD, Perez-Miller S, Breen H. Order and disorder in mitochondrial aldehyde dehydrogenase. Chem Biol Interact. 2001;130-132(1-3):3–14. doi: 10.1016/s0009-2797(00)00217-9. [DOI] [PubMed] [Google Scholar]
- [36].Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251(3):549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- [37].Farres J, Wang TT, Cunningham SJ, Weiner H. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry. 1995;34(8):2592–2598. doi: 10.1021/bi00008a025. [DOI] [PubMed] [Google Scholar]
- [38].Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Strickland KC, Holmes RS, Oleinik NV, Krupenko NI, Krupenko SA. Phylogeny and evolution of aldehyde dehydrogenase-homologous folate enzymes. Chem Biol Interact. 2011 doi: 10.1016/j.cbi.2010.12.025. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1(2):171–176. [PubMed] [Google Scholar]
- [41].Garcia-Martinez LF, Appling DR. Characterization of the folate-dependent mitochondrial oxidation of carbon 3 of serine. Biochemistry. 1993;32(17):4671–4676. doi: 10.1021/bi00068a027. [DOI] [PubMed] [Google Scholar]
- [42].Galper JB, Darnell JE. The presence of N-formyl-methionyl-tRNA in HeLa cell mitochondria. Biochem Biophys Res Commun. 1969;34(2):205–214. doi: 10.1016/0006-291x(69)90633-0. [DOI] [PubMed] [Google Scholar]
- [43].Halbreich A, Rabinowitz M. Isolation of Saccharomyces cerevisiae mitochondrial formyltetrahydrofolic acid:methionyl-tRNA transformylase and the hybridization of mitochondrial fMet-tRNA with mitochondrial DNA. Proc Natl Acad Sci U S A. 1971;68(2):294–298. doi: 10.1073/pnas.68.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]