Abstract
The Aldo Keto Reductases (AKRs) are a superfamily of enzymes that catalyze the reduction of biogenic and xenobiotic aldehydes and ketones. AKR1B family has 2 known members in humans and 3 in rodents. Two novel gene loci, hereafter referred to as AKR1B15 in human and Akr1b16 in mouse have been predicted to exist within the AKR1B clusters. AKR1B15 displays 91% and 67% sequence identity with human genes AKR1B10 and AKR1B1, respectively while Akr1b16 shares 82–84% identity with murine Akr1b8 and Akr1b7. We tested the hypothesis that AKR1B15 and Akr1b16 genes are expressed as functional proteins in human and murine tissues, respectively. Using whole tissue mRNA, we were able to clone the full-length open reading frames for AKR1B15 from human eye and testes, and Akr1b16 from murine spleen, demonstrating that these genes are transcriptionally active. The corresponding cDNAs were cloned into pET28a and pIRES-hrGFP-1α vectors for bacterial and mammalian expression respectively. Both genes were expressed as 36 kDA proteins found in the insoluble fraction of bacterial cell lysate. These proteins, expressed in bacteria showed no enzymatic activity. However, lysates from COS-7 cells transfected with AKR1B15 showed a 4.8-fold (with p-nitrobenzaldehyde) and 3.3-fold (with DL-glyceraldehyde) increase in enzyme activity compared with untransfected COS-7 cells. The Akr1b16 transcript was shown to be ubiquitously expressed in murine tissues. Highest levels of transcript were found in heart, spleen, and lung. From these observations we conclude that the predicted AKR1B15 and 1b16 genes are expressed in several murine and human tissues. Further studies are required to elucidate their physiological roles.
Keywords: AKR1B15, Akr1b16, Aldo-Keto Reductase, gene expression, RT-PCR, aldehyde
1. Introduction
Aldo Keto Reductases (AKRs) are multifunctional enzymes that catalyze the reduction of several endogenous and xenobiotic aldehydes and ketones. The AKR superfamily comprises 15 families containing 151 members which are further classified into subfamilies. To date, 13 human AKRs have been described. These enzymes utilize a variety of substrates, including steroid hormones, prostaglandins, retinals, lipid aldehydes and sugars and they prefer use of NADPH over NADH as the reducing cofactor [1–2]. The AKRs are generally cytosolic and monomeric (35–37kDA) proteins although some have been shown to exist as dimers (AKR7) or in association with membrane proteins [3–5]. Although most AKRs are enzymatically active, some of these proteins have been reported to possess little or no activity with typical AKR substrates (e.g., MVDP, AKR1C12-C13, Kvβ) or they have been shown to play a structural role (e.g., rho-crystallins) [6–7].
Mammalian AKRs have been classified into 3 families: AKR1, AKR6 and AKR7. Of these, the AKR1 family is the largest and has been further divided into subfamilies A through E. The most widely expressed member of the AKR1B subfamily, AKR1B1, - human aldose reductase, has received the most attention, and its role in the development of diabetic complications has been studied since the 1960-s [8]. Several studies have shown that inhibition of AKR1B1 could prevent, delay or even reverse tissue injury induced by hyperglycemia [9]. The AKR1B1 catalyzes reduction of several substrates of high physiological significance including glucose, AGE precursors, HNE and oxidized phospholipids [10–11]. It also plays an essential role in vascular smooth muscle cell proliferation in arterial lesions [12].
Identification of human aldose reductase was later followed by the identification of 3 other murine AKR1B proteins. The murine Akr1b3 was identified as a homolog of human aldose reductase, however, 2 other members seem specific to rodents: Akr1b7 (mouse vas deferens protein, MVDP) [13], and Akr1b8 (mouse fibroblast growth factor regulated protein (FR-1) [14]. The second human AKR1B gene, small intestine reductase, AKR1B10, was identified later [15–16], and appears to be an ortholog of neither 1b7, nor 1b8. The AKR1B10 is expressed mainly in small intestine, colon, liver, thymus [15] and adrenal gland [16]. AKR1B10 shares 71% amino acid sequence identity with aldose reductase but differs from it in substrate specificity and tissue distribution. AKR1B10 has higher Km and kcat values for glyceraldehyde and methylglyoxal, and is not an efficient glucose reductase. However, AKR1B10 demonstrates a higher catalytic efficiency in reducing all-trans-retinal, and a number of xenobiotics including the drugs daunorubicin, oracin, and dolacetron. AKR1B10 is also induced by cigarette smoke condensate [15, 17–19]. The abundance of the AKR1B10 is markedly increased in lung and hepatic carcinomas (squamous cell and adenocarcinomas) [15] as well as in colorectal and uterine cancers [20].
Currently, it seems that AKR1B family has 2 known members in humans but 3 in rodents. However, a novel gene locus, tcag7.1260 (hereafter referred to as AKR1B15) has been predicted to cluster with human members of the AKR1B subfamily, AKR1B1 and AKR1B10 on chromosome 7 [2, 21]. This gene is predicted to have 10 exons and an open reading frame that displays 91% and 67% sequence identity with AKR1B10 and AKR1B1 genes, respectively, and is also homologous to the murine Akr1b8 and Akr1b7 genes (81% and 76% respectively). Also, cross-genome comparison of human and murine AKR genes reveals the existence of an unknown murine AKR gene, 2310005E10Rik, hereafter referred to as Akr1b16, which has an open reading frame with 82–84% amino acid identity with other AKR1B proteins. No protein product has yet been demonstrated for the AKR1B15 and Akr1b16 genes. We therefore tested the hypothesis that AKR1B15 and Akr1b16 genes are expressed into functional proteins (AKR1B15 and Akr1b16) in human and murine tissues, respectively.
2. Materials and Methods
2.1. Materials
Restriction enzymes Nde1 (catalog number; R6801), Xho1 (R6161) and Not1 (R 6431), and AMV reverse transcriptase (M510A) were obtained from Promega. pET28a (+) (69864-3) and pIRES-GFP1α (240031) vectors were obtained from Novagen and Stratagene, respectively. Mammalian COS-7 cells (CRL-1651™) were obtained from American Type Culture Collection. Kanamycin (K4378), NADPH (N5130), isopropyl-1-thio-β-D-galactopyranoside - IPTG (I5502), para-nitrobenzaldehyde (N9015), glyceraldehyde-3-phosphate (G5001), Triton X-100 (T8532) and protease inhibitor cocktail (P8340) were obtained from Sigma-Aldrich. Anti His-tag antibody (sc-51946) was obtained from Santa Cruz Biotechnologies.
2.2. mRNA isolation and reverse transcriptase PCR
Whole human tissue mRNA from different tissues was obtained from Clontech (cat. no; 636643). Whole mouse mRNA was isolated from different tissues of C57BL/6J mice using TRIZOL (15596-026) reagent (Invitrogen) and the concentration accurately determined by measuring absorbance at 260nm using a Nano drop (Thermo scientific). A 20μl reverse transcription reaction mixture containing 1μg mRNA, 10 units AMV reverse transcriptase, 0.4μM poly T primer (dT18), 0.2mM dNTPs (cat. no; U1511), and 20 units RNasin (cat. no; N2611, Promega) was subjected to the following conditions in a thermal cycler (BioRad); 70°C for 2min, 42°C for 1 h and 94°C for 5 min. After cDNA synthesis, 2μl of cDNA was used for amplification of GOI by PCR. A 25μl PCR containing 2μl cDNA template, 12.5μl PCR master mix (cat. no; M750B, Promega), 0.5μl forward and reverse primer each (0.4μM final), and 9.5μl dH2O was subjected to the following thermal conditions; 1 cycle of 94°C for 3min, 28 cycles of (94°C for 45 s, 50°C for 45 s, 72°C for 45 s) and 1 cycle of 72°C for 7 min.
2.3. Cloning and production of recombinant proteins
The primers used for cloning whole length AKR1B15 and Akr1b16 and to detect Akr1b8, AKR1B10 and Akr1b16 in different human and mouse tissues are listed in Table I (primers for cloning whole length AKR1B15 and 1b16 have restriction enzyme sites underlined). Primers used for detecting genes in tissues were designed such that they amplified only short portions of the genes of interest. All primers were purchased from integrated DNA technology (IDT, USA)
Table I.
Primers used for cloning whole length AKR1B genes and for detecting these genes in different tissues
| Genes | Primers |
|---|---|
| Akr1b8 | 5′-TCAGCCCACGAGGCTTCCTTC-3′ (sense) |
| 5′-CTCCGGAGTCGCATTTGCTCGCA-3′ (anti sense) | |
| AKR1B10 | 5′-TGGCCACGTTTGTGGAGCTCAGTA-3′ (sense) |
| 5′-AACGTTGCTTTTCCACCGATGGC-3′ (anti sense) | |
| Akr1b16 | 5′-AATTAGTGACAAACATTGCCC-3′ (sense) |
| 5′-AGATGCGCTTGCCTGGAGGACA-3′ (anti sense) | |
| AKR1B15 (whole length) | 5′-AAG AAG CGG CCG CAC CAT GGG CAG CAG CCA T-3′ (sense) |
| 5′-AGATCCCTCGAGTCAATATTCTGCATCGAA-3′ (anti sense) | |
| Akr1b16 (whole length) | 5′-AAG AAG CGG CCG CAC CAT GGG CAG CAG CCA T-3′ (sense) |
| 5′-AGATCCCTCGAGTCAGTATTCCGCATGGAA-3′ (anti sense) |
Recombinant AKR1B15 and Akr1B16 were produced as 6 histidine N-terminal-tagged proteins. Purified PCR fragments were digested and inserted into NdeI/XhoI sites (in pET28a (+) vector) and NotI/XhoI sites (in pIRES-hrGFP-1α vector). DNA sequencing was used to confirm that the inserted coding sequence had no mutations. For bacterial expression, 1L culture volumes were inoculated with BL21-DE3 bacterial culture containing pET28a (+)/AKR1B15 or pET28a (+)/Akr1B16 expression plasmids and then allowed to grow to OD600 = 0.6. The cultures were then induced with 1mM IPTG and grown overnight at 25°C. For determining solubility of expressed proteins, 500μl culture was sonicated and then centrifuged for 10 min at 13000 rpm. The resulting pellet was dissolved in SDS sample buffer and corresponding amounts loaded on a 12.5% SDS gel alongside the supernatant. For large scale purification, cells were lysed by sonication in 2% sarkosyl (L-5125, Sigma). After centrifugation at 12000xg for 30 min to remove debris, the supernatant was incubated with 50% slurry of nickel resin (cat. no; 30210, Qiagen) at 4°C overnight. The protein was eluted with stepwise gradient of 50–300 mM imidazole (cat. no; I5513, Sigma Aldrich) and small aliquots of different fractions were loaded on a 12.5% SDS gel, electrophoresed and stained with Coomassie blue to ascertain purity.
For mammalian protein expression, COS-7 cells in 10 cm dishes were transfected with 3μg of either pIRES-hr-GFP/AKR1B15, Akr1b16 or AKR1B10 with the use of lipofectamine 2000 (cat no; 11668, Invitrogen). Transfected cells were kept for 48h at 37°C in 5% CO2 incubator. GFP was used for monitoring transfection efficiency. To obtain cytosolic and membrane fractions, the cells were trypsinized and then sonicated in 20 mM phosphate buffer (pH7.4) containing 1:100 protease inhibitor cocktail. The lysates were centrifuged at 100,000xg for 1h. The resultant supernatant was designated as the soluble fraction and the pellet was redissolved in 0.5% triton for Western blot and activity measurement.
2.4. Western Blotting
For Western blotting, 50μg of protein (measured using the Lowry’s assay) were applied to each lane of a 12.5% Bis-Tris gel and electroblotted onto a PVDF membrane. The membrane was then incubated with 1:2000 dilution of anti Histag antibody (Santa Cruz Biotech) overnight at 4°C. After several washes in TBS-Tween, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG. Immunoreactive bands were detected with enhanced chemiluminescent detection reagent (cat. no; RPN 2132, GE Healthcare).
2.5. Enzyme assays
The reaction mixture for enzyme activity measurements contained 100 mM sodium phosphate buffer (pH 7.0), 175μM NADPH, 150μg of enzyme with the appropriate amount of substrate. The reaction was carried out at room temperature, and NADPH consumption was monitored by recording absorbance at 340nm in a spectrophotometer. Identical conditions were used for measuring AKR1B10, AKR1B15 and AKR1B16.
3. Results
3.1. New predicted AKR1B genes exist in human and mouse genomes
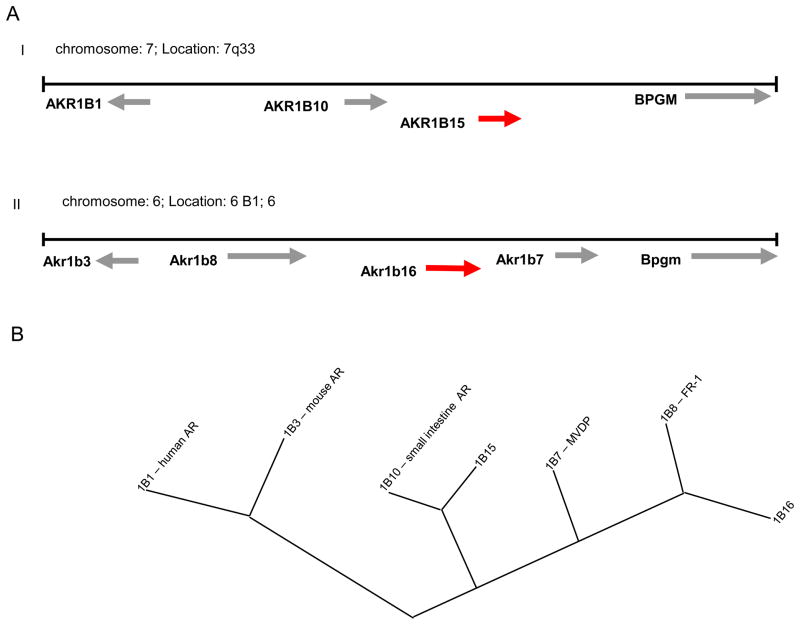
Examination of the human and murine genome databases at the NCBI Genbank revealed existence of two yet unknown predicted genes within AKR1B clusters on human chromosome 7 and murine chromosome 6, respectively (Fig. 1A). The spatial arrangement and orientation of genes on their respective chromosomes is similar between human, and mouse AKR genes. The aldose reductase gene is located on the (−) strand of the chromosome, and the three remaining genes (two in human) clustered on the strand opposite to the 5′ end of the aldose reductase gene. The new human gene designated as AKR1B15 shares 95% sequence identity with AKR1B10 on the cDNA level. Similarly, in the murine genome, a predicted gene, Akr1b16 shares 89% sequence identity with Akr1b7 and 82% identity with Akr1b8 in the coding region of the cDNA. Like other AKRs, both genes consist of 10 coding exons.
Figure 1. Position of novel AKR1B genes in human and mouse genomes.
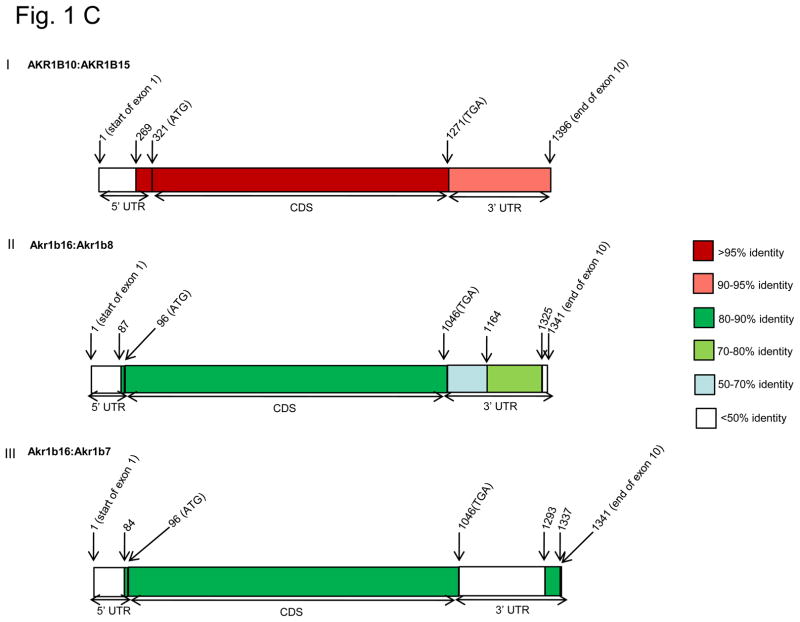
A, Locus maps for human chromosome 7q33-35 (I) and mouse chromosome 6B1 (II) showing the positions of human AKR1B15 and mouse Akr1b16 genes; B, phylogenetic tree of human and mouse AKR1B members showing relatedness of AKR1B proteins; C, Heat diagrams showing degree of identity along the mRNA sequence of AKR1B10 with AKR1B15 (I) and Akr1b16 with Akr1b7 (II) and 1b8 (III).
A comparison of the conceptually translated protein sequences within AKR1B family showed that aldose reductase (AKR1B1, AKR1B3) is the most distant member and forms a separate branch on the evolutionary tree (Fig. 1B). The novel human protein, AKR1B15 shares the highest homology (91% identity) with AKR1B10 from the same species (Table II and Fig. 2) suggesting that it is a result of a current duplication event. In the cross-species comparison, AKR1B15 shows the highest identity with FR1 (AKR1B8, 81%), followed by the novel murine protein AKR1B16 (80%) and MVDP (AKR1B7, 76%). Murine AKR1B16 displays the most homology with AKR1B7 (84%, same species), and AKR1B10 (82%) in human. It still shares high identity (80%) to AKR1B15. Homologene database designates Akr1b16 (2310005E10Rik) as a murine ortholog of AKR1B10, and Akr1b8 as an ortholog of AKR1B15.
Table II.
Protein sequence identities for human and mouse AKRs.
| AKR1B1 | 100 | ||||||
| AKR1B10 | 70 | 100 | |||||
| AKR1B15 | 67 | 91 | 100 | ||||
| Akr1b8 | 70 | 82 | 80 | 100 | |||
| Akr1b7 | 71 | 79 | 76 | 82 | 100 | ||
| Akr1b3 | 85 | 70 | 67 | 69 | 69 | 100 | |
| Akr1b16 | 70 | 82 | 80 | 82 | 84 | 69 | 100 |
| AKR1B1 | AKR1B10 | AKR1B15 | Akr1b8 | Akr1b7 | Akr1b3 | Akr1b16 |
Figure 2.
A comparison of the protein sequence of AKR1B10 with AKR1B15.
In the untranslated region of the cDNA, the two human genes 1B10 and 1B15 share 91% identity at the 3′-end of the coding sequence (Fig. 1CI). At the 5′-untranslated region (UTR) 52 nucleotides adjacent to the translation start site align with each other, while the upstream sequences are completely divergent, suggesting that the promoters of the two genes are quite different; predicting significant differences in the expression and the tissue distribution of 1B10 and 1B15. In the murine genome, the 3′-untranslated region of Akr1b16 shares 71% identity with Akr1b8, but not 1b7, whereas 5′-UTRs of the three genes are completely divergent (Fig. 1CII & III).
A comparison of 5′- and 3′-untranslated regions of AKR1B10 with the murine database revealed that its 3′-UTR aligns only with Akr1b16 3′-UTR (68 %), but not with 1b7 or 1b8. The 31 nucleotides upstream of the translation start site of AKR1B10 (5′-UTR) share 88% identity with 1b7 and 1b8 and 82 % with 1b16. Murine 1b16 untranslated regions share 68 % identity with 1B10 and 65% with 1B15 on the 3′ end; and 82% identity with 1B10 and 78% with 1B15 on the 5′-end. It is thus likely that the novel murine AKR is an ortholog of 1B10. However, based on homology alone, it is unclear whether human AKR1B15 is an ortholog of murine 1b7 or 1b8.
3.2. Cloning of the full-length novel AKR1B mRNA transcripts
To establish that the predicted human and murine genes are expressed at their full length in vivo, we attempted to amplify their entire coding regions from total human and murine RNA. Using primers identical to the sequences of predicted but not homologous AKR1B genes, we amplified the entire coding regions for the AKRs 1B15 and 1b16 from the cDNA from human eye and testes, and murine spleen, respectively. Sequencing of the entire inserts produced sequences that were 100% identical to that reported in the Genbank. Moreover, we obtained and sequenced the clone for Akr1b16 from ATCC and found 4 nucleotide mismatches resulting in 2 amino acid substitutions (pos. 197 Thr in ATCC, Ile in NCBI; pos. 243. Lys in ATCC, Arg in NCBI). As our clone and the sequence reported in the Genbank relate to C57BL/6J, whereas that of ATCC is from the Czech II strain, there seems to be strain-related polymorphism in the murine Akr1b16 gene.
3.3. Expression of novel AKR1B proteins in bacterial and mammalian systems
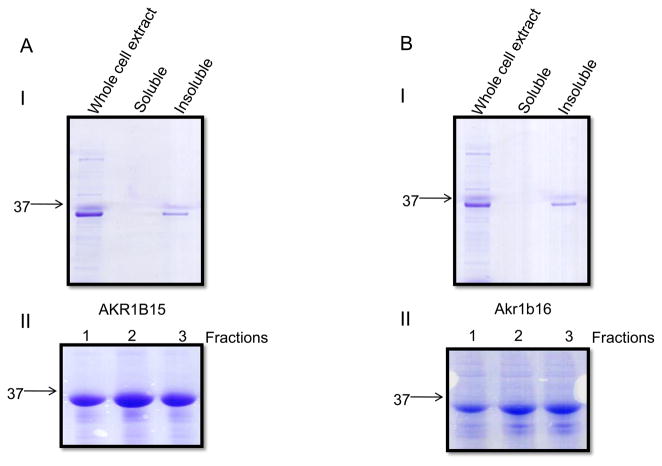
To establish whether new AKRs possess enzymatic activity AKR1B15 and B16 proteins were cloned into the bacterial expression vector, pET28 and expressed with an N-terminal His-tag. Surprisingly, both proteins were found in the insoluble fraction of bacterial cell lysate (Fig. 3A and B). Pellets of protein extracts were solubilized using the Sarkosyl-Triton detergent system and purified to homogeneity over the Ni-affinity column. The detergent concentration was gradually decreased in the course of purification process (see Materials and Methods, Fig. 3AII and 3BII). After dialysis into a detergent-free solution the proteins remained in solution. However, these proteins displayed no enzymatic activity using NADPH (175 μM) as cofactor and the typical AKR substrates DL-glyceraldehyde, p-nitrobenzaldehyde or methylglyoxal. No activity was detected in the crude bacterial extract. In contrast, AKR1B10 expressed from an identical construct in the same bacterial strain was localized to the soluble fraction of the protein extract and exhibited oxidoreductase activity consistent with literature data (not shown). The same expression system is routinely used in our laboratory for the expression of at least 10 other well characterized AKR1, 6, and 7 proteins and it produces active soluble enzymes. To rule out His-tag interference we also cloned, expressed and purified AKR1B15 with N-terminal GST-tag and without any tags at all. In all cases, the expressed protein was found in the insoluble fraction, and no activity was detected. Attempts to refold AKR1B15 from inclusion bodies were made using 96 different buffer conditions in the pH range 7.0–8.5 available with an iFOLD system (Novagen). Nevertheless, no enzyme activity was detected under any of these conditions.
Figure 3. Bacterial expression of human and mouse AKR1B proteins.
Coomassie blue staining following SDS-PAGE of soluble and insoluble fractions of bacterial extracts overexpressing A, AKR1B15 and B, AKR1B16. The lower panels (II) show fractions of the purified protein eluting from Ni-affinity column with 100mM imidazole.
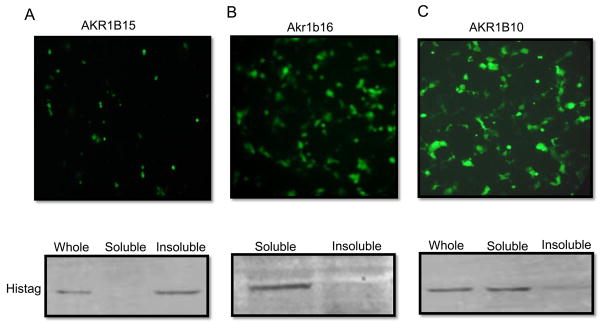
To investigate whether localization to the insoluble fraction is an artifact of the bacterial expression system or an intrinsic property of these proteins, we constructed mammalian expression vectors for AKRs 1B10, 1B15 and 1B16 with N-terminal His-tags and determined their localization and enzymatic activity in the crude extracts of transfected COS-7 cells. AKR1B15 was found in the microsomal fraction (100,000xg pellet) whereas, AKRs 1B10 and 1B16 were present in the supernatant (Fig. 4). As most AKRs (with the exception of AKR6 family) are found in the cytosol, microsomal localization might be due to association with another as yet unknown, membrane protein.
Figure 4. Mammalian expression of AKR1B proteins.
COS-7 cells were transfected with the pIRES-hrGFP vector containing inserts for A, AKR1B15; B, AKR1B16; and C, AKR1B10. The vector also contained the expression cassette for GFP expressed through IRES independently of the AKRs. Upper panel shows the expression of GFP in transfected cells prior to their lysis as an indication of transfection efficiency. Lower panels shows a representative Western blot of soluble and microsomal fractions performed with His-tag antibody.
Catalytic activity was measured in the soluble and microsomal fractions of the protein extracts from transfected and untransfected COS-7 cells using p-nitrobenzaldehyde and DL-glyceraldehyde. As shown in Table III, the soluble fraction of the AKR1B10-transfected cells exhibited 21-fold higher activity compared with untransfected cells with p-nitrobenzaldehyde as a substrate. Lower, but measurable activity, was detected in both the pellet and supernatant of AKR1B15-transfected cells, indicating that this novel protein might indeed possess oxidoreductase enzymatic activity. No activity was found in the AKR1B16-transfected cells despite its cytosolic localization by Western blot.
Table III.
Enzymatic activity of soluble and insoluble protein extracts of COS-7 cells transfected with AKR1B expression vectors
| Soluble protein fraction with 500μM p-nitrobenzaldehyde | ||
|---|---|---|
| Activity (μmol/min/μg)a | Fold increase vs untransfected | |
| Untransfected COS7 cells | 3.0×10−5 | 1 |
| AKR1B15 transfected COS7cells | 9.4×10−5 | 3.1 |
| AKr1b16 transfected COS7cells | 2.90×10−5 | 1 |
| AKR1B10 transfected COS 7 cells | 6.2×10−4 | 21 |
| Insoluble fraction with 500μM p-nitrobenzaldehyde | ||
| Activity (μM/min/μg) | Fold increase vs untransfected | |
| Untransfected COS7 cells | 1.3×10−5 | 1 |
| AKR1B15 transfected COS7cells | 6.3×10−5 | 4.8 |
| Akr1b16 transfected COS 7 cells | 1.4×10−5 | 1 |
| Insoluble fraction with 10mM DL-glyceraldehyde | ||
| Activity (μM/min/μg) | Fold increase vs untransfected | |
| Untransfected COS7 cells | 3.8×10−5 | 1 |
| AKR1B15 transfected COS7cells | 6.9×10−5 | 1.8 |
| Ar1b16 transfected COS7cells | 3.6×10−5 | 1 |
160μg of supernatant (soluble protein fraction) or 100,000xg pellet dissolved in 0.1% triton (insoluble fraction) were used for activity measurements in the presence of 175μM NADPH and respective substrates in 0.1M phosphate buffer pH7.0. The rate of decrease in absorbance at 340nm was recorded and activity reported as μM/min/μg protein.
3.4. Tissue Distribution of Akr1b16
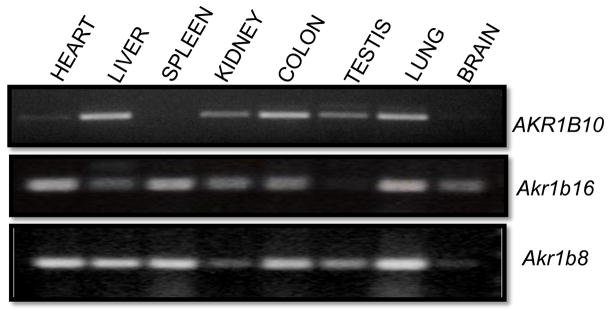
AKR1b16 is designated as a murine ortholog of AKR1B10 in the NCBI databases. The question of orthology is important because it could lead to a clear identification of the murine ortholog of AKR1B10, and thereby enable the use of mouse knockout and transgenic technologies to investigate the physiological function of AKR1B10. To compare tissue distribution of the two transcripts we performed PCR on a panel of tissues using primers specific for AKR1B10, 1b16, and 1b8, respectively. The primers were designed on different exons to verify that the correct RNA splicing occurs. As shown in Fig. 5, Akr1b16 is ubiquitously expressed in murine tissues with the exception of testes. Highest transcript levels were found in heart, spleen, and lung. This pattern is similar to the also ubiquitously expressed Akr1b8, which is highly expressed in the lung, with lowest levels found in the brain and the kidney. In contrast, the levels of the AKR1B10 transcript have been found to be very low in the heart and the spleen, but the gene is highly expressed in the liver, colon and lung. Although tissue distribution may differ somewhat between species, neither the tissue distribution nor enzymatic activity data support a clear assignment of orthology between AKR1B10 and any of the murine Akr1b genes.
Figure 5. Tissue distribution of AKR1B10, Akr1b16, and Akr1b8.
Total RNA (1ug) from human and murine tissues was reverse transcribed into cDNA. RT-PCR was performed using specific primers for the indicated AKR1B genes.
4. Discussion
In this report, we describe the identification and expression of two, as yet unknown, members of the AKR superfamily of proteins. Computational analysis of assembled genomic sequences indicates that these two genes exist in human and murine genomes. Based on predicted sequences, we designed PCR primers and were able to amplify from the total RNA of human and murine tissues correctly spliced transcripts with sequences identical to those predicted by computational analysis. Hence, we demonstrate that these are active genes and that their products are expressed in vivo.
Expression of AKR1B15 using either bacterial or mammalian expression systems produced an insoluble protein which showed little to no catalytic activity with typical AKR substrates such as p-nitrobenzaldehyde, methylglyoxal and DL-glyceraldehyde. Reasons for this are unclear; AKR1B15 shares 91% sequence identity with the AKR1B10 protein, which is an active cytosolic AKR that is readily expressed in soluble form in bacterial and mammalian systems. Hydrophobicity plots of the two proteins showed no significant differences that could explain microsomal localization of AKR1B15. Also, the Tyr-Lys-Asp-His catalytic tetrad common to the enzymes of the AKR family is conserved in AKR1B15. The only significant difference between the two sequences is a stretch of 6 amino acid residues (299–304) close to the C-terminus of the protein (Fig. 2). It is well known that the C-terminus determines the substrate specificity of the AKRs and is the most divergent region in the superfamily [22–24]. Of particular interest is that Cys299, which plays important role in the physiological regulation of aldose reductase activity [25] and is conserved among all other AKR1B members, is substituted with Phe in AKR1B15. Thus, it is possible that AKR1B15 possesses all the necessary catalytic machinery, but displays substrate specificity different from other AKR1Bs due to the sequence diversity at its C-terminus. A wider screen of carbonyl and other compounds may prove helpful in identification of possible substrates for this enzyme. Alternatively, AKR1B15 may not have a catalytic role at all and it is a structural protein that binds to other proteins, similar to, members of the AKR6 family (Kvβ proteins) that act as nucleotide sensors in voltage gated potassium channels [26].
AKR1B16 shares less homology with other murine proteins in its cluster than human AKR1B15 and 1B10. It displays 84% sequence identity with AKR1B7 and 82% identity with AKR1B8. As with the human proteins, the major region of diversity lies at the C-terminal end of the proteins at amino acid residues 299–316. In this region of 18 residues, AKR1B16 has only 6 common residues with AKR1B8, and 9 with AKR1B7. Additional region of diversity between AKRs 1B16 and 1B8, but not 1B7 is around amino acid residue 120 in the loop between strand β6 and helix α4 [27]. This loop is long in all AKR1 enzymes and contains several active site residues indispensable for catalytic activity, such as His110 (AR numbering). It is interesting that the AKR1B16 protein is more homologous to AKR1B7 than AKR1B8, whereas the 3′-untranslated region of its RNA shares no homology with that of 1b7, but significant homology with 1b8. Hence, this gene might have evolved as a duplication event of Akr1b8, but may resemble AKR1B7 in its functional properties as a protein. Expression of Akr1b16 in mammalian expression system yielded soluble protein, albeit with no measurable enzymatic activity. It is noteworthy that the catalytic activity of AKR1B7 is poor in comparison with other AKRs and the range of substrates is rather narrow [28]. Thus, as with human AKR1B15, other substrates, such as e.g. retinoids and prostaglandins will have to be tested to identify the enzymatic properties and the physiological roles of these novel AKRs.
PCR analysis of the Akr1b16 mRNA indicated ubiquitous tissue distribution at quite significant levels. This is consistent with the presence of expressed sequence tags in the Genbank database. In addition, Akr1b16 has been detected in the mitochondria of 12 out of 14 murine tissues as part of the mitochondrial proteome in the course of high throughput proteomic analysis [29]. Although sequence analysis returned only 26 % probability of the mitochondrial targeting of AKR1B16 protein, its detection in the mitochondria indicates that this new AKR may play a yet to be appreciated role in mitochondrial function.
5. Conclusion
Using RT-PCR amplification followed by cloning and sequencing we show that two predicted genes AKR1B15 and Akr1b16 are expressed in human and murine tissues, respectively. The corresponding human protein AKR1B15 possesses weak oxidoreductase activity. Murine AKR1b16 is widely distributed across murine tissues. With the addition of these new genes, the AKR1B subfamily now consists of 3 genes in human (AKRs 1B1, 1B10, and 1B15) and 4 genes in mouse (AKRs 1b3, 1b7, 1b8, and 1b16) genome. Apart from aldose reductases (1B1 and 1b3), the rest of the genes share >80% sequence identity with each other and form a separate phylogenetic cluster. Such high degree of identity makes it difficult to assign orthology between mouse and human genes within this cluster, even though such assignment is important to enable the application of methods of mouse genetics to the studies of physiological function of human AKR1B10 and 1B15 genes.
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL-89372 and RR024489, and EY005856. The technical assistance of Terry Griest and Theresa Harter is gratefully acknowledged.
Footnotes
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Penning TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys. 2007;464(2):241–50. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40(4):553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gulbis JM, et al. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science. 2000;289(5476):123–7. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- 4.Kelly VP, et al. Novel homodimeric and heterodimeric rat gamma-hydroxybutyrate synthases that associate with the Golgi apparatus define a distinct subclass of aldo-keto reductase 7 family proteins. Biochem J. 2002;366(Pt 3):847–61. doi: 10.1042/BJ20020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozma E, et al. The crystal structure of rat liver AKR7A1. A dimeric member of the aldo-keto reductase superfamily. J Biol Chem. 2002;277(18):16285–93. doi: 10.1074/jbc.M110808200. [DOI] [PubMed] [Google Scholar]
- 6.Fujii Y, et al. Purification and characterization of rho-crystallin from Japanese common bullfrog lens. J Biol Chem. 1990;265(17):9914–23. [PubMed] [Google Scholar]
- 7.Weng J, et al. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281(22):15194–200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabbay KH, Merola LO, Field RA. Sorbitol pathway: presence in nerve and cord with substrate accumulation in diabetes. Science. 1966;151(707):209–10. doi: 10.1126/science.151.3707.209. [DOI] [PubMed] [Google Scholar]
- 9.Gabbay KH. Aldose reductase inhibition in the treatment of diabetic neuropathy: where are we in 2004? Curr Diab Rep. 2004;4(6):405–8. doi: 10.1007/s11892-004-0047-z. [DOI] [PubMed] [Google Scholar]
- 10.Vander Jagt DL, et al. Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal, and diabetic complications. J Biol Chem. 1992;267(7):4364–9. [PubMed] [Google Scholar]
- 11.Srivastava S, et al. Aldose reductase-catalyzed reduction of aldehyde phospholipids. J Biol Chem. 2004;279(51):53395–406. doi: 10.1074/jbc.M403416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramana KV, et al. Aldose reductase mediates mitogenic signaling in vascular smooth muscle cells. J Biol Chem. 2002;277(35):32063–70. doi: 10.1074/jbc.M202126200. [DOI] [PubMed] [Google Scholar]
- 13.Martinez A, et al. Androgen regulation of the mRNA encoding a major protein of the mouse vas deferens. Mol Cell Endocrinol. 1990;72(3):201–11. doi: 10.1016/0303-7207(90)90144-w. [DOI] [PubMed] [Google Scholar]
- 14.Donohue PJ, et al. A delayed-early gene activated by fibroblast growth factor-1 encodes a protein related to aldose reductase. J Biol Chem. 1994;269(11):8604–9. [PubMed] [Google Scholar]
- 15.Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273(19):11429–35. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- 16.Hyndman DJ, Flynn TG. Sequence and expression levels in human tissues of a new member of the aldo-keto reductase family. Biochim Biophys Acta. 1998;1399(2–3):198–202. doi: 10.1016/s0167-4781(98)00109-2. [DOI] [PubMed] [Google Scholar]
- 17.Crosas B, et al. Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases: consequences for retinoid metabolism. Biochem J. 2003;373(Pt 3):973–9. doi: 10.1042/BJ20021818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin HJ, et al. Purification and characterization of akr1b10 from human liver: role in carbonyl reduction of xenobiotics. Drug Metab Dispos. 2006;34(3):464–70. doi: 10.1124/dmd.105.007971. [DOI] [PubMed] [Google Scholar]
- 19.Nagaraj NS, et al. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol Lett. 2006;165(2):182–94. doi: 10.1016/j.toxlet.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshitake H, et al. Aldo-keto reductase family 1, member B10 in uterine carcinomas: a potential risk factor of recurrence after surgical therapy in cervical cancer. Int J Gynecol Cancer. 2007;17(6):1300–6. doi: 10.1111/j.1525-1438.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 21.Scherer SW, et al. Human chromosome 7: DNA sequence and biology. Science. 2003;300(5620):767–72. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski OA, Gabbay KH, Bohren KM. The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity. Biochemistry. 1996;35(45):14276–80. doi: 10.1021/bi9619740. [DOI] [PubMed] [Google Scholar]
- 23.Rees-Milton KJ, et al. Aldehyde reductase: the role of C-terminal residues in defining substrate and cofactor specificities. Arch Biochem Biophys. 1998;355(2):137–44. doi: 10.1006/abbi.1998.0721. [DOI] [PubMed] [Google Scholar]
- 24.Bohren KM, Grimshaw CE, Gabbay KH. Catalytic effectiveness of human aldose reductase. Critical role of C-terminal domain. J Biol Chem. 1992;267(29):20965–70. [PubMed] [Google Scholar]
- 25.Kaiserova K, et al. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281(22):15110–20. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 26.Tipparaju SM, et al. Differential regulation of voltage-gated K+ channels by oxidized and reduced pyridine nucleotide coenzymes. Am J Physiol Cell Physiol. 2005;288(2):C366–76. doi: 10.1152/ajpcell.00354.2004. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DK, et al. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992;257(5066):81–4. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 28.Lefrancois-Martinez AM, et al. Product of side-chain cleavage of cholesterol, isocaproaldehyde, is an endogenous specific substrate of mouse vas deferens protein, an aldose reductase-like protein in adrenocortical cells. J Biol Chem. 1999;274(46):32875–80. doi: 10.1074/jbc.274.46.32875. [DOI] [PubMed] [Google Scholar]
- 29.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]