Abstract
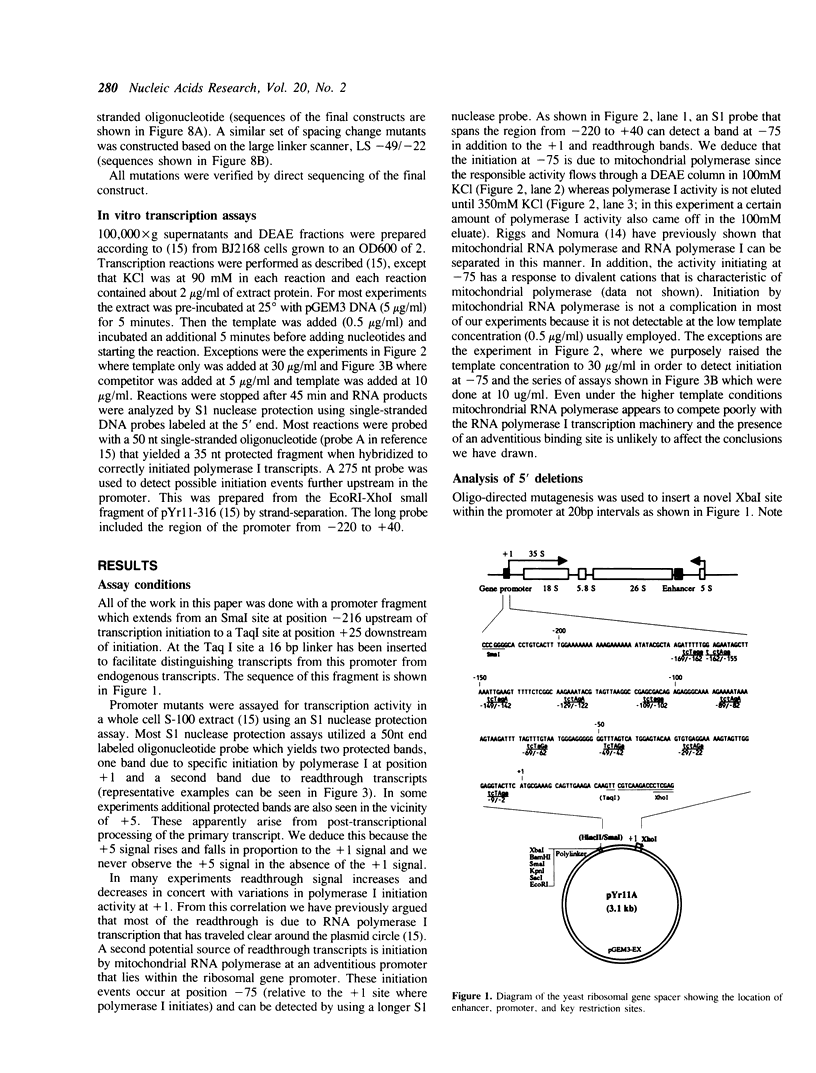
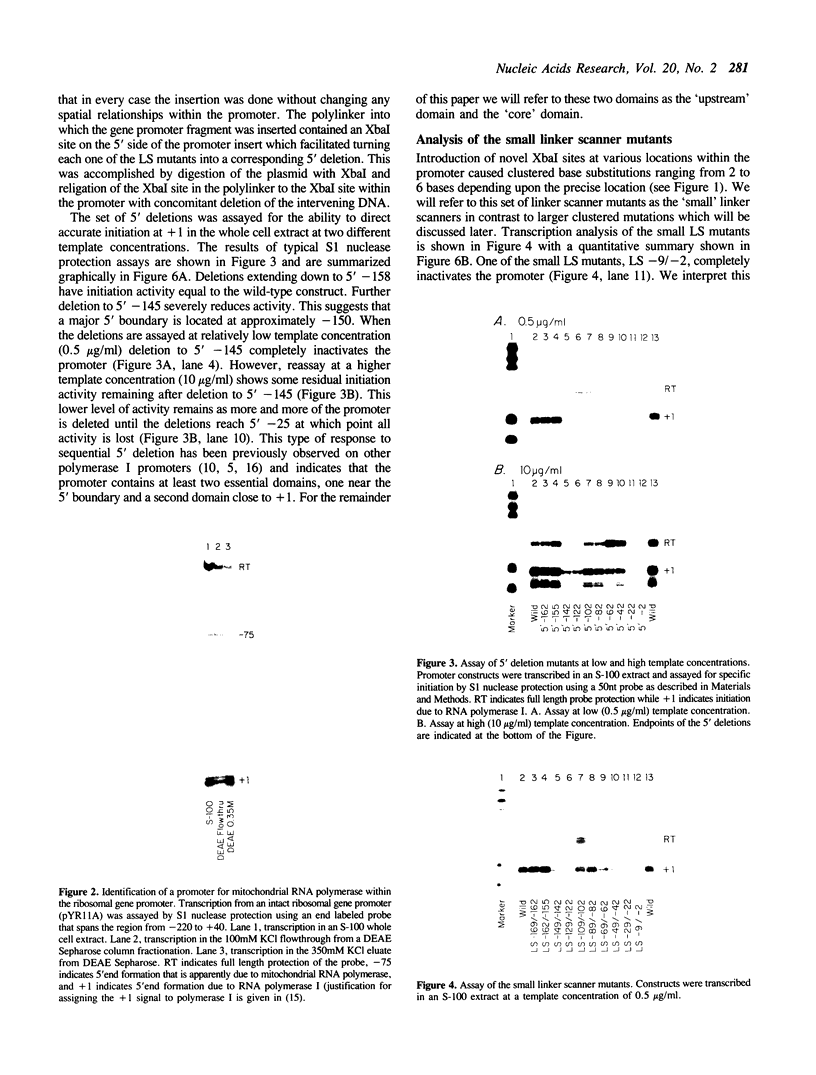
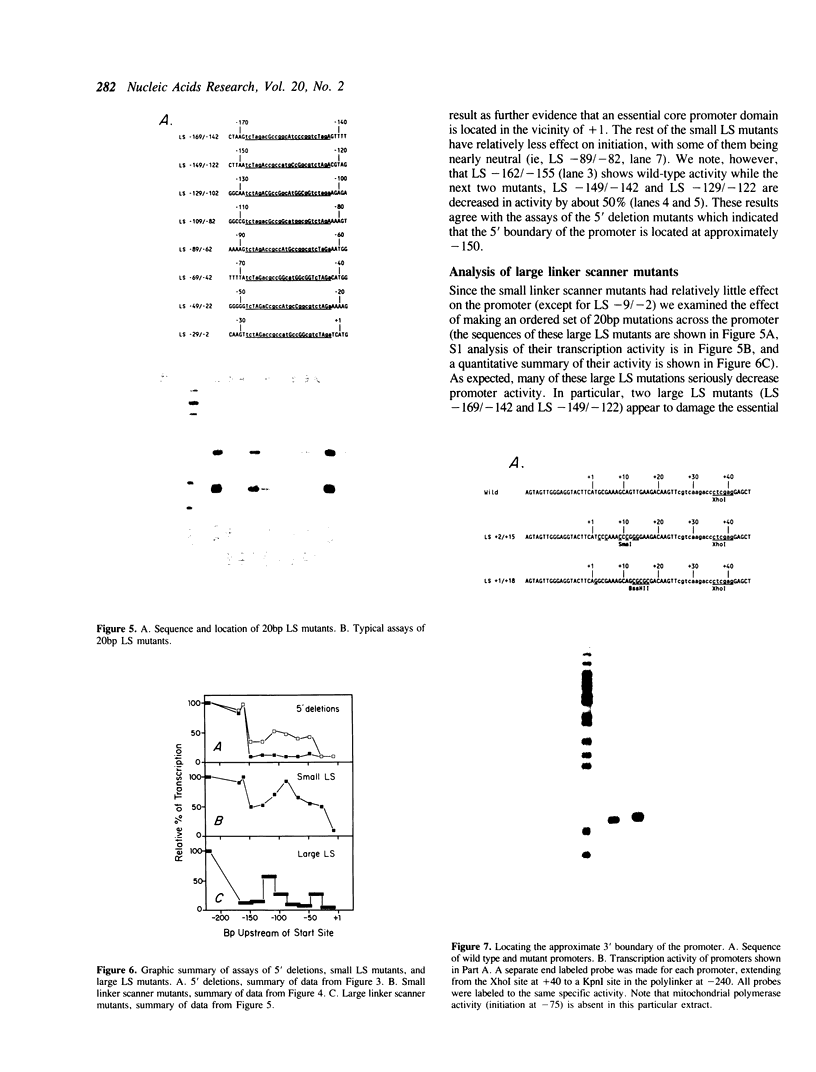
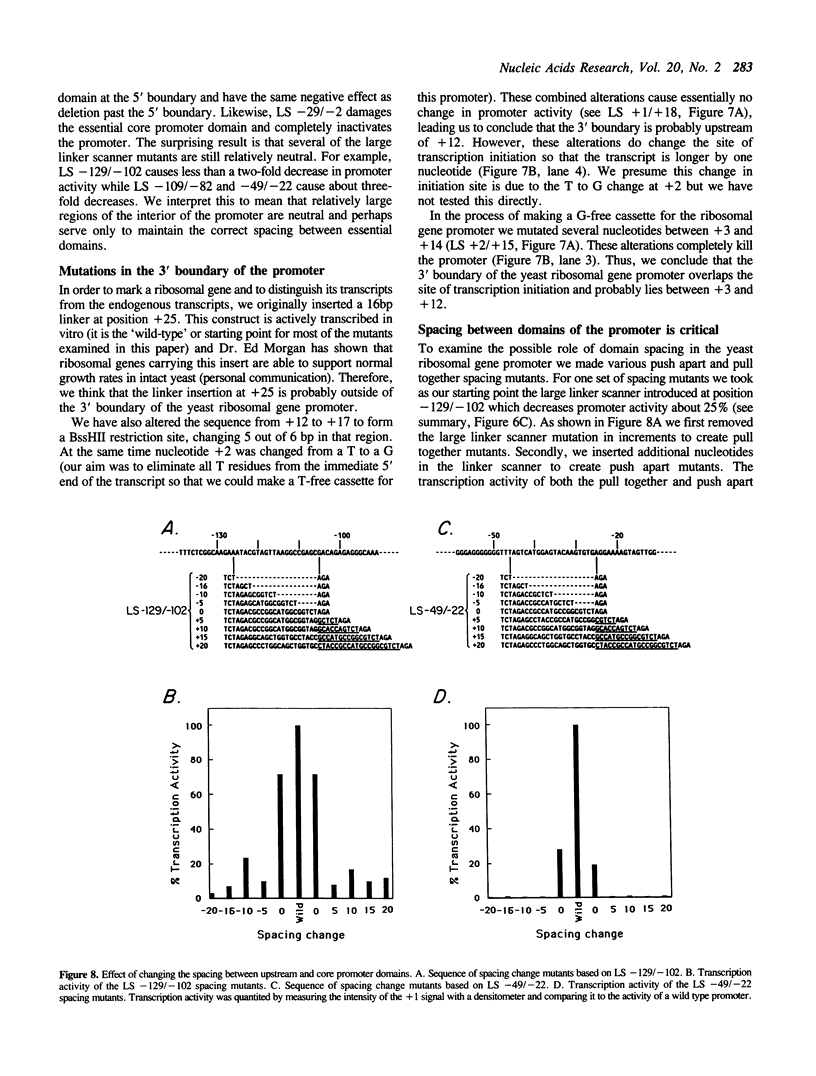
The structure of the ribosomal gene promoter from Saccharomyces cerevisiae has been analyzed in a whole cell in vitro extract. The promoter contains at least two essential domains, an upstream domain located at the 5' boundary near position -150 and a core promoter domain around the site of transcription initiation at +1. The upstream domain augments transcription in vitro but is not absolutely required. Maintenance of correct spacing between the two domains is critical. The in vitro analysis agrees well with prior in vivo analysis and it appears that the yeast promoter has a structure very similar to that of vertebrate ribosomal gene promoters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Cassidy B., Haglund R., Rothblum L. I. Regions upstream from the core promoter of the rat ribosomal gene are required for the formation of a stable transcription initiation complex by RNA polymerase I in vitro. Biochim Biophys Acta. 1987 Jul 14;909(2):133–144. doi: 10.1016/0167-4781(87)90035-2. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Grummt I. Nucleotide sequence requirements for specific initiation of transcription by RNA polymerase I. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6908–6911. doi: 10.1073/pnas.79.22.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiner M. M., Smale S. T., Tjian R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol. 1986 Jan;6(1):227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. L., Sollner-Webb B. The mouse ribosomal DNA promoter has more stringent requirements in vivo than in vitro. Mol Cell Biol. 1990 Sep;10(9):4970–4973. doi: 10.1128/mcb.10.9.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Learned R. M., Tjian R. Analysis of clustered point mutations in the human ribosomal RNA gene promoter by transient expression in vivo. Proc Natl Acad Sci U S A. 1988 Feb;85(3):669–673. doi: 10.1073/pnas.85.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T., Riggs D. L., Heck J. D., Planta R. J., Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991 Oct 11;19(19):5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 1990 Jul;4(7):1240–1251. doi: 10.1101/gad.4.7.1240. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A. F., van Heerikhuizen H., Planta R. J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape L. K., Windle J. J., Sollner-Webb B. Half helical turn spacing changes convert a frog into a mouse rDNA promoter: a distant upstream domain determines the helix face of the initiation site. Genes Dev. 1990 Jan;4(1):52–62. doi: 10.1101/gad.4.1.52. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Pennock D., McStay B., Roan J., Tolentino E., Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987 Sep 25;15(18):7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
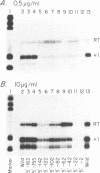
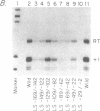
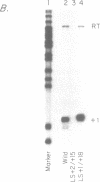
- Riggs D. L., Nomura M. Specific transcription of Saccharomyces cerevisiae 35 S rDNA by RNA polymerase I in vitro. J Biol Chem. 1990 May 5;265(13):7596–7603. [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle J., Sollner-Webb B. Upstream domains of the Xenopus laevis rDNA promoter are revealed in microinjected oocytes. Mol Cell Biol. 1986 Apr;6(4):1228–1234. doi: 10.1128/mcb.6.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto O., Takakusa N., Mishima Y., Kominami R., Muramatsu M. Determination of the promoter region of mouse ribosomal RNA gene by an in vitro transcription system. Proc Natl Acad Sci U S A. 1984 Jan;81(2):299–303. doi: 10.1073/pnas.81.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]