SUMMARY
SETTING
Although considerable effort has been put into the development and evaluation of new diagnostics for tuberculosis (TB) and multidrug-resistant TB (MDR-TB), little attention has thus far been paid to the technical aspects of initiating quality-assured routine service use. For implementation of the microscopic-observation drug susceptibility (MODS) methodology in the Peruvian reference laboratory network, a laboratory accreditation process was devised; MODS results from an expert reference laboratory (Universidad Peruana Cayetano Heredia [UPCH]) were used as the standard against which implementing laboratory MODS results were judged to ensure that, prior to use for patient care, implementing laboratories achieved the same high performance with MODS as previously demonstrated in the research laboratory.
OBJECTIVE
To evaluate the validity of MODS-based accreditation and the concordance of MODS drug susceptibility testing (DST) with molecular testing.
DESIGN
Head-to-head comparison of MODS DST results from implementing Peruvian regional reference laboratories and the accrediting expert MODS laboratory (UPCH) with GenoType® MTBDRplus DST.
RESULTS
The concordance of phenotypic MODS rifampicin (RMP) DST with GenoType MTBDRplus was respectively 97.4%, 97.9% and 97.1% for the two implementing regional laboratories and UPCH, and respectively 94.7%, 95.7% and 94.6% for isoniazid (INH) DST.
CONCLUSION
High and consistent levels of MODS/MTBDRplus concordance for INH and RMP DST confirm the validity of the use of rapid methods as reference standards for accreditation.
Keywords: tuberculosis, MODS, MTBDRplus, laboratory accreditation
IN THE PAST DECADE, there has been an explosion of interest in the development and evaluation of new diagnostic tools for the diagnosis of tuberculosis (TB) and the rapid identification of multidrug-resistant TB (MDR-TB).1 Some of these are now reaching the implementation stage and, although guidelines are available to help policy makers navigate setting appropriate test selection, resource implications, procurement and advocacy,2 little attention has thus far been paid to the technical aspects of initiating routine service use and quality assurance for these new tests.
The microscopic-observation drug susceptibility (MODS) assay is a low-cost, non-proprietary phenotypic test for rapid TB and MDR-TB diagnosis which is being implemented in the National TB Control Programme and Regional TB Laboratory Network of Peru. MODS utilises microscopic observation of liquid cultures for rapid detection of TB growth and direct drug susceptibility testing (DST) for isoniazid (INH) and rifampicin (RMP) (standard operating procedures available at www.modsperu.org).
In the absence of an internationally accepted procedure for the accreditation of TB laboratories wishing to perform MODS, an accreditation process was developed by a team of experts in quality assurance and TB microbiology from the National TB Reference Laboratory (NRL) at the Instituto Nacional de Salud, Ministry of Health and the Laboratorio de Investigación de Enfermedades Infecciosas at the Universidad Peruana Cayetano Heredia (UPCH), where the MODS assay was developed. The purpose of this accreditation process was to ensure that, prior to use for patient care, implementing laboratories achieved the same high performance with MODS that was previously demonstrated in the research laboratory where it was developed.3 The focus of the accreditation process is thus on sensitivity of culture and DST performance.
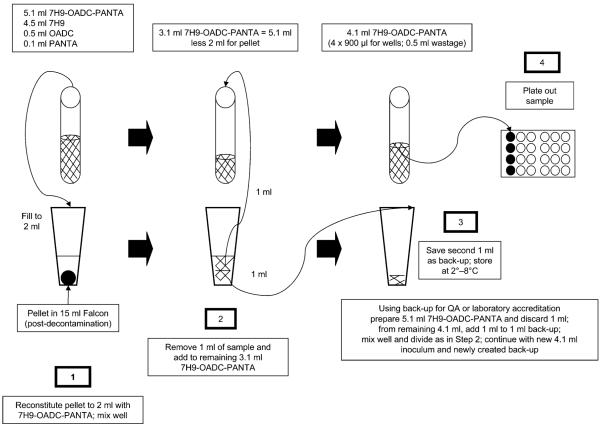
In brief, to be accredited for MODS use, implementing laboratories were required to perform MODS on 120 samples from TB suspects presenting to government health centres for diagnosis, of which at least 100 had to be positive for acid-fast bacilli on Ziehl-Neelsen sputum smear microscopy. An aliquot of each decontaminated sputum sample created during the standard MODS procedure (Figure 1) was stored at 4°C. Lots of these aliquots were then transferred to the UPCH research laboratory, where blinded retesting by MODS was performed. Results of culture (positive/negative/contaminated/indeterminate) and DST (susceptible/resistant) were sent by each laboratory independently to the NRL, where data were collated and analysed. Accreditation was granted if the results from the implementing laboratory agreed with the reference laboratory, based on predetermined standards (a description of this process and these data will be reported separately).
Figure 1.
MODS procedure and generation of back-up aliquot. OADC = oleic acid-albumin-dextrose-catalase; PANTA = polymyxin B, amphotericin B, nalidixic acid, trimethoprim, azlocillin; QA = quality assurance; MODS = microscopic-observation drug susceptibility.
Using MODS results obtained by the expert reference laboratory as the standard against which MODS performance in the implementing laboratory is judged has the advantage that the accreditation process can be completed relatively swiftly, without the several months of delay inherent in the use of conventional indirect DST. However, there was some concern that a second test modality for evaluating implementing laboratory performance might be more appropriate.
In 2008, the World Health Organization (WHO) endorsed the use of line-probe assays for TB DST4 based on evidence of their reliability, sensitivity and specificity in high-,5,6 middle-7 and lower-income8 settings. Where available, such molecular tools might be suitable alternatives for use in the MODS accreditation process, similarly circumventing the long turnaround times inherent in conventional DST.
To investigate the validity of the novel MODS-based accreditation process for evaluating the performance achieved by the regional laboratories i mplementing MODS, a second round of blinded retesting of all isolates was undertaken using a WHO approved molecular method (GenoType® MTB-DRplus line-probe assay, Hain Lifescience GmbH, Nehren, Germany) in an unrelated national TB reference laboratory. This furthermore permitted concurrent evaluation of the use of the MTBDRplus assay as a potential alternative method for rapid accreditation of MODS-implementing laboratories.
METHODS
Setting
Two urban regional TB reference laboratories in Lima Sur and Callao, Peru, serving populations of respectively 1 930 000 and 864 000, participated in the MODS accreditation process during March 2008–January 2009. Both succeeded in obtaining formal accreditation by the Peruvian NRL of the Instituto Nacional de Salud based on the results of their accreditation process.
External testing strategy
All UPCH mycobacterial strains produced from samples submitted by the implementing laboratories for the initial accreditation process were stored in 1000 μl aliquots in Middlebrook 7H9 with 10% glycerol at −70°C. Prior to transportation to the UK Health Protection Agency (HPA) National Mycobacteria Reference Laboratory (NMRL) for line-probe assay testing, all samples were inactivated as follows: in a Class II biological safety cabinet the frozen aliquots of mycobacterial strains in Middlebrook 7H9 with 10% glycerol were thawed and 500 μl were each transferred into a sterile screw-cap microcentrifuge tube. After centrifugation at approximately 10 000 g for 15 min, the supernatant was discarded and the pelleted bacteria were resuspended in 300 μl of polymerase chain reaction (PCR) water (Sigma, St Louis, MO, USA) by vortexing and then heat-inactivated by incubation in a heater block at 95°C for 20 min. To ensure all cultures were inactivated, mycobacterial DNA extraction in the UK HPA laboratory was performed by further incubation for 20 min at 95°C in a covered boiling water bath, followed by incubation at room temperature for 15 min in an ultrasonic bath. Samples were centrifuged at 13 000 g for 5 min, and 5 μl (20–100 ng DNA) of the supernatant was used directly per 50 μl PCR reaction and tested for mutations associated with INH and RMP resistance by GenoType MTBDRplus. The staff performing the GenoType MTBDRplus assay were blinded to phenotypic DST results from Peru.
Data analysis
Three INH and RMP susceptibility determinations were available for each strain—phenotypic MODS results from 1) the implementing regional reference laboratory, 2) the UPCH research laboratory and 3) genotypic results from the UK HPA NMRL. Concordance of MODS testing by each laboratory with GenoType MTBDRplus testing was assessed for each drug using McNemar’s χ2 test for discordant pairs, percentage agreement and kappa (κ) values to determine degree of agreement beyond chance.
Ethics review
Ethical review was not requested for this study which used anonymised, unlinked strains of M. tuberculosis from our laboratory strain bank.
RESULTS
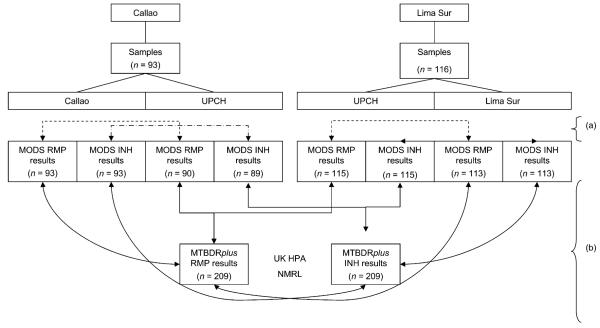
Of the 209 strains of M. tuberculosis sent for MTB-DRplus testing, 116 came from Lima Sur and 93 from Callao; 12 of the 836 INH or RMP MODS results (1.4%) were not available due to culture contamination (Figure 2).
Figure 2.
Sample and strain testing flow chart: (a) denotes accreditation process analyses (not reported here); (b) denotes evaluations reported here: 93 culture-positive samples from Lima Sur were split and processed in MODS at both Lima Sur and UPCH, and 116 culture-positive samples from Callao were split and processed in MODS at both Callao and UPCH. The comparison of these parallel RMP and INH MODS DST results was the basis for the accreditation process (reported elsewhere and indicated by (a) in the figure); the strains derived from the positive MODS cultures at UPCH were sent to the UK for MTBDRplus testing. The analysis reported here is the comparison of this molecular testing with the results obtained by the implementing laboratory (Lima Sur or Callao) and UPCH by direct MODS DST of the parent sample. Of the 836 MODS results, 12 (1.4%) were unavailable due to contamination of the well—3 RMP and 3 INH from Lima Sur, and 4 RMP and 5 INH from UPCH. UPCH = Universidad Peruana Cayetano Heredia; MODS = microscopic-observation drug susceptibility; RMP = rifampicin; INH = isoniazid; UK HPA NMRL = United Kingdom Health Protection Agency National Mycobacteria Reference Laboratory; DST = drug susceptibility testing.
Rifampicin DST
GenoType MTBDRplus identified RMP resistance mutations in 18.2% of the isolates. The concordance of phenotypic MODS RMP DST with GenoType MTBDRplus was 97.4%, 97.9% and 97.1% for the two implementing regional laboratories and UPCH, respectively (Table).
Table.
Rifampicin and isoniazid phenotypic and genotypic DST concordance*
| MODS |
||||||
|---|---|---|---|---|---|---|
| Lima Sur |
Callao |
UPCH |
||||
| Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | |
| Rifampicin | ||||||
| MTBDRplus | ||||||
| Susceptible | 98 | 1 | 69 | 1 | 165 | 2 |
| Resistant | 2 | 12 | 1 | 22 | 4 | 34 |
| Percentage agreement | 97.35 | 97.85 | 97.07 | |||
| kappa† | 0.874 | 0.942 | 0.901 | |||
| P value (McNemar’s χ2)‡ | 0.56 | 1.00 | 0.41 | |||
| P value (Exact McNemar)‡ | 1.00 | 1.00 | 0.69 | |||
| Isoniazid | ||||||
| MTBDRplus | ||||||
| Susceptible | 93 | 2 | 64 | 1 | 153 | 3 |
| Resistant | 4 | 14 | 3 | 25 | 8 | 40 |
| Percentage agreement | 94.69 | 95.70 | 94.61 | |||
| kappa† | 0.792 | 0.896 | 0.845 | |||
| P value (McNemar’s χ2)‡ | 0.41 | 0.32 | 0.13 | |||
| P value (Exact McNemar)‡ | 0.69 | 0.63 | 0.23 | |||
4 missing values for rifampicin UPCH analyses due to contamination of drug-containing wells in UPCH MODS assay.
Kappa value interpretation: 0.61–0.80 = substantial agreement, 0.81–1.0 = almost perfect agreement.9
P values relate to the null hypothesis that there is no difference between MODS and MTBDRplus.
5 missing values for isoniazid UPCH analyses due to contamination of drug-containing wells in UPCH MODS assay.
DST = drug susceptibility testing; MODS = microscopic-observation drug susceptibility; UPCH = Universidad Peruana Cayetano Heredia.
Isoniazid DST
GenoType MTBDRplus identified INH resistance mutations in 23.0% of the isolates: 79% of these had mutations only in katG, 17% had mutations only in inhA, and 4% had mutations in both genes. The concordance of phenotypic MODS INH DST with GenoType MTBDRplus was 94.7%, 95.7% and 94.6% for the two implementing regional laboratories and UPCH, respectively (Table).
Discrepant analysis
For seven patients with strains that had been defined as phenotypically susceptible in both MODS assays (regional reference laboratory and UPCH) and an eighth defined as susceptible in MODS at UPCH (no regional reference laboratory result due to contamination), GenoType MTBDRplus identified strain INH resistance due to a mutation in the inhA gene only. All 39 strains expressing a mutation in katG (associated with high-level INH resistance) were identified as phenotypically resistant in both MODS assays.
DISCUSSION
This is the first report comparing the results of direct phenotypic DST in the MODS assay with indirect genotypic DST by the GenoType MTBDRplus test. The two principal findings of this investigation are 1) very high levels of concordance between the two methodologies for INH and RMP DST (over 94.5% and 97.0%, respectively) and 2) that this concordance was consistently achieved across both research and routine clinical reference laboratories.
These findings have three immediate implications. First, they extend the evidence base for the reliability of the MODS assay in rapid detection of drug resistance3,10–16 by indicating excellent agreement with a WHO-endorsed DST methodology.4 Second, they demonstrate the highly accurate performance that can be achieved in routine clinical service laboratories starting to implement MODS. Finally they validate the accreditation procedure put in place by the NRL, in which parallel MODS testing performed by an expert laboratory is utilised as the reference methodology.
The median turnaround time for conventional solid medium-based DST can run to 5 months in overloaded reference laboratories.17 The use of conventional methods as the reference standard for the accreditation of laboratories implementing rapid diagnostics creates lengthy delays and could extend the duration of the accreditation process beyond half a year. The data presented here indicate that an accreditation procedure utilising parallel rapid testing in an expert laboratory is feasible and reliable. In Peru, the MODS assay was used; however, these data also demonstrate that rapid molecular tests such as the MTBDRplus line-probe assay could also be considered; batching strains is logistically helpful and all strains can be run within a day or two after completion of the final MODS culture in the implementing laboratory, saving a week over comparator MODS testing and a minimum of 3 months (and more often up to 5 months) over conventional phenotypic DST.
The central theme underlying this work is the importance of establishing a pragmatic, efficient and effective MODS quality assurance system. Laboratory accreditation is the first step in this quality assurance (QA) process and is a critical prerequisite to ensuring that service laboratory performance meets expected standards before any new tests are used for patient care; here the proposed accreditation process for laboratories implementing MODS proved reliable. However, regardless of the reference standard employed, accreditation in no way obviates the need for ongoing QA for tests employed in clinical practice. The best approaches to monitoring the quality of rapid TB culture and DST have yet to be determined. At present, laboratories are developing strategies tailored to their needs and resources. The MODS QA programme (available for download at www.modsperu.org) is used to monitor performance in sample collection and delivery, sample decontamination, culture sensitivity and specificity, direct DST (RMP and INH) and result delivery; it should be an integral element of implementation of MODS in all laboratories that have achieved accreditation.
Acknowledgements
The authors thank P Navarro and W Solano for valuable assistance in the UPCH laboratory; L Asencios and G Obregon for important contributions to the design of the accreditation plan; A Valencia and N Trejos for crucial advocacy and logistical help; W Loayza and L Garay for institutional support in the regional laboratories of Callao and Lima Sur; S Lopez, E Sanchez, M Huayta, R Salazar and C Solis for logistical field support. DAJM is supported by The Wellcome Trust (078067/Z/05/Z) and is also grateful for support from the National Institute for Health Research Biomedical Research Centre funding scheme and mentor support from B Gilman and J Friedland.
References
- 1.Stop TB Partnership Retooling Task Force and New Diagnostic Working Group . New laboratory diagnostic tools for tuberculosis control. World Health Organization; Geneva, Switzerland: [Accessed August 2010]. 2008. http://apps.who.int/tdr/svc/publications/non-tdr-publications/diagnostic-tool-tb. [Google Scholar]
- 2.Stop TB Partnership Retooling Task Force . New technologies for tuberculosis control: a framework for their adoption, introduction and implementation. World Health Organization; Geneva, Switzerland: 2007. WHO/HTM/STB/2007.40. [Google Scholar]
- 3.Moore DA, Evans CA, Gilman RH, et al. Microscopic observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355:1539–1550. doi: 10.1056/NEJMoa055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strategic and Technical Advisory Group for Tuberculosis . Report on conclusions and recommendations of 7th meeting of Strategic and Technical Advisory Group for Tuberculosis (STAG-TB) World Health Organization; Geneva, Switzerland: 2007. [Google Scholar]
- 5.Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007;45:2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sam IC, Drobniewski F, More P, Kemp M, Brown T. Mycobacterium tuberculosis and rifampin resistance, United Kingdom. Emerg Infect Dis. 2006;12:752–759. doi: 10.3201/eid1205.041339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolayevskyy V, Balabanova Y, Simak T, Malomanova N, Fedorin I, Drobniewski F. Performance of the GenoType (R) MTBDRplus assay in the diagnosis of tuberculosis and drug resistance in Samara, Russian Federation. BMC Clin Pathol. 2009;9:2. doi: 10.1186/1472-6890-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard M, Albert H, Coetzee G, O’Brien R, Bosman ME. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am J Respir Crit Care Med. 2008;177:787–792. doi: 10.1164/rccm.200709-1436OC. [DOI] [PubMed] [Google Scholar]
- 9.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 10.Caviedes L, Lee TS, Gilman RH, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J Clin Microbiol. 2000;38:1203–1208. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore DA, Mendoza D, Gilman RH, et al. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J Clin Microbiol. 2004;42:4432–4437. doi: 10.1128/JCM.42.10.4432-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mengatto L, Chiani Y, Imaz MS. Evaluation of rapid alternative methods for drug susceptibility testing in clinical isolates of Mycobacterium tuberculosis. Mem Inst Oswaldo Cruz. 2006;101:535–542. doi: 10.1590/s0074-02762006000500009. [DOI] [PubMed] [Google Scholar]
- 13.Shiferaw G, Woldeamanuel Y, Gebeyehu M, Girmachew F, Demessie D, Lemma E. Evaluation of microscopic observation drug susceptibility assay for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2007;45:1093–1097. doi: 10.1128/JCM.01949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ejigu GS, Woldeamanuel Y, Shah NS, Gebyehu M, Selassie A, Lemma E. Microscopic-observation drug susceptibility assay provides rapid and reliable identification of MDR-TB. Int J Tuberc Lung Dis. 2008;12:332–337. [PubMed] [Google Scholar]
- 15.Bwanga F, Hoffner S, Haile M, Joloba ML. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect Dis. 2009;9:67. doi: 10.1186/1471-2334-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devasia RA, Blackman A, May C, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis: an assessment of MGIT 960, MODS and nitrate reductase assay and fluoroquinolone cross-resistance. J Antimicrob Chemother. 2009;63:1173–1178. doi: 10.1093/jac/dkp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagui M, Perales MT, Asencios L, et al. Timely diagnosis of MDR-TB under program conditions: is rapid drug susceptibility testing sufficient? Int J Tuberc Lung Dis. 2006;10:838–843. [PMC free article] [PubMed] [Google Scholar]