Abstract
Introduction:
Genome-wide association studies have linked single-nucleotide polymorphisms (SNPs) in the CHRNA5/A3/B4 gene cluster with heaviness of smoking. The nicotine metabolite ratio (NMR), a measure of the rate of nicotine metabolism, is associated with the number of cigarettes per day (CPD) and likelihood of cessation. We tested the potential interacting effects of these two risk factors on CPD.
Methods:
Pretreatment data from three prior clinical trials were pooled for analysis. One thousand and thirty treatment seekers of European ancestry with genotype data for the CHRNA5/A3/B4 SNPs rs578776 and rs1051730 and complete data for NMR and CPD at pretreatment were included. Data for the third SNP, rs16969968, were available for 677 individuals. Linear regression models estimated the main and interacting effects of genotype and NMR on CPD.
Results:
We confirmed independent associations between the NMR and CPD as well as between the SNPs rs16969968 and rs1051730 and CPD. We did not detect a significant interaction between NMR and any of the SNPs examined.
Conclusions:
This study demonstrates the additive and independent association of the NMR and SNPs in the CHRNA5/A3/B4 gene cluster with smoking rate in treatment-seeking smokers.
Introduction
Heaviness of smoking is a heritable trait, with heritability estimates of approximately 50%–86% (Broms, Silventoinen, Madden, Heath, & Kaprio, 2006; Koopmans, Slutske, Heath, Neale, & Boomsma, 1999; Vink et al., 2004). Genome-wide association studies and candidate gene studies have found an association between heaviness of smoking and single-nucleotide polymorphisms (SNPs) found on chromosome region 15q25 in the α5-α3-β4 nicotinic receptor gene cluster (N. L. Saccone et al., 2009, 2010; Thorgeirsson et al., 2010). SNPs in this region are in strong linkage disequilibrium (LD), and Saccone et al. recently demonstrated at least two statistically distinct loci (marked by CHRNA5 rs16969968 and CHRNA3 rs578776, respectively) associated with smoking quantity (Saccone et al., 2010).
Another factor that has been shown to be associated with cigarettes per day (CPD) is the rate of nicotine metabolism as measured by the nicotine metabolite ratio (NMR; Benowitz, Pomerleau, Pomerleau, & Jacob, 2003; Johnstone et al., 2006). The NMR is the ratio of 3′-hydroxycotinine to cotinine and is a measure of the rate of nicotine clearance (Dempsey et al., 2004). Nicotine is metabolized to cotinine primarily by the liver enzyme CYP2A6. Cotinine is in turn metabolized to 3′-hydroxycotinine exclusively or nearly exclusively by the same enzyme. As a ratio of metabolite to parent drug, the NMR is a measure of enzymatic activity through that pathway (i.e., CYP2A6 activity). The activity of CYP2A6 is determined in part by genetics. The CYP2A6 gene is polymorphic, and various alleles are associated with reduced activity; however, known CYP2A6 genotypes account for only a modest proportion of the heritability of nicotine clearance rates (Swan et al., 2005). The NMR is significantly associated with CYP2A6 genotype (Malaiyandi et al., 2006) and has the additional advantage of reflecting environmental influences on CYP2A6 activity (Benowitz, Hukkanen, & Jacob, 2009). Therefore, the NMR may better reflect individual variation in nicotine metabolism as it affects smoking behavior (Ray, Tyndale, & Lerman, 2009).
Although both 15q25 genetic variants and NMR have been associated independently with CPD, to our knowledge, no studies have examined them in combination. Classic pharmacogenetic studies have shown that both variation in drug metabolism and drug targets are important to consider (Evans & Johnson, 2001). In this study, we examined potential interactions between the NMR and three of the SNPs in the CHRNA5-CHRNA3-CHRNB4 gene region that have been previously associated with CPD: CHRNA5 rs16969968, CHRNA5 rs578776, and CHRNA3 rs1051730 (Saccone et al., 2010; TAG Consortium, 2010; Thorgeirsson et al., 2010). Although rs16969968 and rs1051730 are in strong LD and may mark the same genetic locus, some studies have reported rs1051730 as a stronger signal (TAG Consortium, 2010; Thorgeirsson et al., 2010); we therefore decided to include both in our analysis.
While the precise functional consequences of SNPs in this region are unknown, the nonsynonymous SNP rs16969968 codes an amino acid change (aspartate to asparagine) at codon 398 of the CHRNA5 gene. This amino acid is highly conserved across species, and the presence of the A allele reduces the maximal receptor response to an agonist in vitro (Bierut et al., 2008), while the G allele of rs578776 has been associated with reduced mRNA levels in vivo (Wang et al., 2009); these are the same alleles associated with higher CPD in smokers. A recent paper by Jackson et al. (2010) showed that mice lacking the α5 receptor subunit were less sensitive to the initial effects of nicotine but continued to find nicotine rewarding at higher doses in a conditioned place-preference test. Taken together, these data suggest that the α5 subunit may be mediating receptor sensitivity and that a loss of function of the α5 subunit may decrease sensitivity to the adverse effects of nicotine at high doses. Thus, it is possible that smokers with the CHRNA5-A3 polymorphisms that decrease receptor response may smoke more because they experience fewer adverse effects. Moreover, these SNP effects on CPD may be more pronounced among faster metabolizers who exhibit more rapid nicotine clearance (i.e., a SNP by NMR interaction). Specifically, we expected that the faster metabolizers who also have the putative risk allele for the CHRNA5-A3 SNPs would smoke more CPD as compared to the other groups. Because our data were generated from three studies with different treatments and study designs, we examined the association of NMR and SNP genotype on pretreatment smoking rate only.
Methods
The study sample was composed of participants from three smoking cessation clinical trials: (a) open-label trial of nicotine patch versus nicotine nasal spray (n = 285; Lerman et al., 2004), (b) placebo-controlled trial of bupropion (n = 317; Collins et al., 2004), and (c) placebo-controlled trial of extended duration nicotine patch versus standard duration (n = 428; Schnoll et al., 2010). Analysis of pretreatment data was carried out on participants of European ancestry who had genotypes for the SNPs rs578776 and rs1051730 and complete baseline data for NMR and CPD. Within this subsample, 13 individuals had participated in more than one of the studies and were excluded, leaving a final sample of 1,030. Genotypes on the third SNP, rs16969968, were not collected in the bupropion trial, and 36 participants in the other two studies had missing data for the SNP, leaving 677 participants available for analyses involving rs16969968.
In the overall sample, 47.6% of participants were female, 40.9% were college graduates, and the mean age was 44.98 (SD = 11.01). At baseline, the mean CPD was 22.63 (SD = 9.32), mean nicotine dependence (FTND) score was 5.38 (SD = 2.20), and mean NMR was 0.416 (SD = 0.217).
Genotyping for SNPs rs16969968, rs578776, and rs1051730 was performed using the TaqMan SNP Genotyping Assays in a 384-well microplate format along with SNP-specific control samples (Applied Biosystems). The genotype distribution for the three SNPs was as follows: For rs16969968, 97 participants (14.3%) were of the A/A genotype, 325 (48.0%) were A/G, and 255 (37.7%) were G/G; for rs578776, 49 (4.8%) were A/A, 367 (35.6%) were A/G, and 614 (59.6%) were G/G; and for rs1051730, 152 (14.8%) were A/A, 491 (47.7%) were A/G, and 387 (37.6%) were G/G.
Pairwise LD (|D′| and r2) was determined using Haploview (Barrett, Fry, Maller, & Daly, 2005) from the subset of 677 participants who had data for all three SNPs. For rs16969968 and rs578776, |D′| = 1.0 and r2 = .182; for rs578776 and rs1051730, |D′| = 0.99 and r2 = .182; and for rs16969968 and rs1051730, |D′| = 0.98 and r2 = .957 (Supplementary Figure S1), which are consistent with previous studies (Bierut et al., 2008; N. L. Saccone et al., 2009). Genotype frequencies were in Hardy–Weinberg equilibrium.
NMR data were determined by liquid chromatography—tandem mass spectrometry (Dempsey et al., 2004) using blood samples collected at the pretreatment visit in each of the three studies. Participants were not given specific instructions regarding smoking behavior prior to collection; however, all participants reported smoking at least 10 CPD. The range of NMR values in this dataset was 0.01–2.08, consistent with previous studies (Johnstone et al., 2006; Swan et al., 2009). Previous studies at our center observed significant differences in smoking cessation between smokers in the lowest quartile of NMR and smokers in all other quartiles (Lerman et al., 2010; Schnoll et al., 2009). Based on those studies, we defined slow metabolizers as those in the lowest quartile (NMR < 0.27) and normal metabolizers as those in the top three quartiles (NMR > 0.27).
Chi-square tests and one-way ANOVAs were used to check for differences by genotype group on sex, education, and age. These variables did not differ by genotype on any of the three SNPs (p values > .05). Three linear regression models of CPD were then estimated. In each model, the predictors were sex, age, education (college graduate vs. other), NMR, and a set of two indicator variables denoting genotype on one of the three SNPs of interest. For each SNP, the reference category was the genotype with the lowest projected risk (i.e., associated with lower CPD) based on previous association studies (S. F. Saccone et al., 2007; Stevens et al., 2008). For rs16969968 and rs1051730, the indicator variables denoted genotypes of A/G and A/A, with G/G the reference category; for rs578776, the indicator variables denoted genotypes of A/G and G/G, with A/A the reference category. In each model, a set of two indicator variables representing the potential interaction of NMR by genotype was also included. All predictors were entered as a block after which the interaction terms were allowed to drop out if nonsignificant (p > .05). To obtain effect sizes for NMR within genotype, regressions of CPD on sex, age, education, and NMR were performed separately within each of the three genotype groups for each of the three SNPs.
Due to the high LD across SNPs, we used latent-class haplotypes to model CPD using the haplo.stats package for the statistical program R (Sinnwell & Schaid, 2009). Models included haplotype class and NMR as predictors of interest and sex, age, and education as covariates to control for error. Haplotype classes were restricted to those with ≥5% frequency, lumping the remainder into “other.” We used the likelihood ratio to test the interaction between haplotype and NMR.
Results
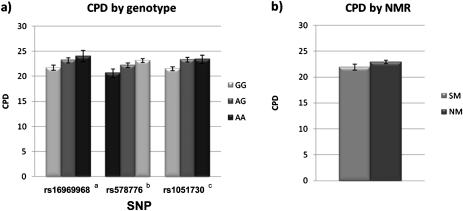
In all three linear regression models, sex, age, and education were significantly associated with CPD (p values < .01; Table 1). Both the NMR and the SNPs rs16969968 and rs1051730 were significant in the regression models of CPD, suggesting additive effects of faster nicotine metabolism and the risk alleles for these genotypes. The interaction terms were nonsignificant in all three models (p values > .40). The models of association of rs578776 and rs1051730 with CPD were repeated in the subsample of 677 participants with nonmissing data on rs16969968; the results were essentially unchanged. As shown in Figure 1a, in all cases, the putative risk allele was associated with higher smoking rates compared with the reference group. NMR accounted for an R2 change of .011 in the model including rs16969968 (.010 for GG, .026 for AG, and .006 for AA), a change of .006 in the model including rs578776 (.010 for AA, .003 for AG, and .011 for GG), and a change of .008 in the model including rs1051730 (.011 for GG, .012 for AG, and .001 for AA).
Table 1.
Linear Regression Models of Baseline Cigarettes Per Day
| Model | Variable | B | SE | β | Significance | R2 changea |
| 1 (R2 = .122) | Sex (female = 1) | −4.409 | 0.685 | −.238 | <.001 | .054 |
| Age | 0.141 | 0.032 | .164 | <.001 | .026 | |
| Education (college graduate = 1) | −2.593 | 0.681 | −.138 | <.001 | .019 | |
| NMR | 4.826 | 1.653 | .109 | .004 | .011 | |
| rs16969968 (A/G = 1) | 1.536 | 0.726 | .083 | .035 | .006 | |
| rs16969968 (A/A = 1) | 2.900 | 1.035 | .110 | .005 | .010 | |
| 2 (R2 = .104) | Sex (female = 1) | −4.419 | 0.568 | −.237 | <.001 | .053 |
| Age | 0.129 | 0.025 | .153 | <.001 | .023 | |
| Education (college graduate = 1) | −2.331 | 0.563 | −.123 | <.001 | .015 | |
| NMR | 3.562 | 1.315 | .083 | .007 | .006 | |
| rs578776 (A/G = 1) | 1.257 | 1.349 | .065 | .352 | .001 | |
| rs578776 (G/G = 1) | 2.340 | 1.317 | .123 | .076 | .003 | |
| 3 (R2 = .107) | Sex (female = 1) | −4.362 | 0.566 | −.234 | <.001 | .052 |
| Age | 0.130 | 0.025 | .153 | <.001 | .023 | |
| Education (college graduate = 1) | −2.281 | 0.562 | −.120 | <.001 | .014 | |
| NMR | 3.986 | 1.313 | .093 | .002 | .008 | |
| rs1051730 (A/G = 1) | 1.714 | 0.603 | .092 | .005 | .007 | |
| rs1051730 (A/A = 1) | 2.022 | 0.847 | .077 | .017 | .005 |
Note. NMR = nicotine metabolite ratio.
R2 change = change in model R2 when the given predictor is removed.
Figure 1.
Cigarettes per day by genotype and NMR. (a) Cigarettes per day (CPD) by genotype alone. Legend: arisk allele = A, brisk allele = G, crisk allele = A. (b) CPD by nicotine metabolite ratio (dichotomous). Legend: SM = slow metabolizers, NM = normal metabolizers.
We also examined a dichotomous division of NMR into “slow metabolizers” (NMR < 0.27) versus “normal metabolizers” (NMR > 0.27; Schnoll et al., 2009); we observed a significant association between the NMR and CPD, but the interaction terms were nonsignificant (p values > .20; Figure 1b; Supplementary Table S1). In the exploratory haplotypic analysis, the risk haplotype (rs16969968 = A, rs578776 = G, and rs1051730 = A) was significantly associated with CPD; however, there were no significant interaction effects between any haplotype and NMR (p values > .30; Supplementary Table S2).
Discussion
We hypothesized that slower nicotine metabolism, as measured by the NMR, would decrease the influence of genotypes for rs16969968, rs578776, and rs1051730 on smoking rate. We found significant independent and additive associations of NMR, rs16969968, and rs1051730 with CPD, but the interactions of the NMR with genotype on CPD were not significant. Contrary to previous studies, we did not observe a significant association of rs578776 with CPD in this sample of treatment seekers.
Although the precise functional consequences of SNPs at rs16969968, rs578776, and rs1051730 have not yet been established, all three SNPs have documented associations with heaviness of smoking in nontreatment-seeking smokers (Berrettini et al., 2008; TAG Consortium, 2010; Thorgeirsson et al., 2010). As demonstrated in prior published studies, rs16969968 has been associated with reduced receptor response to an agonist (Bierut et al., 2008) and rs578776 with reduced mRNA levels (Wang et al., 2009). It is also possible, due to the high LD across the CHRNA5-CHRNA3-CHRNB4 region, that these SNPs could serve as markers of yet undiscovered functional variants.
Contrary to our hypothesis, we did not observe an interaction between the NMR and CHRNA5-A3 SNPs on CPD. Positron emission tomography imaging has demonstrated that low levels of nicotine (i.e., smoking one cigarette after extended abstinence) can desensitize most of the available α4β2 nAChRs in a smoker's brain (Brody et al., 2006). Therefore, it is possible that the higher nicotine levels in the slower metabolizers might cause prolonged α4β2 desensitization, which in turn might minimize the importance of receptor sensitivity, explaining the lack of interaction between the NMR and CHRNA5-A3 variants. Another possible explanation may be that our study population is restricted to treatment-seeking smokers who report smoking at least 10 CPD. Because slower nicotine metabolism has been associated with smoking fewer CPD (Benowitz et al., 2003), by excluding individuals who smoke fewer than 10 CPD, our sample may not represent the full variation in CPD among slow metabolizers in the general smoking population.
In addition, differences in nicotine content between cigarette brands and individual differences in smoking topography may affect an individual's exposure to nicotine in ways that are not reflected in a measure of CPD (Pérez-Stable, Benowitz, & Marin, 1995). Slow nicotine metabolizers have been shown to take smaller cigarette puffs but not differ in the number of puffs taken compared with intermediate and normal metabolizers (Strasser, Malaiyandi, Hoffman, Tyndale, & Lerman, 2007; Strasser et al., in press). Cotinine, the primary metabolite of nicotine, has a half-life of 15–20 hr and may be a more precise measure of total nicotine exposure (Pérez-Stable et al., 1995). A recent investigation by Keskitalo et al. (2009) found an association between rs1051730 and cotinine level; the effect size of rs1051730 on cotinine level was 2.3 times greater than that on CPD in their study. An exploratory analysis of pretreatment cotinine level in our sample revealed a significant (p = .03) interaction between rs1051730 genotype and NMR (results not shown). The risk allele at rs1051730 (i.e., less-sensitive receptors) was associated with greater baseline cotinine levels in both the slow and normal metabolizers; however, this association was more pronounced among slow metabolizers [one-way analysis of variance, F(2 df) = 10.76, p = .00003] as compared with normal metabolizers [F(2 df) = 5.49, p = .004]. However,although rs1051730 is in tight LD with rs16969968 (which alters receptor function), no functional consequences have been noted with this SNP. Cotinine levels are dependent upon time from last cigarette and gradually rise throughout the day (Hukkanen, Jacob, & Benowitz, 2005), and the time of day of cotinine collection was not standardized among the participants in our sample. Additionally, since the NMR is calculated as the ratio of 3′-hydroxycotinine to cotinine, this may be a confounding factor in a statistical model that uses cotinine as an outcome. For these reasons, these cotinine results should be interpreted with caution.
In the recent meta-analysis by Saccone et al. (2010) a third genetic locus (marked by CHRNA5 rs588765) was found to be significantly associated with smoking rate after controlling for rs16969968. It is a limitation of our study that data for rs588765 were not collected for our sample. Another potential limitation is due to the high LD observed between rs16969968 and rs1051730; we might only be assessing two independent signals from the SNPs assessed in our sample. Our sample of 1,030 individuals gave us adequate power to detect an R2 change in an interactive model (SNP × NMR) of .006, which would correspond to effect sizes ranging from onefold to threefold larger than we observed. Thus, it is possible that we were unable to identify smaller interaction effects that may exist. Additionally, our sample is restricted to treatment-seeking smokers who reported smoking at least 10 CPD at pretreatment.
In conclusion, we confirm the association between rs16969968 and rs1051730 and the number of cigarettes smoked per day in a population of treatment-seeking smokers; this was also seen for NMR. The risk genotypes and fast nicotine metabolism appear to have additive effects on cigarettes smoked per day. To our knowledge, although multiple studies have established independent relationships between the NMR and CPD and between genotype at CHRNA5/CHRNA3/CHRNB4 and CPD, no other studies have looked at these two systems in combination. Understanding the interactions between pharmacodynamic and pharmacokinetic risk factors as they affect complex disorders such as nicotine addiction is important to advance our knowledge of nicotine dependence and develop better treatments for the future.
Supplementary Material
Supplementary Figure S1 and Tables S1and S2 can be found online at http://www.ntr.oxfordjournals.org
Funding
This work was supported by grants from the National Cancer Institute to CL (P50 CA143187 and U01 020830) and grants from the National Institute on Drug Abuse (DA 02277 to NB and U01 DA020830.)
Declaration of Interests
Dr. CL has served as a consultant for and/or received research support from Pfizer, AstraZeneca, Novartis, GlaxoSmithKline, and Targacept. Dr. RFT owns shares and participates in Nicogen Research Inc., a company focused on novel smoking cessation treatment approaches. No Nicogen funds were used in this work, and no other Nicogen participants reviewed the manuscript. Dr. RFT has also consulted for Novartis. Dr. NB is a paid consultant for several pharmaceutical companies that market smoking cessation medications and has been a paid expert witness against tobacco companies in litigation related to nicotine addiction. Dr. RR has received a research grant from Pfizer. This research was not supported by industry funds. The other authors declare no conflicts of interest.
Supplementary Material
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. Retrieved from http://bioinformatics.oxfordjournals.org/content/21/2/263.long. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Hukkanen J, Jacob P., III Nicotine chemistry, metabolism, kinetics and biomarkers. Handbook of Experimental Pharmacology. 2009;192:29–60. doi: 10.1007/978-3-540-69248-5_2. doi:10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Pomerleau OF, Pomerleau CS, Jacob P., III Nicotine metabolite ratio as a predictor of cigarette consumption. Nicotine & Tobacco Research. 2003;5:621–624. doi: 10.1080/1462220031000158717. doi:10.1080/1462220031000158717. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Molecular Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. doi:10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. American Journal of Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. doi:10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody AL, Mandelkern MA, London ED, Olmstead RE, Farahi J, Scheibal D, et al. Cigarette smoking saturates brain alpha 4 beta 2 nicotinic acetylcholine receptors. Archives of General Psychiatry. 2006;63:907–915. doi: 10.1001/archpsyc.63.8.907. Retrieved from http:// archpsyc.ama-assn.org/cgi/content/full/63/8/907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: A study of Finnish adult twins. Twin Research and Human Genetics. 2006;9:64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- Collins BN, Wileyto EP, Patterson F, Rukstalis M, Audrain-McGovern J, Kaufmann V, et al. Gender differences in smoking cessation in a placebo-controlled trial of bupropion with behavioral counseling. Nicotine & Tobacco Research. 2004;6:27–37. doi: 10.1080/14622200310001656830. Retrieved from http:// www.informaworld.com/smpp. [DOI] [PubMed] [Google Scholar]
- Dempsey D, Tutka P, Jacob P, III, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clinical Pharmacology and Therapeutics. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. doi:10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Evans WE, Johnson JA. Pharmacogenomics: The inherited basis for interindividual differences in drug response. Annual Review of Genomics and Human Genetics. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. Retrieved from http://www.annualreviews.org. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacological Reviews. 2005;57:79–115. doi: 10.1124/pr.57.1.3. doi:10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- Jackson KJ, Marks MJ, Vann RE, Chen X, Gamage TF, Warner JA, et al. Role of α5 nicotinic acetylcholine receptors in pharmacological and behavioral effects of nicotine in mice. Journal of Pharmacology and Experimental Therapeutics. 2010;334:137–146. doi: 10.1124/jpet.110.165738. doi:10.1124/jpet.110.165738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone E, Benowitz N, Cargill A, Jacob R, Hinks L, Day I, et al. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clinical Pharmacology and Therapeutics. 2006;80:319–330. doi: 10.1016/j.clpt.2006.06.011. doi:10.1016/j.clpt.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Broms U, Heliövaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Human Molecular Genetics. 2009;18:4007–4012. doi: 10.1093/hmg/ddp322. doi:10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans JR, Slutske WS, Heath AC, Neale MC, Boomsma DI. The genetics of smoking initiation and quantity smoked in Dutch adolescent and young adult twins. Behavior Genetics. 1999;29:383–393. doi: 10.1023/a:1021618719735. Retrieved from http://web.ebscohost.com. [DOI] [PubMed] [Google Scholar]
- Lerman C, Jepson C, Wileyto EP, Patterson F, Schnoll R, Mroziewicz M, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clinical Pharmacology and Therapeutics. 2010;87:553–557. doi: 10.1038/clpt.2010.3. doi:10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman C, Kaufmann V, Rukstalis M, Patterson F, Perkins K, Audrain-McGovern J, et al. Individualizing nicotine replacement therapy for the treatment of tobacco dependence: A randomized trial. Annals of Internal Medicine. 2004;140:426–433. doi: 10.7326/0003-4819-140-6-200403160-00009. Retrieved from http:// www.annals.org/content/140/6/426.long. [DOI] [PubMed] [Google Scholar]
- Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Molecular Psychiatry. 2006;11:400–409. doi: 10.1038/sj.mp.4001794. doi:10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- Pérez-Stable EJ, Benowitz NL, Marín G. Is serum cotinine a better measure of cigarette smoking than self-report? Preventive Medicine. 1995;24:171–179. doi: 10.1006/pmed.1995.1031. doi:10.1006/pmed.1995.1031. [DOI] [PubMed] [Google Scholar]
- Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: Role of genetic variation in nicotine-metabolizing enzymes. Journal of Neurogenetics. 2009;23:252–261. doi: 10.1080/01677060802572887. doi:10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An T, Cannon DS, Chen X, Cichon S. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genetics. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. doi:10:1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. American Journal of Medical Genetics Part B Neuropsychiatric Genetics. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. doi:10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human Molecular Genetics. 2007;16:36–49. doi: 10.1093/hmg/ddl438. doi:10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoll RA, Patterson F, Wileyto EP, Heitjan DF, Shields AE, Asch DA, et al. Effectiveness of extended-duration transdermal nicotine therapy: A randomized trial. Annals of Internal Medicine. 2010;152:144–151. doi: 10.7326/0003-4819-152-3-201002020-00005. Retrieved from http://www.annals.org/content/152/3/144.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: A validation study. Pharmacology, Biochemistry, and Behavior. 2009;92:6–11. doi: 10.1016/j.pbb.2008.10.016. doi:10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnwell JP, Schaid DJ. Statistical analysis of haplotypes with traits and covariates when linkage phase is ambiguous. Comprehensive R Archive Network; 2009. Retrieved from http://cran.r-project.org/ [Google Scholar]
- Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiology, Biomarkers. & Prevention. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. Retrieved from http:// cebp.aacrjournals.org/content/17/12/3517.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser AA, Benowitz NL, Pinto A, Tang KZ, Hecht SS, Carmella SG, et al. Nicotine metabolite ratio predicts smoking topography and carcinogen biomarker level. Cancer Epidemiology, Biomarkers & Prevention. 2011;20:234–238. doi: 10.1158/1055-9965.EPI-10-0674. doi:10.1158/1055-9965.EPI-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser AA, Malaiyandi V, Hoffman E, Tyndale RF, Lerman C. An association of CYP2A6 genotype and smoking topography. Nicotine & Tobacco Research. 2007;9:511–518. doi: 10.1080/14622200701239605. doi:10.1080/14622200701. [DOI] [PubMed] [Google Scholar]
- Swan GE, Benowitz NL, Lessov CN, Jacob P, III, Tyndale RF, Wilhelmsen K. Nicotine metabolism: The impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenetics and Genomics. 2005;15:115–125. doi: 10.1097/01213011-200502000-00007. Retrieved from http://ovidsp.tx.ovid.com/sp-3.2.4aAccession: 01213011-200502000-00007. [DOI] [PubMed] [Google Scholar]
- Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3’hydroxycotinine to cotinine in plasma and urine. Pharmacogenetics and Genomics. 2009;19(5):388–398. doi: 10.1097/FPC.0b013e32832a404f. doi:10.1097/FPC.0b013e32832a404f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAG Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42:441–447. doi: 10.1038/ng.571. doi:10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature Genetics. 2010;42:448–453. doi: 10.1038/ng.573. doi:10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Beem AL, Posthuma D, Neale MC, Willemsen G, Kendler KS, et al. Linkage analysis of smoking initiation and quantity in Dutch sibling pairs. Pharmacogenomics Journal. 2004;4:274–282. doi: 10.1038/sj.tpj.6500255. [DOI] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Human Molecular Genetics. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. doi:10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.