Abstract
Purpose
Cataracts are the most common cause of blindness worldwide. Inherited cataract is a clinically and genetically heterogeneous disease. Here we report a novel mutation in the paired-like homeodomain 3 (PITX3) gene segregating in a four generation English family with an isolated autosomal dominant posterior polar cataract.
Methods
A genome-wide linkage was performed by means of single nucleotide polymorphism (SNP) and microsatellite markers. Linkage analyses were performed with the GeneHunter and MLINK programs. Direct sequencing of PCR products was performed to detect mutation in the gene, using the BigDye version 3.1 and analyzed using Sequence analysis version 5.2.
Results
Genome-wide linkage analysis with SNP markers, identified a disease-haplotype interval on chromosome 10q. Two point positive logarithm of odds (LOD) scores was obtained with markers D10S205 (Z=3.10 at θ=0.00), flanked by markers D10S1709 and D10S543, which harbors the homeobox gene PITX3. Sequence analysis of PITX3 revealed a 1-bp deletion that cosegregated with all the affected members of this family which resulted in a frameshift in codon 181 and likely to produce an aberrant protein consisting of 127 additional residues.
Conclusions
The 542delC is a novel mutation in PITX3 causing an isolated posterior polar cataract.
Introduction
Bilateral congenital cataract is the most common cause of treatable childhood blindness. Cataracts are phenotypically and genotypically heterogeneous, most show autosomal dominant inheritance with complete penetrance [1,2]. Less frequently, autosomal recessive and X-linked inheritance patterns are seen. There has been significant progress in identifying the molecular genetic basis of human cataract. Many genes have been implicated including those encoding the transparent intracellular lens proteins (crystallins), membrane gap junction proteins (connexins), water channel proteins (aquaporins), solute carrier protein (SLC16A12) various cytoskeletal proteins (e.g., phakinin, filensin, vimentin), transmembrane proteins (transmembrane protein 114 [TMEM114], lens intrinsic membrane protein [LIM2], chromatin modifying protein-4B [CHMP-4B], and EPH receptor A2 [EPHA2]) and transcription factors [3].
Transcription factors play an important role in the embryological development of the lens including the interaction between the embryonic surface ectoderm and the budding optic vesicle. This interaction is critical for normal lens induction [4]. Mutations in several transcription factor genes notably paired box 6 (PAX6), forkhead box E3 (FOXE3), eyes absent homolog 1 (Drosophila; EYA1), and v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian; MAF), and paired-like homeodomain 3 (PITX3) have been implicated in both congenital cataract and anterior segment mesenchymal dysgenesis (ASMD) [5-10]. Here we report a novel 1-bp deletion (542delC) mutation in PITX3 in a family with an isolated autosomal dominant posterior polar cataract.
Methods
Phenotyping
The family in this study was identified through the proband attending the cataract clinic at Moorfields Eye Hospital, London, UK. The local ethics committee approval was obtained for the studies and all individuals taking part in the study gave written informed consent. Both affected and unaffected family members underwent full ophthalmic examination, with careful slit lamp examination. In this pedigree all the affected individuals were diagnosed as having isolated posterior polar cataract.
Genotyping and linkage analysis
Genomic DNA was extracted from EDTA-sequestered blood samples using the Nucleon II DNA extraction kit (Scotlab Bioscience, Strathclyde, Scotland, UK).
Genotype data of the individual family members were generated using the GeneChip Human Mapping 50K Array Xba 240 and Assay Kit from GeneChip Human Mapping 100k Set (Affymetrix, High Wycombe, UK). Initial checks of the results were performed with GeneChip Command Console Viewer (v1.1.0.845). Genotyping Console (v3.0.2) assigned individuals’ genotypes. Alohomora version 0.30 (Max Delbrück Center for Molecular Medicine, Berlin, Germany) was used to prepare the raw genotype data for linkage analysis and for PedCheck (version1.1, Jeff O’Connell; University of Pittsburgh, Pittsburgh, PA.) to detect and remove Mendelian Errors (ME) from the data. Genehunter (version 2.1_r5 beta) was used to perform the subsequent parametric linkage analysis with dominant inheritance and full penetrance, of the disease allele with a frequency of 0.0001 in the general population.
The region showing significant logarithm of odds (LOD) score was refined using markers from Marshfield, GDB Human genome database and Ensemble databases. Analysis was performed using GeneMapper (version 4.0, Applied biosystems, Warrington, UK) on a ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Warrington, UK). A full penetrance and a gene frequency of 0.0001 were used for the cataract locus. Two-point linkage analysis was performed using the MLINK component of the LINKAGE program package version 5.10. The pedigree and haplotype data was managed by Cyrillic software (version 2.1.3).
Sequence analysis
Genomic DNA from all the individuals was amplified using PCR Reddy Mix (AB gene; Thermo Scientific, Epsom, UK) and PITX3-specific primers (Table 1). Samples were processed through 30 cycles of amplification consisting of 30 s at 94 °C, 30 s at 60 °C, and 45 s at 72 °C. The final step was lengthened to 5 min. Direct sequencing of PCR products was performed using the BigDye version 3.1 (Applied Biosystems) on a ABI 3730 DNA Analyzer and analyzed using Sequence analysis version 5.2.
Table 1. Primers for PITX3.
| Set | Forward primer | Reverse primer | Annealing temp (°C) | Product size (bp) |
|---|---|---|---|---|
| Exon 1 |
ccggctgggggtggcagtacgcgg |
ggtccagcaatagctcctcggccc |
60 |
237 |
| Exon 2 |
cagctttacggctggggttgag |
ggatgaagctgttatgtcctgcac |
60 |
296 |
| Exon 3 |
gggagccagcgagtggcttaggag |
gggtggaaccgctggcctccg |
60 |
374 |
| Exon 4 | gtctctagccacctcatctc | ctggggcgggagcaagccagtc | 60 | 808 |
Results
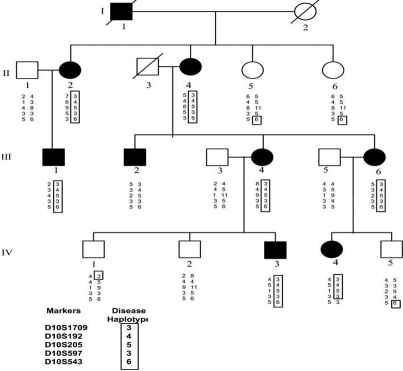
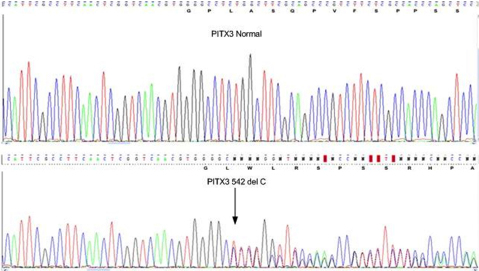
A four-generation family with posterior polar cataract, comprising 16 members of the pedigree (Figure 1) including 8 affected individuals, 5 unaffected individuals, and 3 spouses were genotyped with SNP markers using GeneChip Human Mapping 50K Array. Linkage analysis identified a likely disease-haplotype interval on chromosome 10q (rs911579-8Mb-rs2286396). Further, microsatellite markers were used to narrow down the region on chromosome 10q25. Two point positive LOD score was obtained with markers D10S205 (Z=3.10 at θ=0.00), flanked by markers D10S1709 and D10S543 (Table 2). This area encompasses the homeobox gene PITX3 between markers D10S192 and D10S205. PITX3 comprises four exons and encodes a protein of 302 amino acid residues. Sequence analysis of this gene revealed, in exon 4, a 1-bp deletion (542delC; Figure 2) that cosegregated with all the affected members of PPC family. It resulted in a frameshift in codon 181 and likely produced an aberrant protein consisting of 127 additional residues. This mutation found in PITX3 affected the region outside the homeodomain in the COOH-terminal end of the protein and result primarily in posterior polar cataract. This change was not seen in 200 healthy individuals.
Figure 1.
Abridged pedigree of the posterior polar cataract family used in this study showing the segregation of five chromosome 10q markers listed in descending order. Squares and circles symbolize males and females respectively. Open and filled symbols indicate unaffected and affected individuals. The disease haplotype is shown in the box.
Table 2. Two-Point LOD scores for linkage between the PITX3 locus and 10q25 markers.
|
|
Distance |
Z at θ= |
||||||
|---|---|---|---|---|---|---|---|---|
| Marker | cM | 0.0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 |
| D10S1709 |
4.52 |
1.80 |
1.79 |
1.71 |
1.55 |
1.14 |
0.64 |
0.20 |
| D10S192 |
1.19 |
0.52 |
0.51 |
0.45 |
0.40 |
0.30 |
0.24 |
0.14 |
| D10S205 |
3.33 |
3.10 |
3.05 |
2.83 |
2.53 |
1.90 |
1.21 |
0.52 |
| D10S597 |
0.00 |
0.82 |
0.81 |
0.74 |
0.65 |
0.47 |
0.27 |
0.09 |
| D10S543 | −4.44 | −1.52 | −0.79 | −0.47 | −0.20 | −0.10 | −0.07 | |
Figure 2.
Sequence analysis of PITX3 with normal and 1-bp deletion fragment showing a frame shift in an affected individual.
Discussion
Posterior polar cataract (PPC) is a clinically distinct opacity that is located at the back of the lens and, because of its proximity to the optical center of the eye, can have a marked effect on visual acuity. Previously, PPCs have been described in association with mutation in five genes (EPHA2 on 1p36, CRYAB on 11q22-q22.3, CHMP4B on chromosome 20p12, CRYBA1/A3 on 17q12, and PITX3 on 10q25) [11-14]. PITX3 encodes a paired-like class of homeobox transcription factor, a member of the PITX family, which also includes PITX1 and PITX2. PITX2 and PITX3 are involved in eye development and are expressed in cornea, lens, and retina [15]. Mutations in PITX2 have been linked to Rieger syndrome causing glaucoma and mild craniofacial dysmorphism in humans [16]. In the aphakia mouse mutant, two deletions in the promoter of the homeobox transcription factor Pitx3 lead to loss of its function and to arrest of eye development at the lens stalk stage [17]. Mutations in the homologus human PITX3 gene have been demonstrated to cause cataracts and anterior segment dysgenesis.
So far three different mutations in PITX3 have been reported in man. The first mutation was a COOH-terminal 17-bp insertion (657ins17) that resulted in a frame shift and abnormal configuration of nearly one third of the protein. This mutation was found in a large family with anterior segment ocular dysgenesis and cortical cataracts [9]. Several recent studies have shown a recurrence of the same 17-bp insertion mutation in number of families of different ethnic backgrounds affected with congenital posterior polar cataract that, in some cases, included anterior segment defects [10]. The second mutation was a serine to asparagine substitution in the NH2-terminal region of the protein (S13N) [9]. An additional COOH-terminal single-nucleotide deletion, 650delG was identified in two families affected with posterior polar cataract; this mutation is predicted to result in a truncation of the normal protein around the same site two amino acids upstream as the recurrent 17-bp insertion [10,18]. Homozygous mutation for 650delG has been found in two siblings from consanguineous marriage causing microphthalmia and central nervous system defects [18].
The PITX3 protein mutantions S13N and G219fs have been shown to alter the DNA-binding profiles and transactivation activities and there is a partial loss-of-function in both mutants with the G219fs form being more severely affected. The G219fs mutation was found in multiple families affected with congenital cataracts along with anterior segment malformations in many members. These findings suggested that the presence/severity of anterior segment defects in families affected with G219fs may be determined by secondary factors that are expressed in the developing anterior segment structures and may modify the effect(s) of this mutation [19]. PITX3 is expressed in the developing lens, skeletal muscle, and dopaminergic neurons of the substantia nigra in the brain. Recently PITX3 polymorphisms have been shown to be associated with Parkinson disease [20].
Here we report a novel mutation (542delC) in PITX3 causing an isolated posterior polar cataract in an English pedigree. This 1-bp deletion mutation in exon 4 of PITX3, resulted in a frameshift in codon 181 that may lead to the production of an aberrant protein consisting of 127 additional residues at the COOH-terminal region. This region is thought to be involved in complex protein–protein interactions, imparting specificity and efficiency to homeoprotein function [19]. This mutation does not affect the homeodomain region of the protein but highlights the significance of the COOH-terminal region, which already have been associated with the disease.
Acknowledgments
We would like to acknowledge funding from the Wellcome Trust project grant 063969/Z/01, EU project “PYTHIA” (FP7-ICT2–224030), and NIHR (Moorfields Eye Hospital Biomedical Research Centre). We would like to thank the members of the family for taking part in this study.
References
- 1.Ionides A, Francis P, Berry V, Mackay D, Bhattacharya SS, Shiels A, Moore AT. Clinical and genetic heterogeneity in autosomal dominant congenital cataract. Br J Ophthalmol. 1999;83:802–8. doi: 10.1136/bjo.83.7.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graw J. The genetic and molecular basis of congenital eye defects. Nat Rev Genet. 2003;4:876–88. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 3.Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–15. [PMC free article] [PubMed] [Google Scholar]
- 4.Ogino H, Yasuda K. Sequential activation of transcription factors in lens induction. Dev Growth Differ. 2000;42:437–48. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- 5.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–72. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 6.Semina EV, Brownell I, Mintz-Hittner HA, Murray JC, Jamrich M. Mutations in the human forkhead transcription factor FOXE3 associated with anterior segment ocular dysgenesis and cataracts. Hum Mol Genet. 2001;10:231–6. doi: 10.1093/hmg/10.3.231. [DOI] [PubMed] [Google Scholar]
- 7.Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9:363–6. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GC. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- 9.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 10.Berry V, Yang Z, Addison PK, Francis PJ, Ionides A, Karan G, Jiang L, Lin W, Hu J, Yang R, Moore A, Zhang K, Bhattacharya SS. Recurrent 17 bp duplication in PITX3 is primarily associated with posterior polar cataract (CPP4) J Med Genet. 2004;41:e109. doi: 10.1136/jmg.2004.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Hua R, Xiao W, Burdon KP, Bhattacharya SS, Craig JE, Shang D, Zhao X, Mackey DA, Moore AT, Luo Y, Zhang J, Zhang X. Mutations of the EPHA2 receptor tyrosine kinase gene cause autosomal dominant congenital cataract. Hum Mutat. 2009;30:E603–11. doi: 10.1002/humu.20995. [DOI] [PubMed] [Google Scholar]
- 12.Berry V, Francis P, Reddy MA, Collyer D, Vithana E, MacKay I, Dawson G, Carey AH, Moore A, Bhattacharya SS, Quinlan RA. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–5. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Ji B, Wan C, He G, Zhang J, Zhang M, Feng G, He L, Gao L. A splice site mutation in CRYBA1/A3 causing autosomal dominant posterior polar cataract in a Chinese pedigree. Mol Vis. 2010;16:154–60. [PMC free article] [PubMed] [Google Scholar]
- 15.Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 16.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–72. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 17.Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–16. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- 18.Bidinost C, Matsumoto M, Chung D, Salem N, Zhang K, Stockton DW, Khoury A, Megarbane A, Bejjani BA, Traboulsi EI. Heterozygous and homozygous mutations in PITX3 in a large Lebanese family with posterior polar cataracts and neurodevelopmental abnormalities. Invest Ophthalmol Vis Sci. 2006;47:1274–80. doi: 10.1167/iovs.05-1095. [DOI] [PubMed] [Google Scholar]
- 19.Sakazume S, Sorokina E, Iwamoto Y, Semina EV. Functional analysis of human mutations in homeodomain transcription factor PITX3. BMC Mol Biol. 2007;8:84. doi: 10.1186/1471-2199-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le W, Nguyen D, Lin XW, Rawal P, Huang M, Ding Y, Xie W, Deng H, Jankovic J. Transcription factor PITX3 gene in Parkinson’s disease. Neurobiol Aging. 2011;32:750–3. doi: 10.1016/j.neurobiolaging.2009.03.015. [DOI] [PubMed] [Google Scholar]