Abstract
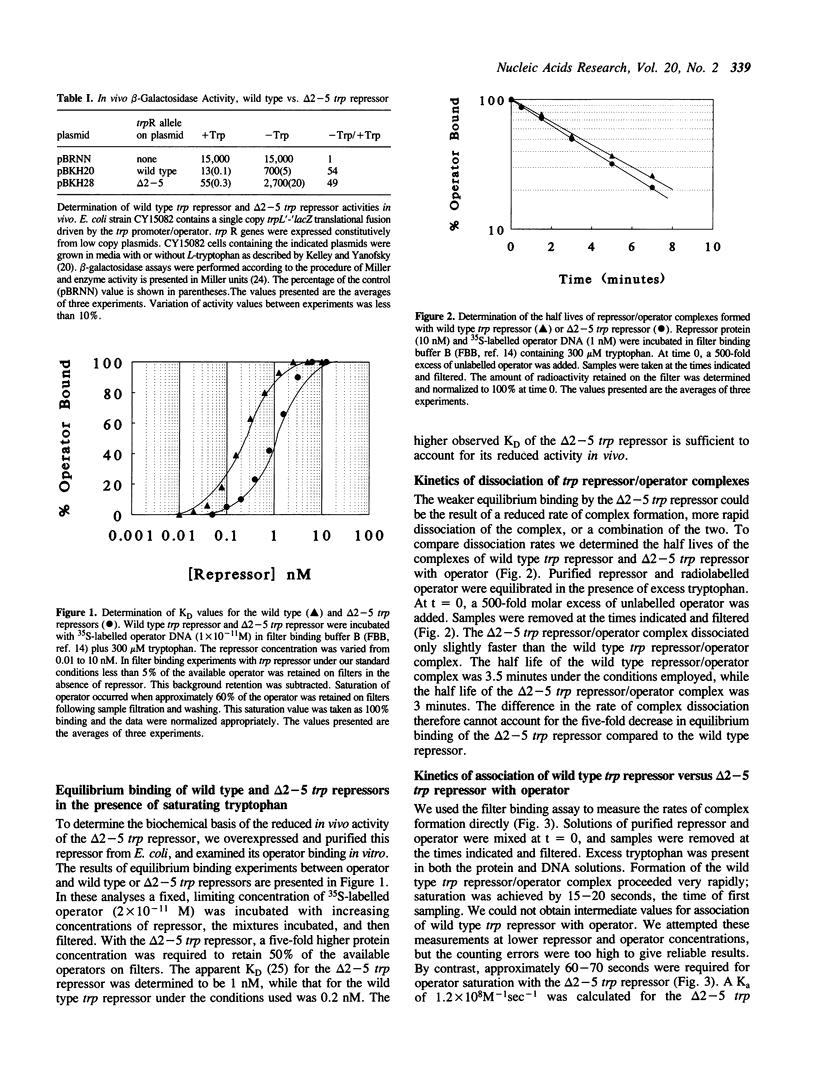
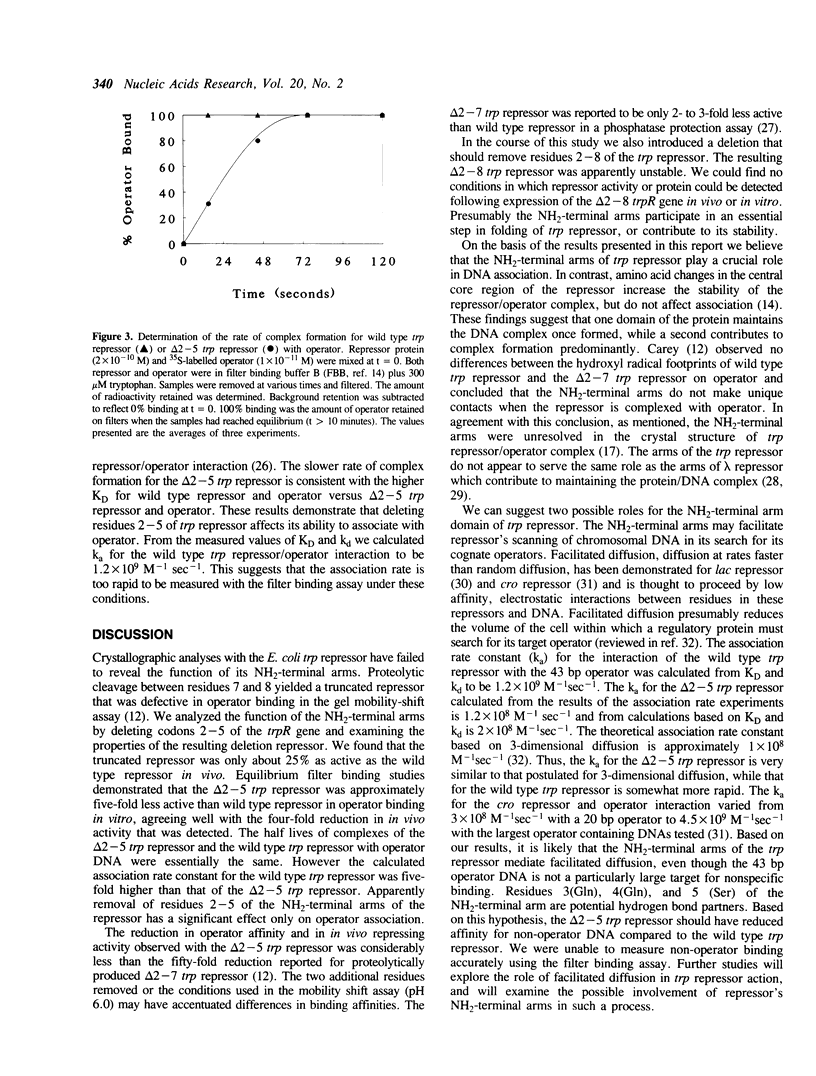
The 3-dimensional structure of the trp repressor, aporepressor, and repressor/operator complex have been described. The NH2-terminal arms of the protein, comprising approximately 12-14 residues, were not well resolved in any of these structures. Previous studies by Carey showed that the arms are required for full in vitro repressor activity. To examine the roles of the arms more fully we have removed codons 2-5 and 2-8 of the trpR gene and analyzed the resulting truncated repressors in vivo and in vitro. The delta 2-5 trp repressor was found to be approximately 25% as active as the wild type repressor in vivo. In in vitro equilibrium binding experiments, the delta 2-5 trp repressor was shown to be five-fold less active in operator binding. The rate of dissociation of the complex formed between the delta 2-5 trp repressor and operator was essentially the same as the rate of dissociation of the wild type trp repressor/operator complex. However association of the delta 2-5 trp repressor with operator was clearly defective. Since the NH2-terminal arms of the trp repressor appear to affect association predominantly they may play a role in facilitating non-specific association of repressor with DNA as repressor seeks its cognate operators. The delta 2-8 trp repressor was unstable in vivo and in vitro, suggesting that some portion of the NH2-terminal arm is required for proper folding of the remainder of the molecule.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass S., Sugiono P., Arvidson D. N., Gunsalus R. P., Youderian P. DNA specificity determinants of Escherichia coli tryptophan repressor binding. Genes Dev. 1987 Aug;1(6):565–572. doi: 10.1101/gad.1.6.565. [DOI] [PubMed] [Google Scholar]
- Bennett G. N., Yanofsky C. Sequence analysis of operator constitutive mutants of the tryptophan operon of Escherichia coli. J Mol Biol. 1978 May 15;121(2):179–192. doi: 10.1016/s0022-2836(78)80004-7. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci U S A. 1988 Feb;85(4):975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J. trp repressor arms contribute binding energy without occupying unique locations on DNA. J Biol Chem. 1989 Feb 5;264(4):1941–1945. [PubMed] [Google Scholar]
- Eliason J. L., Weiss M. A., Ptashne M. NH2-terminal arm of phage lambda repressor contributes energy and specificity to repressor binding and determines the effects of operator mutations. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2339–2343. doi: 10.1073/pnas.82.8.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Yanofsky C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7117–7121. doi: 10.1073/pnas.77.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatwole V. M., Somerville R. L. Cloning, nucleotide sequence, and characterization of mtr, the structural gene for a tryptophan-specific permease of Escherichia coli K-12. J Bacteriol. 1991 Jan;173(1):108–115. doi: 10.1128/jb.173.1.108-115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt B. K., Yanofsky C. Enhanced operator binding by trp superrepressors of Escherichia coli. J Biol Chem. 1990 May 15;265(14):7853–7858. [PubMed] [Google Scholar]
- Joachimiak A., Kelley R. L., Gunsalus R. P., Yanofsky C., Sigler P. B. Purification and characterization of trp aporepressor. Proc Natl Acad Sci U S A. 1983 Feb;80(3):668–672. doi: 10.1073/pnas.80.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Mutational studies with the trp repressor of Escherichia coli support the helix-turn-helix model of repressor recognition of operator DNA. Proc Natl Acad Sci U S A. 1985 Jan;82(2):483–487. doi: 10.1073/pnas.82.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. G., Takeda Y., Matthews B. W., Anderson W. F. Kinetic studies on Cro repressor-operator DNA interaction. J Mol Biol. 1987 Jul 5;196(1):149–158. doi: 10.1016/0022-2836(87)90517-1. [DOI] [PubMed] [Google Scholar]
- Klig L. S., Carey J., Yanofsky C. trp repressor interactions with the trp aroH and trpR operators. Comparison of repressor binding in vitro and repression in vivo. J Mol Biol. 1988 Aug 20;202(4):769–777. doi: 10.1016/0022-2836(88)90557-8. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Sigler P. B. The stereochemistry and biochemistry of the trp repressor-operator complex. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):113–126. doi: 10.1016/0167-4781(90)90047-6. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Q., Joachimiak A., Sprinzl M., Sigler P. B. The structural basis for the interaction between L-tryptophan and the Escherichia coli trp aporepressor. J Biol Chem. 1987 Apr 5;262(10):4922–4927. [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. High level production and rapid purification of the E. coli trp repressor. Nucleic Acids Res. 1986 Oct 24;14(20):7851–7860. doi: 10.1093/nar/14.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Otwinowski Z., Joachimiak A., Lawson C. L., Sigler P. B. The three-dimensional structure of trp repressor. 1985 Oct 31-Nov 6Nature. 317(6040):782–786. doi: 10.1038/317782a0. [DOI] [PubMed] [Google Scholar]
- Squires C. L., Lee F. D., Yanofsky C. Interaction of the trp repressor and RNA polymerase with the trp operon. J Mol Biol. 1975 Feb 15;92(1):93–111. doi: 10.1016/0022-2836(75)90093-5. [DOI] [PubMed] [Google Scholar]
- Staacke D., Walter B., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. How Trp repressor binds to its operator. EMBO J. 1990 Jun;9(6):1963–1967. doi: 10.1002/j.1460-2075.1990.tb08324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Gunsalus R. P., Brown K. D., Yanofsky C. Structure and regulation of aroH, the structural gene for the tryptophan-repressible 3-deoxy-D-arabino-heptulosonic acid-7-phosphate synthetase of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):47–73. doi: 10.1016/0022-2836(81)90334-x. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Berg O. G. Facilitated target location in biological systems. J Biol Chem. 1989 Jan 15;264(2):675–678. [PubMed] [Google Scholar]


