Abstract
Objective
To identify serum markers of subsequent spontaneous preterm birth (SPTB) in asymptomatic women prior to labor.
Study design
Serum proteomics was applied to sera from 80 pregnant women sampled at 24 weeks and an additional 80 pregnant women sampled at 28 weeks. Half had uncomplicated pregnancies and half SPTB.
Results
Three specific peptides arising from inter-alpha-trypsin inhibitor heavy chain 4 protein were significantly reduced in women at 24 and 28 weeks having subsequent SPTB. The most discriminating peptide had a sensitivity of 65.0% and specificity of 82.5%, OR=8.8, CI: 3.1-24.8. A combination of the 3 new biomarkers and 6 previously studied biomarkers increased sensitivity to 86.5% with a specificity of 80.6% at 28 weeks.
Conclusions
Three novel serum markers of SPTB have been identified using serum proteomics. Using a combination of these new markers with additional markers, women at risk of SPTB can be identified weeks prior to SPTB.
Keywords: Proteomics, Mass Spectrometry, Preterm Birth, Biomarker, Amino Acid Sequencing
INTRODUCTION
Spontaneous preterm birth (SPTB) is the leading cause of perinatal morbidity and mortality in the United States.(1-2) Despite the magnitude of the problem and the substantial research efforts of many investigators, completely efficacious therapies for the treatment or prevention of SPTB have yet to be developed. Indeed, the rate of SPTB has not changed in decades.(3) A major obstacle to the development of an effective treatment for preterm labor is a limited understanding of the molecular events required to initiate and maintain term and preterm labor.
Several proteins present in maternal serum or cervical secretions have been proposed as markers that may predict SPTB. We have previously evaluated a large number of potential markers in a prospectively collected cohort and shown that a screening test consisting of three serum markers (CRH, AFP, alkaline phosphatase) and two cervical secretion markers (fetal fibronectin and ferritin) provided increased sensitivity, specificity and odds ratio. (4) However, none of the current SPTB markers alone or in combination provides adequate specificity or sensitivity to be used in clinical prediction.
Recent advances in technology allow for the evaluation of a large, unbiased portion of the complement of peptides and/or proteins present in maternal serum. Serum proteomic analysis, consisting of chromatographic separation followed by mass spectrometry to identify peptides and proteins by mass, can provide an extensive inventory of peptides and/or proteins present at any given time. Previous studies have attempted to use proteomic patterns to identify patients with early ovarian, breast and prostate cancers.(7) The use of proteomic analysis to identify phenotypic molecular characteristics of women who experience SPTB or infection has been attempted in amniotic fluid,(6-9) and cervical secretions (8, 10, 11) but serum proteomic analysis has not been reported.
SPTB is well suited for a proteomic approach given likely serologic changes that precede its clinical manifestations by weeks. We hypothesize that proteomic differences exist in maternal serum several weeks prior to the onset of clinical symptoms in women destined to develop SPTB. Our aim was to use serum proteomics to differentiate women having a subsequent SPTB from those having term deliveries. Moreover, we hoped to identify all peptides that are found to be increased or decreased in the serum of women who go on to have a SPTB as compared with those who deliver at term.
MATERIALS AND METHODS
Patient Population
This study represents a nested case-control study that used samples and data that were collected during the National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network Preterm Prediction Study (12). The Preterm Prediction Study, conducted between 1992 and 1994, was a multicenter observational investigation of 2929 symptom-free women evaluated longitudinally to determine risk factors for spontaneous preterm birth. Women were enrolled in this study without regard to specific risk factors for spontaneous preterm birth. Extensive information and/or biologic specimens were collected at each of 4 study visits, beginning at approximately 22 to 24 weeks’ gestation and occurring at 2-week intervals. The overall study population and the methods used in the Preterm Prediction Study have been previously described in detail. Gestational age was based on the last menstrual period if the last menstrual period–derived gestational age was confirmed within 10 days by the earliest ultrasonographic evaluation. A spontaneous preterm birth was defined as a preterm birth < 35 weeks gestation occurring as the result of the spontaneous onset of labor or spontaneous rupture of membranes.
Serum was collected at 24 and 28 weeks gestation and pregnancy outcomes were obtained. Participating women provided voluntary, informed consent. The original study protocols as well as these secondary analyses were approved by the representative institutional review boards. For this study, serum from 40 subjects who experienced a subsequent SPTB and 40 subjects having uncomplicated pregnancies were obtained at 24 wks gestation and submitted to proteomic analysis. Additionally, serum from a separate group of 40 subjects who experienced a subsequent SPTB and 40 subjects having uncomplicated pregnancies that ultimately delivered at term after spontaneous onset of labor was obtained at a 28 wks gestation visit and was likewise analyzed. Pregnancies complicated by indicated preterm births were excluded from both cases and controls. Cases and controls were randomly selected by the MFMU Network independent statistical core to produce a representative group from among the cohort. Researchers were given two groups of subjects for evaluation but were blinded to case or control status these groups during the proteomic analysis and data evaluation.
Specimen Preparation
Blood was collected and rapidly processed to serum using a serum separator tube. Once obtained the specimen was quickly frozen and maintained at -80°C until analysis. The specimens had not undergone repeated freeze-thaw cycles.
Given our interest in the low molecular weight proteome, a protein precipitation method was selected and high molecular weight, and typically uninformative, proteins were removed by acetonitrile precipitation following an established protocol (1-2) In brief, specimens were thawed on ice and kept at 0°C until the initiation of precipitation. Two volumes of HPLC grade acetonitrile (400 μL) were added to 200 μL serum, the sample was vortexed vigorously and allowed to stand at room temperature for 30 min. Samples were centrifuged for 10 min at 13,400 × g at room temperature. An aliquot of the supernatant (~550 μL) was transferred to a microcentrifuge tube containing 300 μL HPLC grade water. The sample was vortexed and lyophilized to ~200 μL in a vacuum centrifuge (Labconco CentriVap Concentrator, Labconco Corporation, Kansas City, MO). There was complete removal of acetonitrile. Supernatant protein concentration was determined using a Bio-Rad microtiter plate protein assay performed according to manufacturer instructions (Bio-Rad, Hercules, CA). An aliquot of the same supernatant containing 4 μg of apparent total protein was transferred to a new microcentrifuge tube and lyophilized to a volume less than 20 μL. Lyophilized samples were brought to 20 μL with HPLC water and acidified by addition of 20 μL 88% formic acid and 0.50 μg of protein was loaded on the column. If not used immediately, the protein-depleted specimen was kept at -80°C and for the cLC step the specimen was placed at 4°C until introduced onto the column.
Capillary Liquid Chromatography
Capillary liquid chromatography (cLC) was interfaced with a mass spectrometer, allowing for the continuous direct delivery of fractionated, protein-depleted serum to the mass detector. Additional details of the method have been published (14).
Electrospray-Ionization, Time-of-Flight Mass Spectrometry
Effluent from the cLC was directed into a QSTAR® Pulsar i quadrupole orthogonal time-of-flight mass spectrometer through an IonSpray source (Applied Biosystems). Mass spectra were collected every second for m/z 500 to 2500 from 5 to 55 min elution. Data collection and preliminary formatting were accomplished using Analyst QS® software with BioAnalyst add-ons (Applied Biosystems). Specimens from cases and controls were analyzed together in random order.
To reduce data file size, each mass chromatogram was divided into ten 2-minute elution intervals. One reference peak, observable in all specimens, near the center of each interval, and which did not demonstrate differences in abundance between the two groups was used to align time in that elution region. Of the ten elution intervals, the first to be analyzed (and the only one reported here) was the second 2 min window, chosen because more peptides were present. Almost all biological mass spectrometers have some ability to statistically compare groups of specimens for quantitative differences. However, that software was developed for comparisons of at most a few hundred species. Several attempts were made to use software to do comparisons of candidate peaks between cases and controls across all spectra. This was attempted using software from Bioanalyst, Agilent, Mass Finder, Nonlinear, and others. All of the software experienced one or more of the following problems; artifactual peaks, artifactual peak differences, loss of >50% of data. The problems appeared to be due to the observation of 4000-5000 peaks with many have very close m/z values, partial overlap of peaks, and many multiply charged ion envelops. Consequently, the initial review of mass spectra was accomplished by overlaying 2 min summary spectra from cases and controls distinguished by color and visual review.
Evaluation of Candidate Markers
The candidate markers were not defined a priori but were identified only after visual inspection of the actual spectra generated in both cases and controls. Each candidate marker, appearing quantitatively different between groups, was further evaluated. See Figure 1 for an example of actual mass spectra representing apparent differences between cases and controls for a candidate marker. In addition, to reduce non-biological variation, a second peak was also chosen. To be considered the reference peak, the peak had to elute in the same time interval, had to be present in each specimen, had to have a mass to charge ratio very near the candidate peak, but the reference peak had to be consistently quantitatively comparable between cases and controls as determined initially by visual inspection and then by machine software extraction. This reference peak was then used to normalize the candidate peak of interest, correcting for variability in specimen processing, specimen loading, ionization efficiency and instrument performance and allowing comparison across runs performed on different days. Thereafter, the candidate markers were ‘extracted’ by the Analyst® software to determine a quantitative peak height of both candidate and reference peaks in each specimen for all subjects.
Figure 1.
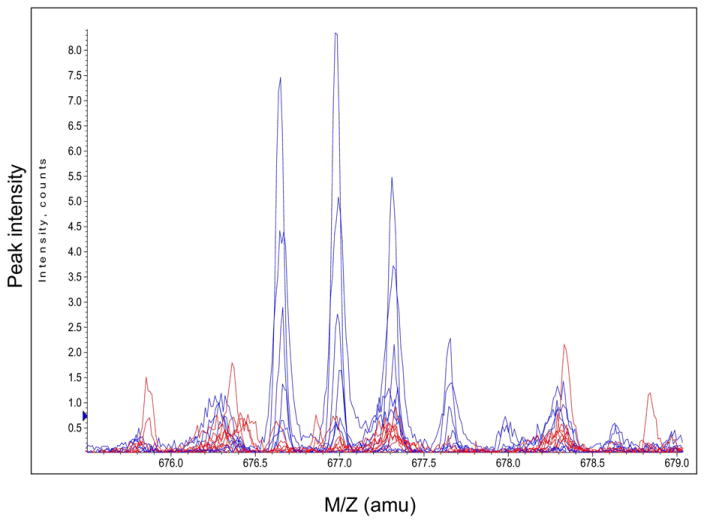
Representative mass spectra from 8 women with later PTB (red) and 8 women with term deliveries (blue). The multiplets centered at 677 m/z represents an example of a peptide marker.
Mass Spectral Data Analysis
The abundance (peak height) of each candidate and its relevant reference peak was quantified (extracted) by the instrument’s software and tabulated as was the calculated ratio of each candidate marker abundance relative to the abundance of the reference within each patient. The log of that ratio was also determined because abundance varied substantially. The data were submitted to statistical analysis. Attempts at statistical analysis of data without visual pre-selection failed both when using the instrument software and when using other software for the analysis. This was likely due to the large number of peaks creating many overlapping envelopes, in particular the presence of multiply charged ions. Problems included overlooking 80% of data in some analyses and in generating artefactual differences that did not exist when single peak analyses were carried out.
MS-MS Amino Acid Sequencing
Candidate markers demonstrating statistically different abundances between cases and controls were further analyzed in an effort to chemically identify the candidate molecule. The approach employed here has been described in detail elsewhere. (12). Frozen supernatant (from the protein reduction step) was thawed and injected for MS-MS analysis of the ion of interest. The selected candidate marker was fragmented using N2 gas collision and the daughter fragments were recorded in the second MS detector. This collision fragment peak list was submitted to Mascot (www.matrixscience.com), a searchable MS database allowing protein/peptide identification. Amino acid sequences were also independently submitted to the short homologous or near homologous protein BLAST search available through the NCBI website as a confirmation.
Previous Biomarker Analysis
Plasma corticotropin releasing factor, defensin, ferritin, lactoferrin, thrombin anti-thrombin complex and tumor necrosis factor α receptor type 1 assays have been previously analyzed by immunoassays and reported.(4) Most of these assays were research assays developed in the laboratory of the relevant investigator. The results from those previous assays were re-evaluated statistically for the subjects included in this study.
Statistical Analysis
Data are expressed as means ± 1 standard deviation for demographic measures and means ± 1 standard error for biomarkers. Species that appeared to be quantitatively different were considered. Only four species were numerically and statistically evaluated. Comparisons of the abundance of a single species for the two study populations were carried out by the Wilcoxon rank sum test. Fisher’s exact test was used for categorical analysis. Comparisons were carried out for each candidate at both 24 and 28 wks gestation. No corrections were made for multiple comparisons. Twenty three additional serum markers were previously assayed in these samples as part of other studies, and comparisons of the abundance of each individual marker were calculated using Wilcoxon rank sum test for the subset of subjects considered in this study. Logistic regression analyses were performed for the three novel biomarkers in combination with the best 6 of the previously tested markers. The combination was used for classification performance by means of receiver operator curves. For all statistical tests, nominal two-sided p-values are reported with statistical significance defined as a p-value < 0.05. SAS 8.2 (SAS Institute, Cary, North Carolina) was used for these analyses.
In calculating the sensitivity and specificity of the three peptide biomarkers, a threshold was established. The requirement was that the numeric threshold chosen provided at least 80% sensitivity. These thresholds at the 28 wk gestation sampling, using the log ratio (biomarker/reference) data, were as follows: biomarker peptide m/z 677 = 0.00, biomarker peptide m/z 857 = -0.347 and biomarker peptide m/z = -0.222.
RESULTS
The demographics of the four groups (Case and Control at the 24 week visit and Case and Control at the 28 week visit) are provided in Table I.
Table I.
Demographics (* p<0.001; p values reported are for comparisons between cases and controls within each gestational age period)
| 24 Weeks Controls N=40 | 24 Weeks Cases N=40 | 28 Weeks Controls N=40 | 28 Weeks Cases N=40 | |
|---|---|---|---|---|
| Maternal Age (year) | 23.2 ± 5.26 | 23.6 ± 5.10 | 24.3 ± 5.67 | 24.2 ± 5.93 |
| GA at Delivery (wk) | 38.9 ± 1.22 | 31.4 ± 2.79* | 38.9 ± 1.16 | 32.3 ± 1.77* |
| Time from sample collection to Delivery (wk) | 15.5 ± 1.19 | 8.1 ± 2.82* | 15.5 ± 1.17 | 9.0 ± 1.89* |
| Parity (% nulliparous) | 30.0 | 32.5 | 30.0 | 32.5 |
| Race (% African American) | 75.0 | 70.0 | 77.5 | 67.5 |
A total of 4000-5000 unique ion peaks were detected. Our initial survey involved only the second of ten time windows.
After visual inspection, four candidate markers were further evaluated to determine if quantitative differences were significant. Of the 4 markers considered further, 3 were independently found to be quantitatively significantly reduced (See Table II). Those species were: an ion at 676.66 m/z with a +3 charge corresponding to a neutral parent mass of 2026.98 Da; an ion at 856.85 m/z with a +5 charge corresponding to a neutral parent mass of 4279.25 Da; and an ion at 860.05 m/z with a +5 charge that corresponded to neutral parent masses of 4295.25 Da. The exact peak height of each of these markers as well as a reference peak nearby was determined by the instrument. Quantitatively the reference peaks did not differ between cases and controls. The reference peak for the biomarker at m/z 676.7 was 673.3 (controls: 37.6 ± 3.4 ion counts vs cases: 47.5 ± 8.3 ion counts, p=0.26). A single reference peak was used for both the biomarkers m/z 857.8 and 860.0 which had an m/z of 843.7 (controls: 118.1 ± 14.3 ion counts vs cases: 115.4 + 14.3 ion counts, p=0.89). These same markers were studied at both 24 and 28 wks gestation visits and were found to be significantly different between cases and controls at both gestational ages. Other potential markers with significant difference in abundance between cases and controls are listed in Table III.
Table II.
Ion abundance of the candidate peaks ratioed to reference and log transformed.
| 24 Weeks Gestation | |||||
|---|---|---|---|---|---|
| Preterm Birth | Control | ||||
| Label | Mean | Std Error | Mean | Std Error | P Value |
| Peak at 677(/673) m/z | 0.198 | 0.076 | 0.503 | 0.094 | 0.007 |
| Peak at 857(/843) m/z | -0.137 | 0.086 | 0.284 | 0.092 | 0.002 |
| Peak at 860(/843) m/z | -0.376 | 0.088 | 0.009 | 0.098 | 0.005 |
| 28 Weeks Gestation | |||||
|---|---|---|---|---|---|
| Preterm Birth | Control | ||||
| Label | Mean | Std Error | Mean | Std Error | P Value |
| Peak at 677(/673) m/z | -0.015 | 0.090 | 0.579 | 0.101 | < 0.001 |
| Peak at 857(/843) m/z | -0.149 | 0.095 | 0.231 | 0.102 | 0.007 |
| Peak at 860(/843) m/z | -0.204 | 0.088 | 0.201 | 0.095 | 0.002 |
Table III.
Comparison of relative abundance of other potential markers in the serum of cases and controls. These markers were analyzed in previous studies using the same sample sets.
| 24 Weeks Gestation | |||||
|---|---|---|---|---|---|
| Preterm Birth | Control | ||||
| Label | Mean | Std Error | Mean | Std Error | P Value |
| Placental growth factor (pg/mL) | 446.8 | 46.0 | 596.7 | 63.4 | 0.05 |
| Thrombin anti-thrombin (μg/mL) | 274.8 | 255.9 | 293.8 | 269.6 | 0.02 |
| 28 Weeks Gestation | |||||
|---|---|---|---|---|---|
| Preterm Birth | Control | ||||
| Label | Mean | Std Error | Mean | Std Error | P Value |
| Corticotropin releasing factor (ng/mL) | 0.359 | 0.019 | 0.284 | 0.012 | <0.001 |
| Defensin (ng/mL) | 612.0 | 106.6 | 427.4 | 92.1 | 0.04 |
| Ferritin (ng/mL) | 19.0 | 3.37 | 10.0 | 1.48 | 0.04 |
| Lactoferrin (ng/mL) | 245.0 | 42.0 | 484.6 | 100.7 | 0.05 |
| Thrombin anti-thrombin (μg/mL) | 546.8 | 366.3 | 835.5 | 453.7 | 0.04 |
| TNF receptor type 1 (pg/mL) | 1114.1 | 119.0 | 880.4 | 33.7 | 0.02 |
Predictive value of the markers
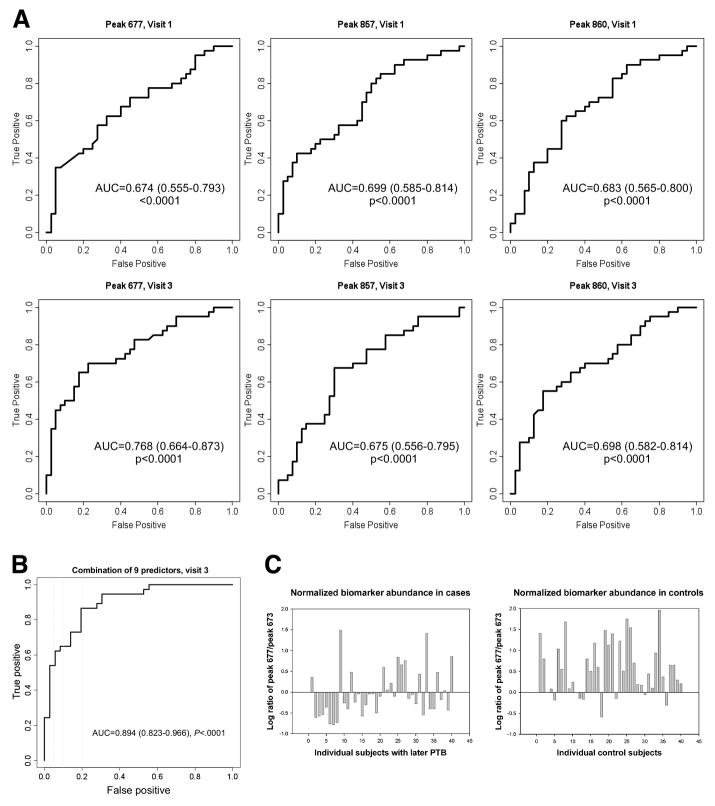
The sensitivity of each of the three biomarkers improved generally from 24 to 28 weeks (At 24 weeks: 677, sensitivity=35.0%, specificity=92.5%, OR=6.64, CI 1.7-25.5; 857, sensitivity=45.0%, specificity=82.5%, OR=3.86, CI 1.4-10.8; 860, sensitivity=45.0%, specificity=80.0%, OR=3.27, CI 1.2-8.8. At 28 weeks: 677, sensitivity=65.0%, specificity=82.5%, OR=8.76, CI 3.1-24.8; 857, sensitivity=37.5%, specificity=80.0%, OR=2.4, CI 0.9-6.6; 860, sensitivity=55.0%, specificity=80.0%, OR=4.89, CI 1.8-13.2). The biomarker at m/z 676.7 was the best single predictor of SPTB at 24 or 28 wks pregnancy. This is shown in Figure 2A. Combination of the 3 markers did not improve the sensitivity and specificity but the inclusion of the 6 best additional markers (7 patients excluded due to missing values) improved the sensitivity to 86.5% with a specificity of 80.6% at 28 weeks (Figure 2B).
Figure 2.
A ROC curves demonstrating the predictive capability of 3 peptide markers to predict subsequent SPTB after sampling at 24 and 28 weeks. Area under the curve and 95% confidence intervals are also included for each marker at each visit.
B ROC curve demonstrating the predictive capability of the combination of 9 predictors including peak 677, peak 857, peak 860, corticotrophin releasing factor, defensin, ferritin, lactoferrin, thrombin antithrombin complex, and tumor necrosis factor – receptor type 1 to predict subsequent SPTB after sampling at 28 weeks. Area under the curve and 95% confidence intervals are also reported.
C. Normalized biomarker abundance was calculated for all individuals at 28 weeks. The graph shows biomarker (m/z 677) abundance in the sera of women having a subsequent SPTB or term delivery. Sixty five percent (26/40) of the women with subsequent SPTB had a value below the threshold whereas only 17.5 (7/40) percent of women with a term delivery had a value below the same threshold.
Peptide Identification
Sequencing by means of a tandem MS-MS with intervening fragmentation allowed for the complete amino acid sequence to be determined by amino acid homology to known peptide or protein sequences. The amino acid sequences are provided in Table IV. The peaks initially assessed represented a +3 charge state for the species at 677 and a +5 charge state for the species at 857 and 860. Molecular ions representing additional charge states (+2 for 677, both +6 and +7 for 857 and 860) were observed and were also quantitatively significantly reduced in the women with subsequent SPTB (Data not shown). When a BLAST search of the individual amino acid sequences was performed using the National Center for Biotechnology Information website, all three peptides were found to be derived from one region of inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4), the common parent protein.
Table IV.
Amino acid sequences for the 3 biomarker peptides
| M/z | Charge | MW | Amino Acid Sequence |
|---|---|---|---|
| 677 | +3 | 2026.98 | qlglpgppdvpdhaayhpf |
| 857 | +5 | 4279.25 | nvhsagaagsrmnfrpgvlssrqlglpgppdvpdhaayhpf |
| 860 | +5 | 4295.25 | nvhsagaagsrm(O)nfrpgvlssrqlglpgppdvpdhaayhpf |
Relationship of Biomarker Abundance as a Function of Time to Delivery
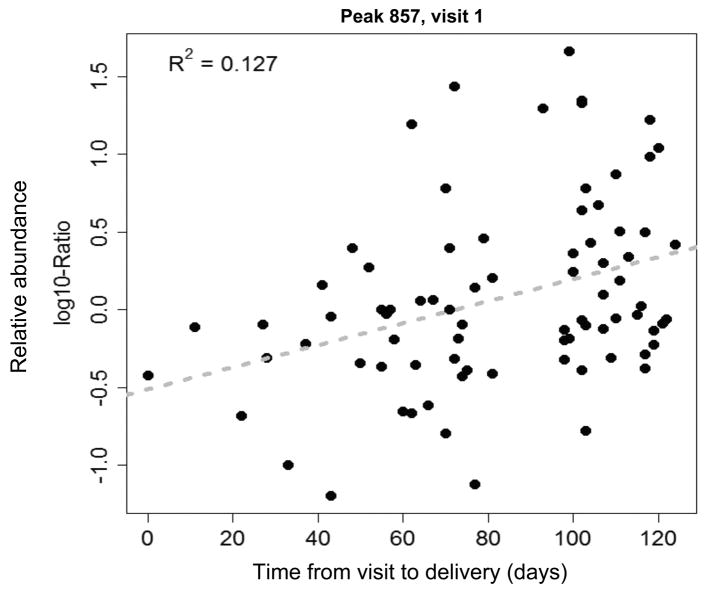
Women in the case group at 24 wks were on average 8.1±2.8 wks away from their eventual preterm delivery, the mean gestational age at delivery being 31.4±2.8 weeks. Women in the case group at 28 wks were on average 4.7±2.0 wks removed from their PTB, the mean gestational age at delivery being 32.3±1.8 weeks. When biomarker abundance was plotted as a function of time to delivery, a significant correlation was found for all three markers at 28 wks gestation (peak 677: R2=0.13, p=0.001, peak 857: R2=0.11, p=0.003, peak 860: R2=0.12, p=0.002) and for two of the markers at 24 wks gestation (peak 857: R2=0.13, p=0.001, peak 860: R2=0.11, p=0.003). See Figure 3 for a representative plot. Correlation for the third marker (peak 677) at 24 wks had a p-value of 0.08 (R2=0.04). In each case abundance of the biomarkers was lower the nearer the delivery. (None of the markers demonstrated a correlation between its abundance and gestational age, as would be expected given the narrow timing of specimen collection.)
Figure 3.
A representative plot of one biomarker’s normalized abundance (m/z 857) as a function of time to delivery (days). The correlation was statistically significant (R=0.11, p=0.001).
Relationship of Biomarker Abundance as a Function of Etiology of PTB
In the subjects sampled at 24 weeks and 28 weeks, chorioamnioitis was confirmed in only 4 of 80 subjects and 2 of 80 subjects respectively. Levels of all 3 biomarkers were markedly reduced in women with confirmed chorioamnioitis, but the number of subjects having this diagnosis was too small for meaningful statistical comparisons. There was no reduction in the abundance of any of the biomarkers with fetal fibronectin positivity.
COMMENTS
We have applied a serum proteomics method utilizing cLC-ESI-TOFMS to the analysis of sera collected from pregnant women at 24 and 28 wks gestation. This method surveys much of the low molecular weight proteome. The specimen employed was protein-depleted serum. Capillary columns are easily fouled by unprocessed serum and sera are typically de-proteinated prior to capillary LC separations. More importantly mass spectrometers have a limited dynamic range which in conjunction with ion suppression in the presence of high abundance serum species, requires the effective removal of highly abundant serum proteins. Given that there are at least 20 highly abundant protein species, removal of albumin and/or immunoglobulins is insufficient to allow for interrogation of the low molecular weight, low abundance proteome.
We have identified 3 peptides within the serum of pregnant women at both 24 and 28 weeks gestation that are significantly decreased in women who experienced a subsequent SPTB. The changes in peptide concentrations at 24 and 28 weeks predated on average the SPTB by a mean of 8.1 wks and 4.7 wks respectively. All three identified peptides came from a single protein and from a highly conserved proline-rich region of that protein that had been processed differently. One of these peptides had an oxidized methionine. The parent compound is termed inter-alpha trypsin inhibitor heavy chain 4 (ITIH4), a glycoprotein that is a kallikrein-sensitive acute phase reactant (15). The intact protein is known to be increased in inflammatory states (15), but little is known about the function of this protein or its peptide fragments, including possible biological activity. Although this is the first study to reports an association between ITIH4 or its fragments and SPTB, peptides derived from ITIH4 that differ from the peptides described here have demonstrated quantitative increases in sera of women with early-stage ovarian cancer (16). Other peptides arising from this same protein, but differing from the peptides described here, appear to be increased in other cancers in a disease specific manner (16). This might suggest differential peptide production having a disease specific pattern.
Early efforts to carry out serum proteomics resulted in methodological controversies. (17) First, the use of computers to evaluate mass spectral data is challenging. No software application has been accepted as fully reliable. Hence, continual reference to the actual mass spectra is critical. Second, day to day variability can be substantial for both the separation step and subsequent mass spectral analysis. Much of the variability can be eliminated or dramatically decreased by means of an internal control. Such standards are often used to correct for inconsistencies in elaborate, multi-step analyses involving human serum. In this study, such controls were employed by utilization of endogenous reference molecules present in all specimens to compensate for variability in specimen processing, chromatography loading and separation, ionization efficiency and instrumental performance. Finally, some have argued that plasma may provide a broader range of proteins than serum. IN the current study only serum was collected and available for analysis. Further, a difference between serum and plasma for the low molecular proteome or peptidome has never been established.
Collectively, these data suggest that the three peptides described here may be useful biomarkers, identifying approximately two thirds of pregnant women who will delivery prematurely, weeks prior to the SPTB. These significant quantitative differences were observed for both sets of women with subsequent SPTB sampled at different gestational ages and were also significantly different for other charge states of the same peptides as observed by MS. In addition, the data demonstrated a significant relationship between biomarker abundance and nearness to delivery, abundance being lower the closer the SPTB. The sensitivity of the most discriminating peptide or for combinations of the 3 was 65.0% with a specificity of 82.5%. However, the data suggest that these markers may become better predictors of SPTB as women near delivery. It is also highly likely that additional biomarkers will be found that increase the sensitivity and specificity of SPTB prediction. For example, when the current three peptides were coupled to 6 previously tested candidate biomarkers, the sensitivity was 86.5% with a specificity of 80.6%.
The data do not allow for any confident differentiation of the biomarkers according to a potential etiology for the SPTB. However, we hypothesize that women with infectious etiology of their preterm birth will have lower levels of these 3 markers. This will be the focus of future investigations.
The specimens utilized in this analysis were part of a multi-center study representing 10 medical centers located across the US and are demographically heterogeneous. However, we recognize that these findings will need to be confirmed in a larger number of specimens, preferably in a prospective fashion. In addition, studies of active preterm and term labor are needed to define whether these changes are observed only 4-8 wks prior to a preterm delivery or whether they are still present at the time of active preterm labor and whether the changes in biomarker abundance are limited to preterm births or also precede term labor.
Acknowledgments
We would like to acknowledge the contributions of Steven W. Graves, Ph.D. who was instrumental in the design of the study, interpretation of the results and completion of the manuscript. All the proteomics research and analysis reported here was done in his laboratory at Brigham Young University. Moreover, portions of this research were supported by funding from the Department of Chemistry and Biochemistry, Brigham Young University including fellowships to Dr. Karen Merrell, a graduate student at the time in the laboratory of Dr. Graves. We also appreciate the significant contributions of; Elizabeth Thom, Ph.D. for protocol/data management and statistical analysis, and Michael W. Varner, M.D. for his help in the design and completion of this study and the preparation of this manuscript.
In addition to the authors, other members of the Eunice Kennedy Shriver National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network are as follows:
University of Alabama at Birmingham – J.C. Hauth, A. Northen, and C. Neely
Wake Forest University Health Sciences – P. Meis, E. Mueller-Heubach, M. Swain, and A. Frye
University of Chicago – A. Moawad, M. Lindheimer, P. Jones and M.E. Lewis Brown
University of Cincinnati – T.A. Siddiqi, N. Elder, T. Coombs, and J. VanHorn
University of Pittsburgh – J.H. Harger, M. Cotroneo, C. Stallings, and J. Roberts
Ohio State University – M.B. Landon, J. Schneider, and C. Mueller
University of Oklahoma – J.C. Carey, A. Meier, and E. Liles
Medical University of South Carolina – R.B. Newman, B.A. Collins, T. Metcalf, and V. Odell
University of Tennessee – B. Sibai, R. Ramsey, and J.L. Fricke
Wayne State University – M. Treadwell, G.S. Norman.
The George Washington University Biostatistics Center – Elizabeth Thom, R. Bain, A. Das, L. Leuchtenburg, and M. Fischer
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD – S. Yaffe, C. Catz, and M. Klebanoff
Supported by grants from the National Institute of Child Health and Human Development (HD21410, HD21414, HD27860, HD27861, HD27869, HD27883, HD27889, HD27905, HD27915, HD27917, HD19897 and HD36801).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldenberg R, Rouse DJ. Prevention of premature birth. N Engl J Med. 1998;339:313–320. doi: 10.1056/NEJM199807303390506. [DOI] [PubMed] [Google Scholar]
- 2.Rush RW, Keirse MJ, Howat P, Baum JD, Anderson AB, Turnbull AC. Contribution of preterm delivery to perinatal mortality. Br Med J. 1976;2:965–968. doi: 10.1136/bmj.2.6042.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoyert DL, Mathews TJ, Menacker F, Stobino DM, Guyer B. Annual summary of vital statistics: 2004. Pediatrics. 2006;117:168–183. doi: 10.1542/peds.2005-2587. Erratum, Pediatrics 2006;117:2338. [DOI] [PubMed] [Google Scholar]
- 4.Goldenberg RL, Iams JD, Mercer BM, Meis PJ, Moawad A, Das A, Miodovnik M, Vandorsten PJ, Caritis SN, Thurnau G, Dombrowski MP. Maternal-Fetal Medicine Units Network. The Preterm Prediction Study: toward a multiple-marker test for spontaneous preterm birth. Am J Obstet Gynecol. 2001;185(3):643–51. doi: 10.1067/mob.2001.116752. [DOI] [PubMed] [Google Scholar]
- 5.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359(9306):572–7. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 6.Gravett MG, Novy MJ, Rosenfeld RG, Reddy AP, Jacob T, Turner M, McCormack A, Lapidus JA, Hitti J, Eschenbach DA, Roberts CT, Jr, Nagalla SR. Diagnosis of intra-amniotic infection by proteomic profiling and identification of novel biomarkers. JAMA. 2004;292:462–9. doi: 10.1001/jama.292.4.462. [DOI] [PubMed] [Google Scholar]
- 7.Buhimschi IA, Christner R, Buhimschi CS. Proteomic biomarker analysis of amniotic fluid for identification of intro-amniotic inflammation. Brit J Obstet Gynaecol. 2005;112:173–81. doi: 10.1111/j.1471-0528.2004.00340.x. [DOI] [PubMed] [Google Scholar]
- 8.Ruetschi U, Rosen A, Karlsson G, Zetterberg H, Rymo L, Hagberg H, Jacobsson B. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. J Proteome Res. 2005;4:2236–42. doi: 10.1021/pr050139e. [DOI] [PubMed] [Google Scholar]
- 9.Buhimschi CS, Bhandari V, Hamar BD, Bahtiyar MO, Zhao G, Sfakianaki AK, Pettker CM, Magloire L, Funai E, Norwitz ER, Paidas M, Copel JA, Weiner CP, Lockwood CJ, Buhimschi IA. Proteomic profiling of the amniotic fluid to detect inflammation, infection, and neonatal sepsis. Plos Med. 2007;4:e18. doi: 10.1371/journal.pmed.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravett MG, Thomas A, Schneider KA, Reddy AP, Dasari S, Jacob T, Lu X, Rodland M, Pereira L, Sadowsky DW, Roberts CT, Jr, Novy MJ, Nagalla SR. Proteomic analysis of cervical-vaginal fluid: identification of novel biomarkers for detection of intra-amniotic infection. J Proteome Res. 2007;6:89–96. doi: 10.1021/pr060149v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira L, Reddy AP, Jacob T, Thomas A, Schneider KA, Dasari S, Lapidus JA, Lu X, Rodland M, Roberts CT, Jr, Gravett MG, Nagalla SR. Identification of novel protein biomarkers of preterm birth in human cervical-vaginal fluid. J Proteome Res. 2007;6:1269–1276. doi: 10.1021/pr0605421. [DOI] [PubMed] [Google Scholar]
- 12.Merrell K, Southwick K, Graves SW, Esplin MS, Lewis NE, Thulin CD. Analysis of low-abundance, low-molecular-weight serum proteins using mass spectrometry. J Biomolec Techniques. 2004;15:235–244. [PMC free article] [PubMed] [Google Scholar]
- 13.Iams J, Goldenberg R, Meis P, Mercer B, Moawad A, Das A, Thom E, McNellis D, Copper R, Johnson F, Roberts J, Miodovnik M, Van Dorsten J, Caritis S, Thurnau G. Bottoms S and the NICHD Maternal Fetal Medicine Units Network. The length of the cervix and the risk of spontaneous premature delivery. N Eng J Med. 1996;334:567–572. doi: 10.1056/NEJM199602293340904. [DOI] [PubMed] [Google Scholar]
- 14.Merrell K, Thulin CD, Esplin MS, Graves SW. Selection of internal serum peptides as standards for cLC-MS time normalization for serum proteomics. J Biomolec Techniques. 2008;19:320–327. [PMC free article] [PubMed] [Google Scholar]
- 15.Piñeiro M, Andrés M, Iturralde M, Carmona S, Hirvonen J, Pyörälä S, Heegaard PM, Tjørnehøj K, Lampreave F, Piñeiro A, Alava MA. ITIH4 (inter-alpha-trypsin inhibitor heavy chain 4) is a new acute-phase protein isolated from cattle during experimental infection. Infect Immun. 2004;72:3777–82. doi: 10.1128/IAI.72.7.3777-3782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song J, Patel M, Rosenzweig CN, Chan-Li Y, Sokoll LJ, Fung ET, Choi-Miura NH, Goggins M, Chan DW, Zhang Z. Quantification of fragments of human serum inter-alpha-trypsin inhibitor heavy chain 4 by a surface-enhanced laser desorption/ionization-based immunoassay. Clin Chem. 2006;52:1045–53. doi: 10.1373/clinchem.2005.065722. [DOI] [PubMed] [Google Scholar]
- 17.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]