Abstract
Epidermal growth factor receptor (EGFR) is overexpressed in many cancer types including ∼30% of breast cancers. Several small molecule tyrosine kinase inhibitors (TKIs) targeting EGFR have shown clinical efficacy in lung and colon cancers, but no benefit has been noted in breast cancer. Thirteen EGFR expressing breast cancer cell lines were analyzed for response to EGFR TKIs. Seven were found to be EGFR TKI resistant; while shRNA knockdown of EGFR determined that four of these cell lines retained the requirement of EGFR protein expression for growth. Interestingly, EGFR localized to plasma membrane lipid rafts in all four of these EGFR TKI resistant cell lines, as determined by biochemical raft isolation and immunofluorescence. When lipid rafts were depleted of cholesterol using lovastatin, all four cell lines were sensitized to EGFR TKIs. In fact, the effects of the cholesterol biosynthesis inhibitors and gefitinib were synergistic. While gefitinib effectively abrogated phosphorylation of Akt and MAPK in an EGFR TKI sensitive cell line, phosphorylation of Akt persisted in two EGFR TKI resistant cell lines; however, this phosphorylation was abrogated by lovastatin treatment. Thus, we have shown that lipid raft localization of EGFR correlates with resistance to EGFR TKI-induced growth inhibition and pharmacological depletion of cholesterol from lipid rafts decreases this resistance in breast cancer cell lines. Furthermore, we have presented evidence to suggest that when EGFR localizes to lipid rafts, these rafts provide a platform to facilitate activation of Akt signaling in the absence of EGFR kinase activity.
Keywords: EGFR, Statins, TKI resistance, Breast cancer, Lipid rafts
Introduction
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase whose function has been implicated in many biological processes. When activated, EGFR stimulates signaling pathways involved in cell growth, survival, and migration. While EGFR contains activating mutations in glioblastomas and lung cancer, overexpression is the primary mechanism by which EGFR contributes to breast cancer growth and progression (Paez et al., 2004; Wong et al., 1992). EGFR over-expression occurs in approximately 30% of all breast cancers which correlates with poor clinical prognosis (Bolla et al., 1990; Sainsbury et al., 1987; Toi et al., 1991). Several small molecule tyrosine kinase inhibitors (TKIs) targeting EGFR have been tested in clinical trials with some clinical success in lung and colon cancers. While EGFR TKIs have shown some clinical efficacy in hormone receptor-positive breast cancer (Cristofanilli et al., 2010; Guix et al., 2008; Polychronis et al., 2005), EGFR TKIs lack efficacy in hormone receptor-negative breast cancer (Blagosklonny and Darzynkiewicz, 2003).
The sub-cellular localization of EGFR determines the signaling pathways stimulated by EGFR activation. In fact, EGFR promotes differential signaling depending on receptor localization to endosomes, at the mitochondria, within the nucleus, or on the plasma membrane. Specifically, EGFR localization to endosomes results in ligand-dependent activation of extracellular signal-regulated kinase (ERK) and p38 mitogen-activated protein kinase (MAPK) pathways (Sadowski et al., 2009), while mitochondrial localization of EGFR has been implicated in modification of cytochrome c oxidase subunit II activity (Boerner et al., 2004). Also, EGFR localizes to the nucleus where it may act as a transcription factor (Lin et al., 2001). Perhaps the most well known localization of EGFR is to the plasma membrane, where it modulates both MAPK and Akt signaling pathways.
The plasma membrane contains discrete heterogeneous microdomains (Maa et al., 1995). These microdomains are less fluid than the surrounding bulk plasma membrane, and are enriched in cholesterol, sphingolipids, and gangliosides. They have been termed lipid rafts, and act as platforms for cellular signaling (Simons and Ikonen, 1997). Levels of lipid rafts are increased in melanomas, prostate, and breast cancers; results that suggest that these structures play a functional role during tumorigenesis (Hazarika et al., 2004; Li et al., 2006). EGFR is one of many proteins shown to exist within lipid rafts, but the effect of EGFR localization to lipid rafts is not well understood. While it has been noted that lipid raft localization of EGFR inhibits ligand binding and subsequent signaling downstream (Chen and Resh, 2002; Roepstorff et al., 2002), other studies have shown that lipid rafts promote EGFR signaling (Peres et al., 2003; Zhuang et al., 2002).
In this manuscript, we have found that lipid raft localization of EGFR plays a role in the response of breast cancer cell lines to EGFR TKI-induced growth inhibition. Specifically, EGFR localization to lipid rafts correlated with EGFR TKI resistance. In addition, reduction of cholesterol from lipid rafts sensitized resistant breast cancer cells to the EGFR TKI gefitinib. Significantly, the effects of cholesterol biosynthesis inhibitors and gefitinib were synergistic. While gefitinib abrogated both Akt and MAPK phosphorylation in EGFR TKI sensitive cells, Akt remained phosphorylated in EGFR TKI resistant cell lines. Lovastatin, a cholesterol biosynthesis inhibitor, was sufficient to diminish this phosphorylation in two of the EGFR TKI resistant cell lines. Thus, our data suggest that lipid rafts provide a platform for activation of Akt in the absence of EGFR kinase activity in cell lines resistant to EGFR TKIs.
Materials and Methods
Reagents
Gefitinib was provided by AstraZeneca (Wilmington, DE). All other reagents were purchased from Sigma or VWR unless otherwise noted.
Cell lines
The SUM series of cell lines were obtained from Dr. Stephen Ethier (Wayne State University/Karmanos Cancer Institute, Detroit, MI). The remaining cell lines were purchased from ATCC (Manassas, VA). The normal growth mediums for each cell line are as follows. SUM 52, SUM 149, SUM 159, SUM 185, SUM 225, and SUM 229 cells are grown in 5%IH media (Ham's F-12 media, supplemented with 5% FBS, 1μg/ml hydrocortisone, and 5μg/ml insulin). SUM 1315 cells are grown in 5%IE media (Ham's F-12 media, supplemented with 5% FBS, 10ng/ml EGF, and 5μg/ml insulin). SUM 44 and SUM 190 cells are grown in SFIH media (Ham's F-12 media, supplemented with 1μg/ml hydrocortisone, 5μg/ml insulin, 5mM ethanolamine, 10mM HEPES, 5μg/ml transferrin, 10nM triiodo-thyronine, 50μM sodium selenite, and 5% BSA). SUM 102 and MCF10A cells are grown in SFIHE media (Ham's F-12 media, supplemented with 1μg/ml hydrocortisone, 5μg/ml insulin, 10ng/ml EGF, 5mM ethanolamine, 10mM HEPES, 5μg/ml transferrin, 10nM triiodo-thyronine, 50μM sodium selenite, and 5% BSA). MCF7, SKBr3, T47D, MDA-MB-231 and MDA-MB-468 cells are grown in DMEM+10%FBS media (DMEM media, supplemented with 10% FBS). BT-20 cells are grown in Eagles+NEAA media (Eagle's MEM with 2mM L-glutamine and Earle's BSS adjusted to contain 1.5g/L sodium bicarbonate, 0.1mM non-essential amino acids, 1mM sodium pyruvate, and 10% FBS). BT-549 cells are grown in RPMI+L-GLUT(2mM) media (RPMI-1640, supplemented to contain 1.5g/L sodium bicarbonate, 4.5g/L glucose, 10mM HEPES, 1mM sodium pyruvate, 0.023 IU/ml insulin, and 10% FBS). HCC 1937 and HCC 1954 cells are grown in RPMI+L-GLUT media (RPMI-1640 media with 2mM L-glutamine adjusted to contain 1.5g/L sodium bicarbonate, 4.5g/L glucose, 10mM HEPES, 1mM sodium pyruvate, and 10% FBS). The SUM and HCC cells are cultured in 10% CO2 and the remaining cells are cultured in 5% CO2. All media are supplemented with 2.5 μg/ml amphotericin B and 25 μg/ml genatimicin.
Immunoblotting
Breast cancer cell lines were plated at a density of 1×106 cells per 100-mm dish and grown for 48 h. Cells were treated with indicated reagents (1.0 μM gefitinib for 30 min or 72 h and/or 1.0 μM lovastatin 72 h) then lysed in CHAPs lysis buffer [10 mmol/L CHAPs, 50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, and 2 mmol/L EDTA with 10 μmol/L sodium orthovanadate and 1× protease inhibitor cocktail]. For immunoblotting, 10 to 100 μg of protein lysate were separated by SDS-PAGE and transferred to Immobilon P. Membranes were blocked in either 5% nonfat dry milk for 1 h at 25°C or overnight at 4°C (phospho-MAPK). Membranes were probed with EGFR (Cell Signaling Technology, Danvers, MA), Akt (Cell Signaling Technology, Danvers, MA), MAPK (Cell Signaling Technology Danvers, MA), phospho-Akt (Ser473; Cell Signaling Technology, Danvers, MA), phospho ERK1/2 (MAPK) (Invitrogen, Carlsbad, CA), transferrin receptor (Invitrogen, Carlsbad, CA), caveolin-1 (Cell Signaling Technology, Danvers, MA), or flotillin (BD Biosciences, San Jose, CA) antibodies. All antibodies were incubated overnight at 4°C, except for phospho-MAPK (2h at room temperature). Membranes were washed with TBS + 0.1% Tween 20 three times for 10 min, followed by incubation with corresponding secondary antibody and another series of three washes. Incubation with enhanced chemiluminescence (GE Healthcare Buckinghamshire, UK) was followed by exposure to film. Experiments were repeated at least three times and quantified using densitometry (NIH Image).
In vitro kinase assays
Cells were washed in PBS and lysed in solubilization buffer (50 mM HEPES, pH 7.5, 10% glycerol, 0.5% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 1 mM PMSF, 50 μg/ml aprotinin, and 400 nM vanadate). Lysates were cleared by centrifugation, quantified, and 0.5 mg of protein was immunoprecipitated using EGFR antibodies (mab108, M. Weber, University of Virginia, Charlottesville, VA). Antibody bound proteins were collected using protein A beads (Upstate Biotechnology, Lake Placid, NY) and washed three times in HTG buffer (20 mM HEPES, pH 7.5, 0.1% Triton X-100, and 10% glycerol). For the kinase assay, 40 μl HTG buffer, 4 μl MnCl2 (of 100 mM stock), and 10 μCi 32P-γATP were incubated for 10 min at 30°C. The beads were pelleted and the supernatant removed and discarded. Sample buffer was added to the pellets, the samples were boiled, and proteins were separated using 7.5% SDS-PAGE. The gels were dried and exposed to film. Each experiment was repeated at least three times.
ShRNA downregulation of EGFR
To downregulate EGFR expression we utilized 21 EGFR-directed shRNA lentiviral constructs from OpenBiosystems (TRCN0000039633, TRCN0000039634, TRCN0000039635, TRCN0000039636, TRCN0000039637, TRCN0000010329, TRCN0000121067, TRCN0000121068, TRCN0000121069, TRCN0000121070, TRCN0000121071, TRCN0000121202, TRCN0000121203, TRCN0000121204, TRCN0000121205, TRCN0000121206, TRCN0000121327, TRCN0000121328, TRCN0000121329, TRCN0000121330, TRCN0000121331). Three constructs were chosen based on their specific reduction in EGFR expression in our cell models. Specifically, EGFR shRNA #1 = TRCN0000121204 and EGFR shRNA #2 = TRCN0000121071 showed pertinent effects in our model system. The lentiviruses were packaged using a third generation lentiviral packaging system developed by Didier Trono and colleagues (Lausanne, Switzerland) and purchased from Addgene (Dull et al., 1998). Specifically, Addgene plasmids pMLDg/pRRE (12251), pRSV-Rev (12253), and pMD2.G (12259) were transfected into HEK293T cells with the lentiviral vectors containing the shRNAs using FUGENE6 (Roche). Cellular supernatant was collected on days 2 and 3 after transfection, pooled, and filtered. The lentivirus was titered using HEK293T cells incubated with increasing concentrations of virus with polybrene and selected for via the puromycin selection on the lentiviral vector. Colonies were counted and used to compare viral preps and between viruses for consistent titers used in experiments. To determine the efficacy of EGFR downregulation in breast cancer cells, equal multiplicity of infection (MOI) of EGFR shRNA virus (or a non-silencing control) was added to the indicated cells in the presence of polybrene. Four days later, cell lysates were collected, separated by SDS-PAGE, and immunoblotted using EGFR antibodies as described above. EGFR was considered knocked down if the densitometric values of at least three experiments demonstrated at least a 50% reduction of EGFR protein expression.
To determine if EGFR downregulation effects cell proliferation in breast cancer cells, the indicated cells were incubated with equal MOI of virus and allowed to proliferate for three days. Puromycin was then added to media to select for cells that contain the lentivirus and cells were allowed to proliferate for an additional eight days. The number of cells was quantified using a Beckman Coulter Counter. Each experiment was repeated at least three times with the following control conditions: no puromycin added to the cells, no viral infection with puromycin selection, and non-silencing control with puromycin selection. The percent of cell growth was determined by using the non-silencing control with puromycin selection as 100% cell growth.
Immunostaining
Anti-EGFR (mab108) was labeled with Alexa-fluor-488 (Invitrogen, Carlsbad, CA). Cells were plated on coverslips at a density of 1.5×105 cells per 35mm dish and grown for 48 h in growth medium. For lipid raft staining, cells were incubated with Alexa-fluor-594 labeled cholera toxin subunit B (Invitrogen, Carlsbad, CA) at 1 μg/ml for 10min on ice prior to fixation (Liu et al., 2007; Roepstorff et al., 2002). Cells were then fixed with formalin, permeabilized with 0.1 % Triton-x 100 (if applicable), blocked in 20% goat serum for 1 h, then incubated with Alexa-fluor labeled antibody for 1 h, washed, and mounted onto slides with Prolong Gold containing DAPI (Invitrogen, Carlsbad, CA). Imaging was performed via confocal microscopy using a Zeiss Axioplan2 apotome microscope fitted with a 63X 1.25 oil immersion lens at the Microscopy and Imaging Resources Laboratory (Wayne State University, Detroit, MI).
Biochemical Raft Isolation
Biochemical lipid raft isolation was performed following established protocols (Macdonald and Pike, 2005). Briefly, cells were plated at a density of 0.5×106 cells in six-100 mm plates and allowed to grow in growth medium for 72 h. Cells were scraped in base buffer [20 mM Tris, pH 7.8, 250 mM Sucrose, 1 mM MgCl2, 1 mM CaCl2, 100 μM sodium orthovanadate] and then lysed in base buffer containing 1× protease inhibitor cocktail (EMD Biosciences, Gibbstown, NJ) by passing through a 22 gauge × 1.5″ needle 40 times. Lysates were centrifuged as described and the first and second post-nuclear supernatants were combined and frozen at -20°C. Samples were thawed and combined with equal volume of 50% Opti-Prep (Greiner Bio One, Monroe, NC) and 0-20% Opti-Prep gradient was applied. Gradients were centrifuged for 90 min at 52,000×g and then fractionated into 16 - 0.56 mL fractions. Fractions were separated via SDS-PAGE, transferred to Immobolin-P (Millipore, Billerica, MA), and immunoblotted utilizing antibodies described above. Fractions were dot blotted with Cholera Toxin Subunit B-HRP (Invitrogen, Carlsbad, CA) to determine GM-1 expression. Incubation with enhanced chemiluminescence (GE Healthcare, Buckinghamshire, UK) was followed by exposure to film. Experiments were repeated at least three times and quantified using densitometry (NIH Image).
Cholesterol Assay
SUM159 breast cancer cells were plated at a density of 0.5×104 cells per well of a 6-well plate then treated with indicated concentrations of methyl-beta cyclodextrin (0.5 mM MBCD 1 h), gefitinib (1.0 μM 72 h), lovastatin (1.0 μM 72 h), atorvastatin (LC Laboratories, Woburn, MA; 1.0 μM 72 h), or NB-598 (1.0 μM 72 h) in growth medium. Cells were then lysed in CHAPS lysis buffer [10 mmol/L CHAPs, 50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, and 2 mmol/L EDTA with 10 μmol/L sodium orthovanadate and 1× protease inhibitor cocktail] and Bradford protein assay (Bio-Rad, Hercules, CA) was performed. Cholesterol was measured utilizing the Amplex Red cholesterol assay kit (Invitrogen, Carlsbad, CA). Briefly, 5 μl of sample was diluted into 45 μl 1× reaction buffer and 50 μl Amplex Red buffer [2 U/mL HRP, 2 U/mL cholesterol oxidase and 0.2 U/mL cholesterol esterase] was added in a 96-well plate. Reactions were incubated at 37°C for 30 min, then excitation was performed at 540/25nm and emission measured at 620/40nm utilizing filters of a Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT). Emission readings were averaged and compared to standard curve, then normalized for protein content.
Growth Assays
Cells were plated at a density of 3.5×104cells per well in 6-well plates on day 0. Every other day, cells were treated with indicated concentrations of gefitinib, CI-1033 or lovastatin (EMD Biosciences, Gibbstown, NJ) alone or in combination in growth medium. On days 1, 4 and 8, cells were counted with a coulter counter (Beckman Coulter, Brea, CA). Graphs represent the mean of three individual experiments performed in triplicate.
MTS Assays
Breast cancer cells were plated at a density of 1-2×103 in 96-well plates, incubated overnight, and then treated with lovastatin or NB-598 for 72 h in growth medium with or without the addition of gefitinib. Twenty microliters of CellTiter 96 Aqueous One Solution Cell Proliferation Assay reagent (Promega, Madison, WI) were added to each well and allowed to incubate at 37°C. Absorbance at 490nm was detected at 2 h using an OpsysMR microplate reader (Dynex, Chantilly, VA). Absorbance units were normalized to the mean of a single dose to compare between experiments. Dose response curves were generated using non-linear sigmoidal dose response curve analyses in GraphPad Prism. Points in the graph represent a mean of three independent experiments performed in triplicate. IC50s were calculated and plotted on isobolograms. IC50 points represent a mean of at least three independent experiments.
Statistics
Student's t-test was performed utilizing the statistical software in GraphPad Prism. P-values of <0.05 were considered statistically significant. To perform synergy analyses, the IC50 gefitinib was calculated for each dose of lovastatin. The combination index (CI-value) was calculated as follows: (IC50 gefitinib at × dose lovastatin)/(IC50 gefitinib alone) + (dose of lovastatin)/(IC50 lovastatin alone).
Results
Resistance of EGFR expressing breast cancer cell lines to EGFR TKIs
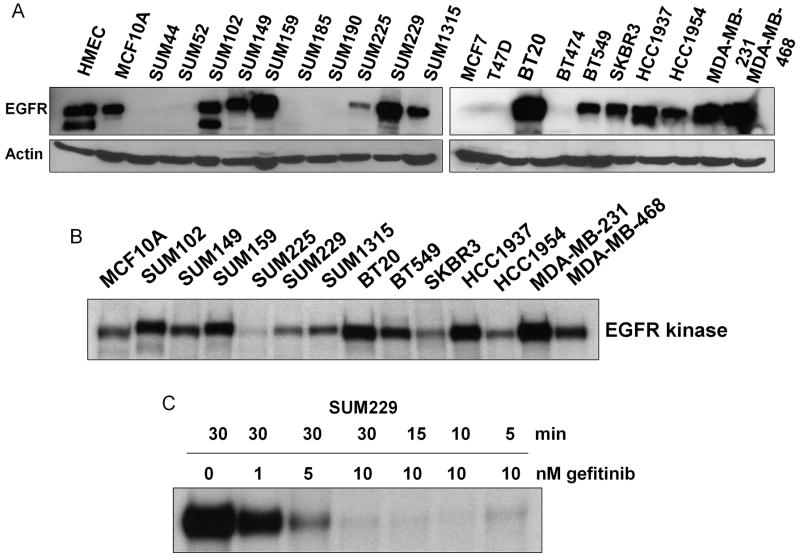
The lack of clinical response of breast cancers to EGFR TKIs prevents the use of an excellent targeted agent for the treatment of this disease. To study mechanisms of resistance to EGFR TKIs in breast cancer, we characterized a panel of twenty breast cancer cell lines for EGFR protein expression (Fig. 1A). Thirteen of the cell lines analyzed expressed EGFR protein. Interestingly, in twelve of the thirteen EGFR expressing cell lines, EGFR was kinase active under normal growth conditions (Fig. 1B). To determine the response of these twelve cell lines to the EGFR TKI gefitinib, we treated the cells with increasing doses of gefitinib, an EGFR TKI, and measured cellular viability via MTS analyses (Table 1). Previous reports in lung cancer cell lines have suggested that an IC50 of 10 μM or less, as determined by MTS analyses, represents sensitivity to gefitinib, while an IC50 value of >10 μM denotes resistance (Helfrich et al., 2006; Noro et al., 2006; Ono et al., 2004; Witta et al., 2006). By these standards, five of the breast cancer cell lines we tested were considered sensitive to gefitinib (Table 1). Seven cell lines, specifically SUM159, SUM229, BT20, BT549, HCC1937, MDA-MB231, and MDA-MB468, had IC50 values for gefitinib >10 μM, suggesting that these cell lines were resistant to EGFR kinase inhibition by gefitinib (Table 1, bold). These designations of sensitivity and resistance are supported by cellular proliferation data showing that physiologically relevant doses of gefitinib decreased proliferation of sensitive cell lines, while proliferation of resistant cell lines continued (Supplemental fig. 1). Breast cancer cells resistant to gefitinib-induced growth inhibition were also shown to be resistant to other EGFR selective TKIs, including the irreversible inhibitor CI-1033 (Supplemental Fig. 2, and data not shown).
Figure 1. EGFR is expressed and kinase active in breast cancer.
Breast cancer cell lines were grown in normal growth medium. (A) Cells were lysed and 100 μg of lysate was separated by SDS-PAGE, transferred to PVDF, and immunoblotted with EGFR or β-actin antibodies. Immunoblots are representative of at least three individual experiments. (B) Cells were lysed in kinase-buffer and immunoprecipitated with EGFR antibodies. Kinase assays were performed with 32P-γATP incorporated into EGFR as the substrate for EGFR kinase activity. Autoradiography was performed at least three individual times. (C) Kinase assays were performed as described after indicated treatment times and doses of gefitinib in SUM 229 cells. Autoradiography was performed at least three individual times.
Table 1. Breast cancer cell lines differ in their sensitivity to EGFR TKIs.
Viability of breast cancer cell lines in the presence of increasing doses of gefitinib was determined as described. Sensitive cell lines are those with <10 μM gefitinib IC50, while resistant (bold) cells lines are those with >10 μM gefitinib IC50.
| Cell Line | Gefitinib IC50 (μM) |
|---|---|
| SUM 102 | 2.24 |
| SUM 149 | 6.79 |
| SUM 159 | 17.91 |
| SUM 229 | 12.12 |
| SUM 1315 | 8.30 |
| BT20 | 13.44 |
| BT549 | 13.81 |
| SKBR3 | 1.861 |
| HCC1937 | 18.24 |
| HCC 1954 | 9.481 |
| MDA-MB 231 | 25.08 |
| MDA-MB 468 | 14.13 |
In order to determine if gefitinib effectively inhibits EGFR kinase activity in these breast cancer cells, in vitro kinase assays were performed. We have previously published that 0.1 μM gefitinib completely abrogates EGFR kinase activity as measured by 32P incorporation into EGFR via autophosphorylation (Mueller et al., 2008). Interestingly, we found that in five of the seven EGFR TKI resistant breast cancer cells, tyrosine phosphorylation was maintained in the absence of EGFR kinase activity which we have evidence to support occurs via transphosphorylation by other activated tyrosine kinases (Mueller et al., 2008). Here, we added to these findings by determining the minimal dose and time of gefitinib required to completely inhibit EGFR kinase activity (Fig. 1C). We found that as little as 10 nM gefitinib for five minutes was sufficient to deplete EGFR kinase activity in these cells. Therefore, EGFR kinase activity was successfully inhibited by the doses of gefitinib utilized in these studies in both EGFR TKI sensitive and resistant cell lines.
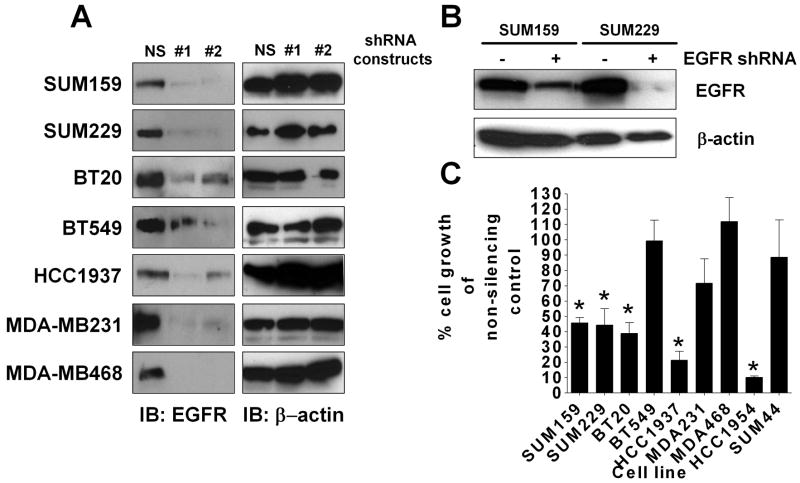
Although EGFR kinase activity is not required for the growth of EGFR TKI resistant cell lines, the previously described maintenance of EGFR phosphorylation in the absence of kinase activity (Mueller et al., 2008) suggests that the protein itself may still be required for proliferation. Thus, to directly determine if proliferation of EGFR TKI resistant cells requires EGFR protein expression, we used EGFR-targeting shRNA lentiviral infection to down-regulate EGFR protein expression. Twenty-one EGFR shRNA constructs were screened for efficiency of knocking down EGFR expression, as measured by immunoblotting. Two EGFR shRNA constructs consistently decreased EGFR protein expression (Fig. 2A). Construct one gave the best knockdown, as there was at least a 50% reduction in EGFR protein of all cell lines tested when compared to the non-silencing shRNA control. In order to determine if knockdown of EGFR was sustained over the period utilized to conduct growth assays, SUM159 and SUM229 cells were infected with EGFR shRNA, and grown with puromycin selection for two weeks. As seen in Figure 2B, EGFR protein expression remained reduced at two weeks in both cell lines, demonstrating that EGFR #1 shRNA sufficiently knocks down EGFR expression over the time period necessary for growth assays to be performed. Additionally, SUM44 cells, which do not express EGFR (Fig. 1A), were utilized as a negative control, and HCC1954 cells which are sensitive to EGFR TKIs (Table 1, Supplemental fig. 1) were utilized as a positive control. Notably, BT549, MDA-MB231, and MDA-MB468 cells continued to grow after a decrease in EGFR protein expression (Fig. 2C). This non-dependence on EGFR protein expression in these three cells lines may be a result of genetic alterations in signaling proteins downstream of EGFR. Specifically, MDA-MB-468 and BT549 cells have lost PTEN expression and MDA-MB-231 cells contain an activating K-Ras mutation (Hollestelle et al., 2007). Conversely, in SUM159, HCC1937, SUM229, and BT20 breast cancer cell lines, knocking down EGFR expression significantly decreased proliferation, suggesting that EGFR protein expression is, at least in part, required for the growth of these cell lines (Fig. 2C, * = p<0.05).
Figure 2. EGFR protein expression is required for growth in four of seven EGFR TKI resistant breast cancer cell lines.
(A) Cells were incubated with equal MOI of virus for four days, lysed, separated by SDS-PAGE, and immunoblotted for EGFR and β-actin. Experiments were performed at least three independent times (B) SUM 159 and SUM 229 cells were grown under selection pressure for two weeks post-infection, then lysed, separated by SDS-PAGE, and immunoblotted for EGFR. Immunoblots are representative of at least three independent experiments (C) Cells were plated at 1,000 cells/well of a 6-well plate and inoculated with EGFR shRNA #1 lentivirus or non-silencing control lentivirus. Three days later, the media was changed to puromycin containing media for eight days. Cells were then counted using a Beckman Coulter Counter. Error bars represent the standard error of the mean of at least three independent experiments. Statistical analyses were performed utilizing Student's t-test, * = p<0.05.
EGFR is localized to lipid rafts in breast cancer cells resistant to EGFR TKI-induced growth inhibition
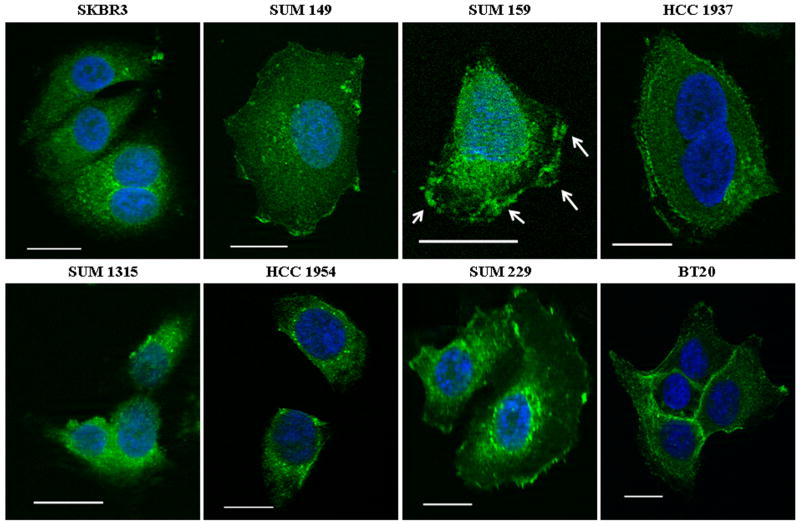
Previous studies have shown that EGFR localization can modulate EGFR signaling (Boerner et al., 2004; Chen and Resh, 2002; Li et al., 2006; Lin et al., 2001; Zhuang et al., 2002). Thus, to determine if the localization of EGFR was mediating the response of cells to EGFR TKIs, immunostaining and confocal microscopy were performed. Cells were stained with Alexa-Fluor 488-labeled EGFR antibodies (Fig. 3; green) and DAPI as a nuclear dye (blue). In two EGFR TKI sensitive cell lines (SKBR3 and SUM1315), EGFR localized entirely within intracellular compartments and the cytosol. However, in two other EGFR TKI sensitive cell lines (SUM149 and HCC1954), as well as all four EGFR TKI resistant cell lines, EGFR localized both within intracellular regions and at the plasma membrane. Interestingly, EGFR staining was not always contiguous around the membrane. The patchy nature of the staining, most prominent in SUM159 cells (Fig. 3; arrows), suggested that EGFR may localize to lipid rafts (Grossmann et al., 2006; Harder et al., 1998).
Figure 3. EGFR is localized to the plasma membrane in breast cancer cell lines.
For each cell line, two hundred thousand cells were plated onto coverslips and cultured in growth medium for 48 h. Cells were fixed, permeabilized, and blocked with 20% goat serum. EGFR was detected with Alexa-fluor 488 labeled EGFR antibody (green) and nuclei were identified with DAPI (blue). Imaging was performed using Zeiss Axioplan2 apotome microscope fitted with a 63X 1.25 oil immersion lens at the Microscopy and Imaging Resources Laboratory (Wayne State University, Detroit, MI). Arrows indicate patchy EGFR staining. Scale bars represent a distance of 50 μm. Images are representative of at least three independent experiments.
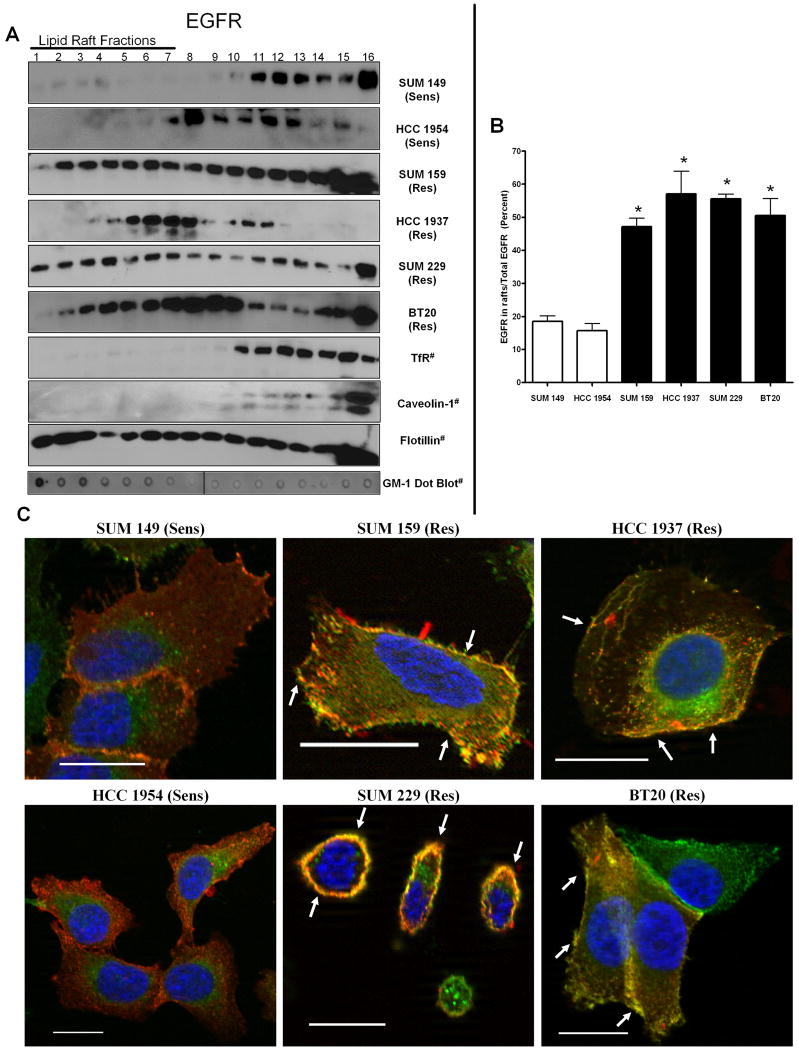
EGFR has been shown to localize within lipid rafts in Hela and CHO cells as well as MDA-MB231 breast cancer cells (Macdonald and Pike, 2005). In order to determine if EGFR was localized to lipid rafts in our panel of EGFR TKI resistant breast cancer cells, we used two methods of identifying these structures: biochemical raft isolation and confocal microscopy. First, a detergent-free Opti-Prep gradient was used to isolate lipid rafts (Macdonald and Pike, 2005). Flotillin, a membrane protein found both within and outside of lipid rafts, was used to show presence of membrane components within all fractions, while transferrin receptor was used as a marker for non-raft containing fractions. In addition, caveolin-1 was used as a marker for lipid containing caveolae (Macdonald and Pike, 2005). These markers, along with dot blotting for the lipid raft specific glycosphingolipid GM-1 (Fig. 4A) indicated fractions 1-7 as lipid raft fractions. When these fractions were immunoblotted using EGFR antibodies, EGFR localization to lipid raft fractions was most prominent in the EGFR TKI resistant cell lines (Figure 4A). As SKBR3 and SUM1315 cell lines showed solely intracellular EGFR staining, these cell lines were excluded from lipid raft analyses. Quantification of the percent of total EGFR that was present in the lipid raft fractions found that the four EGFR TKI resistant breast cancer cell lines contained significantly more EGFR within lipid rafts as compared to the average EGFR content within lipid rafts of two EGFR TKI sensitive cell lines, SUM149 and HCC1954 (Fig. 4B, * = p < 0.05). Taken together, these data suggest that elevated EGFR localization to lipid rafts may correlate with resistance to EGFR TKI-induced growth inhibition.
Figure 4. EGFR localization to lipid rafts correlates with EGFR TKI resistance.
(A) One half million cells were plated, cultured 72 h, detergent-free lysis was performed and lipid rafts were separated by ultracentrifugation (Macdonald and Pike, 2005). Western blotting was performed with EGFR, transferrin receptor, caveolin-1, and flotillin antibodies. Fractions were dot blotted for GM-1 utilizing cholera toxin subunit B-HRP. Fractions 1-7 indicate lipid raft fractions. # indicates that the blots are representative of at least three independent experiments. (B) Densitometry was performed on western blots from A. Bars represent the percent of EGFR in lipid raft fractions (1-7) as compared to the total amount of EGFR present (1-14) from at least three independent experiments. Statistical analyses were performed utilizing a student's t-test, * = p<0.05 compared to SUM149 and HCC1954 cells. (C) Two hundred thousand cells were plated onto coverslips and cultured for 48 h. Coverslips containing cells were then incubated with Alexa-fluor 594 labeled cholera toxin subunit B (red) for 10 min on ice. Following incubation, cells were fixed, blocked in 20% goat serum, and incubated with immunofluorescent EGFR antibodies (extracellular domain epitope, green) and nuclei were stained with DAPI (blue). Imaging was performed using Zeiss Axioplan2 apotome microscope fitted with a 63X 1.25 oil immersion lens at the Microscopy and Imaging Resources Laboratory (Wayne State University, Detroit, MI). Arrows indicate areas of co-localization. Scale bars represent a distance of 50 μm. Images are representative of at least three independent experiments. Sens = EGFR TKI sensitive cell line and Res = EGFR TKI resistant cell line.
While lipid rafts are predominately found within the plasma membrane, there is evidence that they are also present in endosomes, lysosomes, and mitochondria (Galbiati et al., 2001). To determine if EGFR localized specifically within plasma membrane lipid rafts, we used immunofluorescent staining under non-permeabilizing conditions. Cholera toxin subunit B binds specifically to GM-1 and was used to detect localization of lipid rafts and EGFR was detected as described above. In the EGFR TKI resistant cell lines (SUM159, HCC1937, SUM229, and BT20), EGFR (green) co-localized (yellow/orange) with GM-1 (red) at the plasma membrane (Fig. 4C; arrows). In contrast, in the EGFR TKI sensitive cell lines (SUM149 and HCC1954), EGFR and GM-1 did not co-localize (Fig. 4C). These data suggested that EGFR localizes within plasma membrane lipid rafts in breast cancer cells that are resistant to EGFR TKI-induced growth inhibition.
Disruption of lipid rafts sensitizes breast cancer cells to EGFR inhibitors
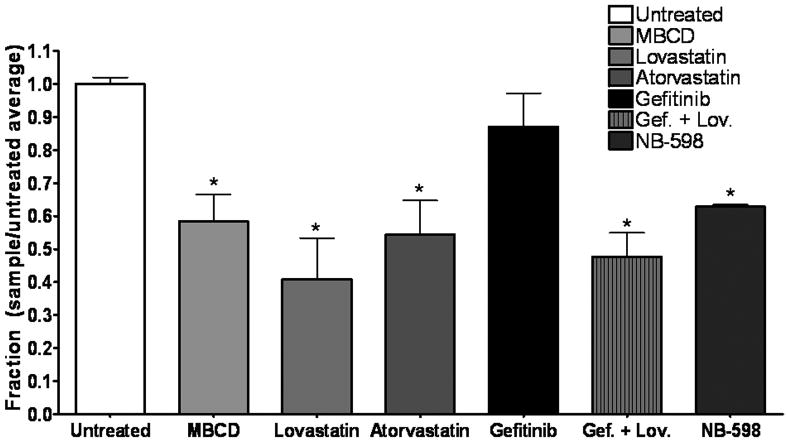
Cholesterol is the primary structural component of lipid rafts (reviewed in (Barenholz, 2002)), thus, to determine if the presence of EGFR in lipid rafts mediates cellular response to EGFR TKIs, we pharmacologically depleted cholesterol from the cells. HMG CoA-reductase inhibitors lovastatin and atorvastatin were used to reduce lipid raft cholesterol content (reviewed in (Simons and Toomre, 2000)). The Amplex Red cholesterol assay, which determines total cellular cholesterol content by measuring the amount of H2O2 produced by the reaction of cholesterol in the sample with cholesterol oxidase and cholesterol esterase enzymes, was utilized to determine the ability of these drugs to reduce cellular cholesterol (Fig. 5). Methyl-β cyclodextrin (MBCD), a cytotoxic cholesterol sequestering agent, reduced cholesterol by 41.5% +/- 8.1%, and was therefore used as a positive control for these experiments. Seventy-two hours of treatment with the HMG CoA reductase inhibitors lovastatin and atorvastatin resulted in depletion of cholesterol content, with a reduction of 59.0% +/-12.4% at 1.0 μM lovastatin and a reduction of 49.6% +/-10.3% at 1.0 μM atorvastatin (Fig. 5). Importantly, gefitinib treatment had no effect on cholesterol content of these cells, and did not alter the ability of lovastatin to reduce total cellular cholesterol (Fig. 5). The levels of cholesterol reduction produced by the statins are comparable with published results (Ehehalt et al., 2008; Sethy-Coraci et al., 2005).
Figure 5. MBCD, lovastatin, atorvastatin, and NB-598 reduce cholesterol in breast cancer cells.
Fifty thousand cells were plated into 6-well plates and treated with 1.0 mM MBCD (1 h), 1.0 μM lovastatin (72 h), 1.0 μM atorvastatin (72 h), 1.0 μM NB-598 (72 h), 1 μM gefitinib (1 h), or a combination of 1.0 μM gefitinib (1 h) and 1.0 μM lovastatin (72 h) (Gef. + Lov.) in growth medium. Lysis was followed by protein quantification and cholesterol was measured using the Amplex Red cholesterol assay kit. Absorbance was converted to μg cholesterol/mL utilizing a cholesterol standard curve, and then samples were normalized to protein concentration for a final value in μg cholesterol/μg protein. Bars represent fraction of cholesterol with untreated samples as 1 (μg cholesterol/μg protein sample)/(μg cholesterol/μg protein untreated). Experiments were repeated at least three times in duplicate. Error bars represent the standard error of the mean. Statistical analyses were performed utilizing Student's t-test, * = p < 0.05 compared to untreated.
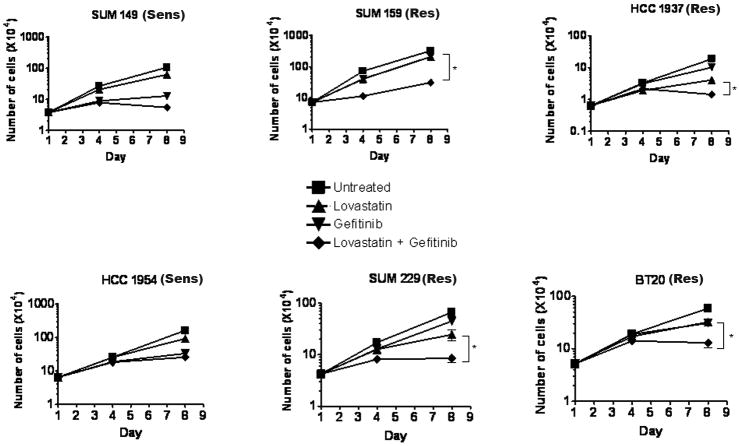
To determine if lovastatin has the ability to sensitize breast cancer cells to gefitinib, cell counting assays were used to measure proliferation. Cells were treated every other day with the drugs and counted on days 1, 4, and 8 (Fig. 6). As described previously, the four EGFR TKI resistant cell lines continued to proliferate in the presence of gefitinib. Interestingly, lovastatin was able to significantly reduce proliferation in the presence of gefitinib when compared to gefitinib or lovastatin treatment alone (Fig. 6; * = p<0.0001, ◆). Taken together, these data suggested that treatment with lovastatin sensitizes EGFR TKI resistant cell lines to gefitinib.
Figure 6. Lovastatin sensitizes EGFR TKI resistant breast cancer cells to gefitinib.
Thirty five thousand cells were plated into 6-well plates and treated for eight days with lovastatin and/or gefitinib in growth medium (HCC1954, SUM149 and SUM159 cell lines were treated with 1.0 μM of both lovastatin and gefitinib, while HCC1937, SUM229, and BT20 cells were treated with 5.0 μM lovastatin and 1.0 μM gefitinib). Cell number was determined on days 1, 4, and 8 using a coulter counter. Experiments were repeated at least three times in triplicate and counts were averaged. Error bars represent the standard error of the mean. Statistical analyses were performed utilizing Student's t-test, * = p < 0.0001 comparing day 8 counts between gefitinib alone or in combination with lovastatin. Sens = EGFR TKI sensitive cell line and Res = EGFR TKI resistant cell line.
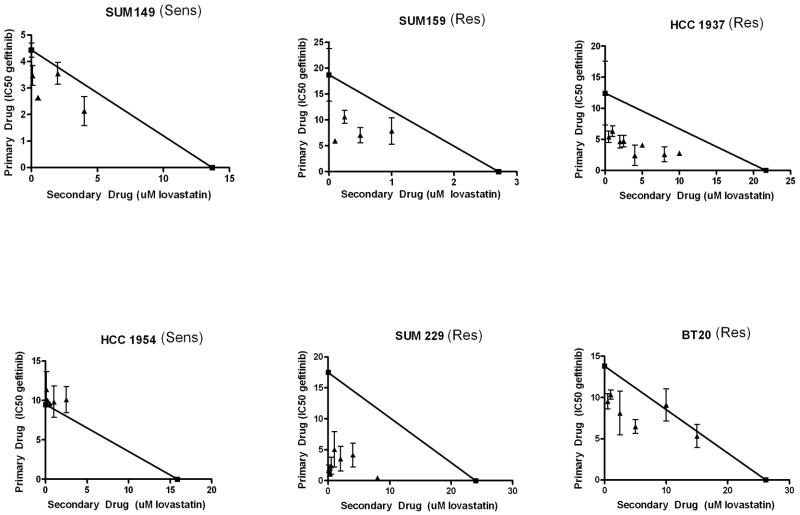
In order to determine if the effects of lovastatin and gefitinib were synergistic in EGFR TKI resistant breast cancer cells, cell viability assays were performed. Briefly, cells were treated for 72 h with the combination of lovastatin and gefitinib prior to performing tetrazolium-based cell viability assays. It can be noted that the IC50 values for cell viability analyses were much higher than doses found to be effective in cellular proliferation assays. While proliferation assays allow for the measurement of the number of cells over time, cell viability assays indicate the metabolic activity of the cell population. The IC50 of gefitinib was calculated at various doses of lovastatin, and then isobolograms were generated (Fig. 7). An additive interaction in SUM149 and HCC1954 cells was calculated from these assays (Fig. 7, points on the line). In contrast, synergistic effects were seen in all four EGFR TKI resistant cell lines (Fig. 7, points below the line). Combination index (CI) values were calculated based on the IC50 values (Table 2). These values were significantly lower than one in all of the EGFR TKI resistant cell lines. These results suggested that the combinatorial inhibition of lipid raft structure and EGFR-kinase activity resulted in a synergistic decrease in cell viability when EGFR is localized to lipid rafts. Therefore, the use of lovastatin and gefitinib in combination may effectively decrease viability and proliferation of breast cancers that contain EGFR within lipid rafts.
Figure 7. Lovastatin and gefitinib act synergistically to reduce breast cancer cell viability.
Two thousand cells were plated onto 96-well plates and treated for 72 h with varying doses of gefitinib and lovastatin alone and in combination in growth medium. Absorbance values were normalized. Non-linear regression (sigmoidal-dose response) curves were generated and IC50 values were calculated. The IC50 for gefitinib was plotted on the y-axis and the IC50 for lovastatin was plotted on the x-axis and a line was drawn between the two points. IC50 values were then calculated for gefitinib in the presence of each dose of lovastatin tested. These new IC50 values for gefitinib were plotted as a function of the dose of lovastatin used to calculate the value (triangles). Points that fall on the line represent additivity, while points that fall below the line represent synergy between the two drugs. Experiments were repeated at least three times in duplicate. Sens = EGFR TKI sensitive cell line and Res = EGFR TKI resistant cell line.
Table 2. Effects of lovastatin and gefitinib are synergistic in EGFR TKI resistant cell lines.
The combination index (CI-value) was calculated as follows: (IC50 gefitinib at X dose of lovastatin)/(IC50 gefitinib alone) + (dose of lovastatin)/(IC50 lovastatin alone). P-values were calculated as a difference between CI-value and one. P-values, calculated by student's t-test, less than 0.05 were considered significant.
| Cell Line | Lovastatin Dose | Combination Index (CI-value) |
p-value (compared to 1) |
|---|---|---|---|
| SUM159 | 0.25 μM | 0.659 +/-0.066 | 0.0262 |
| HCC1937 | 1.0 μM | 0.554 +/-0.069 | 0.0124 |
| SUM229 | 1.0 μM | 0.331 +/- 0.164 | 0.0269 |
| BT20 | 5.0 μM | 0.695 +/- 0.065 | 0.0348 |
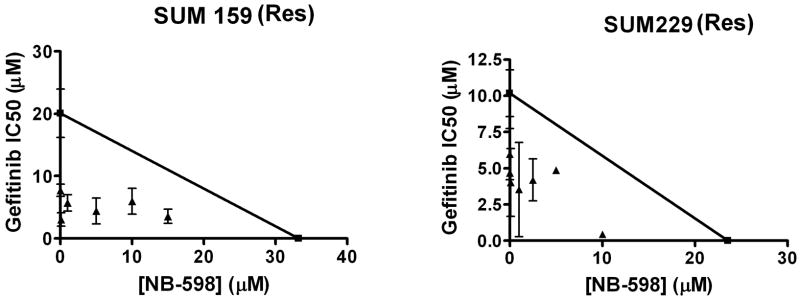
Statin drugs work by inhibiting HMG-CoA reductase. In addition to cholesterol biosynthesis, this enzyme also regulates isoprenoid synthesis. Therefore, in order to determine if the synergistic effect between lovastatin and gefitinib is mediated by the cholesterol depleting effects of the clinically relevant statin drug, the experimental drug NB-598 was used. NB-598 is a squalene monooxygenase inhibitor (Horie et al., 1990), and therefore inhibits cholesterol biosynthesis but not isoprenoid synthesis. First, to determine if NB-598 effectively inhibited cholesterol biosynthesis, SUM159 cells were treated with NB-598 for 72 h prior to assaying cholesterol esterase activity (Fig. 5). NB-598 treatment reduced cholesterol by 37.1% +/- 0.59%, suggesting that NB-598 depleted cholesterol to a level comparable to lovastatin. Therefore, we utilized NB-598 to determine if inhibiting cholesterol biosynthesis in the absence of altering isoprenoid synthesis has the ability to sensitize cells to gefitinib. EGFR TKI resistant breast cancer cells were treated with variable doses of NB-598 alone, or in combination with gefitinib. Cell viability assays were used to determine the IC50 of gefitinib at variable doses of NB-598. As shown in Figure 8, the effects of gefitinib and NB-598 were synergistic. These data suggest that cholesterol depletion alone is sufficient to sensitize EGFR TKI resistant cells to gefitinib.
Figure 8. The effects of NB-598 and gefitinib are synergistic.
Two thousand cells were plated onto 96-well plates and treated for 72 h with varying doses of gefitinib and NB-598 alone and in combination in growth medium. Values were normalized and then plotted. Non-linear regression (sigmoidal-dose response) curves were generated and IC50 values were calculated. The IC50 for gefitinib was plotted on the y-axis and the IC50 for NB-598 was plotted on the x-axis and a line was drawn between the two points. IC50 values were calculated for gefitinib in the presence of each dose of NB-598 tested. These new IC50 values for gefitinib were plotted as a function of the dose of NB-598 used to calculate the value (triangles). Points that fall on the line represent additivity, while points that fall below the line represent synergy between the two drugs. Experiments were repeated at least three times in duplicate. Sens = EGFR TKI sensitive cell line and Res = EGFR TKI resistant cell line.
Akt phosphorylation is abrogated with lipid raft disruption
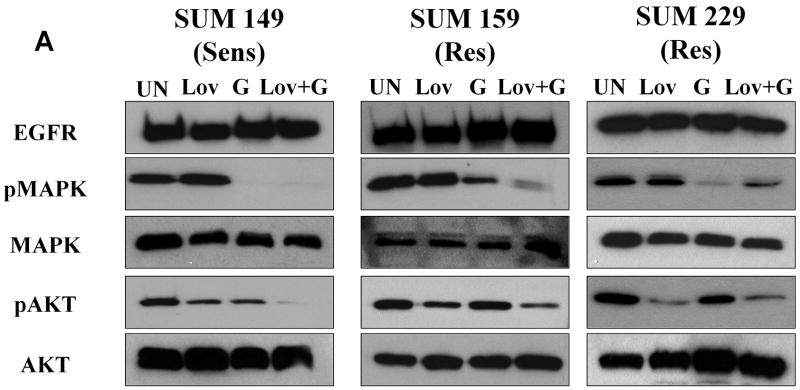
Resistance to EGFR TKIs suggests that inhibiting the EGFR kinase activity is insufficient to turn off growth and survival signaling in these cells. Localization of EGFR to lipid rafts has variable effects on signaling pathways downstream of EGFR (Chen and Resh, 2002; Li et al., 2006), thus we determined what effect depletion of cholesterol had on EGFR signaling in EGFR TKI resistant cells as compared to EGFR TKI sensitive cells. As discussed further below, BT20 cells contain a PIK3CA mutation, and the HCC1937 cell line has loss of PTEN expression, therefore, lovastatin did not affect a change in the phosphorylation of Akt in these cell lines (data not shown). Thus, two EGFR TKI resistant cell lines (SUM159 and SUM229) and one EGFR TKI sensitive cell line (SUM149) were treated with lovastatin and gefitinib alone or in combination and immunoblotting was performed to determine the phosphorylation of two key mediators of EGFR-induced survival and proliferative signaling, Akt and MAPK. Gefitinib treatment resulted in a reduction of MAPK phosphorylation in both the sensitive SUM149 cell line and two gefitinib resistant cell lines (SUM159 and SUM229). In contrast, Akt phosphorylation was inhibited in the EGFR TKI sensitive cell line yet persisted in the presence of gefitinib in EGFR TKI resistant cell lines (Fig. 9, lane 3). This phosphorylation persisted even after 72 h treatment with gefitinib (data not shown). When treated with lovastatin, alone or in combination with gefitinib, Akt phosphorylation was abrogated (Fig. 9, lanes 2 and 4). These data suggested that co-treatment of cells with lovastatin and gefitinib was able to inhibit two major EGFR signaling pathways. Thus, we propose that lipid rafts may provide a platform whereby EGFR may functionally interact with other proteins to activate downstream signaling pathways including Akt which function to modulate the response to EGFR TKIs.
Figure 9. Lovastatin inhibits Akt phosphorylation in EGFR TKI resistant cell lines.
One million cells were plated and allowed to grow for 48 h. Cells were then treated with 1 μM (SUM149 and SUM159) or 5 μM lovastatin (SUM229) for 72 h (LOV) and/or 1 μM gefitinib (G) for 1/2 h. Lysates were prepared and separated by SDS-PAGE. Immunoblotting using EGFR, Akt, MAPK, phospho-Akt, and phospho-MAPK antibodies was performed as described. Immunoblots are representative of at least three independent experiments. Sens = EGFR TKI sensitive cell line and Res = EGFR TKI resistant cell line.
Discussion
We have provided evidence describing a role for lipid rafts in resistance to EGFR TKI-induced growth inhibition using four EGFR expressing breast cancer cell lines which continue to proliferate in the presence of gefitinib, an EGFR TKI. We have shown that seven of thirteen EGFR-expressing breast cancer cell lines are resistant to EGFR TKI-induced growth inhibition, and that four of those cell lines retain the requirement of EGFR protein expression for growth. Also, we have provided evidence that EGFR localization to lipid rafts correlates with EGFR TKI resistance. Further, lovastatin, a HMG CoA reductase inhibitor, as well as NB-598, a squalene monooxygenase inhibitor reduced cholesterol biosynthesis in the EGFR TKI resistant breast cancer cells. In addition, lovastatin sensitized EGFR TKI resistant breast cancer cells to gefitinib-induced growth inhibition. Importantly, this sensitization of EGFR TKI growth resistant cells to gefitinib was determined to be synergistic for both lovastatin and NB-598. Our data suggests that lipid rafts provide a platform to promote survival and growth signaling in the presence of EGFR kinase inhibitors.
Overexpression of EGFR is one mechanism by which EGFR contributes to cancer progression. In fact, overexpression of EGFR occurs in glioblastomas, breast, prostate, ovary, liver, bladder, esophagus, larynx, stomach, colon, and lung cancers (Khazaie et al., 1993). This fairly ubiquitous overexpression suggests that EGFR may be an attractive target for cancer therapeutics. Inhibitors of EGFR kinase activity show clinical efficacy lung, pancreatic, colorectal, and head and neck cancers (Baker, 2004; Cohen et al., 2005; Giusti et al., 2008; Sobrero et al., 2008), however they have proven ineffective in the treatment of breast cancers (Blagosklonny and Darzynkiewicz, 2003; Twombly, 2005). We have provided evidence that EGFR expressing breast cancer cell lines differ in their response to these EGFR TKIs (Table 1). Seven of thirteen breast cancer cell lines were found to be resistant to EGFR TKI-induced growth inhibition using both cellular viability and proliferation assays. Specifically, SUM159, SUM229, BT20, BT549, HCC1937, MDA-MB231, and MDA-MB468 cell lines had IC50 values for gefitinib above 10 μM and continued to proliferate in the presence of 1 μM gefitinib (Table 1, Supplemental Fig. 1). These designations of resistance are consistent with previously published results in other cancer types (Helfrich et al., 2006; Noro et al., 2006; Ono et al., 2004; Witta et al., 2006).
EGFR expressing breast cancers are typically characterized as triple-negative breast cancers, which lack expression of estrogen receptor and progesterone receptor and do not contain HER2 amplification. Therefore, hormone therapy and HER2 targeted antibodies, which are currently in clinical use, are not effective in this population of breast cancer patients. Of the thirteen EGFR expressing breast cancer cell lines that were characterized herein for response to EGFR inhibitors, all thirteen were negative for estrogen and progesterone receptors, and lacked HER2 amplification (Lacroix and Leclercq, 2004; Neve et al., 2006). Taken together, these data support the need for targeted therapeutics for these triple negative, EGFR expressing breast cancers. Unfortunately, despite the expression of EGFR in triple-negative breast cancers, there is a disappointing lack of clinical efficacy of EGFR TKIs. A number of mechanisms have been suggested for resistance to EGFR TKI-induced growth inhibition in other cancers, including EGFR independence, mutations in EGFR and alterations in downstream signaling pathways. We have shown that three of seven EGFR TKI resistant breast cancer cell lines grow independently of EGFR protein expression, while four retain the requirement of EGFR expression for their proliferation (Fig. 2A-C). Mutations of EGFR, such as the VIII or T790M, have been implicated in glioblastomas and non-small cell lung cancers; however, these mutations are rare in breast tumors (Bianco et al., 2005). We have sequenced EGFR in the cell lines we used for our studies and no EGFR mutations were present (R. Haddad, personal communication). Here, we suggest that the localization of EGFR, specifically to lipid rafts, contributes to resistance to EGFR TKI-induced growth inhibition. Our data indicate that localization of EGFR to lipid rafts correlates with resistance to EGFR TKIs (Fig. 4). While EGFR has been suggested to also localize to caveolae (another cholesterol-rich membrane microdomain) (Mineo et al., 1999; Yamabhai and Anderson, 2002), biochemical raft isolation shows EGFR localizes primarily outside of caveolin-1 containing fractions in EGFR TKI resistant breast cancer cell lines (Fig. 4A). Although the majority of EGFR localizes to caveolin-1 negative fractions (fractions 1-7, Fig. 4A), we cannot exclude the possibility that caveolae may also play a role in resistance of these breast cancer cells to EGFR TKIs.
Lipid rafts have been suggested to play a functional role in cancer cell drug resistance. Depletion of lipid rafts through inhibition of fatty acid synthase (FAS) has been found to overcome trastuzumab resistance in breast cancer (Menendez et al., 2005). Specifically Her2/Neu co-localizes with lipid rafts in breast cancer cells, and the lipid environment of Her2/Neu-overexpressing cells influences the dimerization properties and signaling functions of Her2/Neu (Menendez et al., 2005). Furthermore, preclinical data suggest that lipid raft depletion via statins can decrease cell growth and sensitize cells to apoptotic stimuli in a number of cancer models including melanoma, prostate, and HER2-overexpressing breast cancers (Glynn et al., 2008; Herrero-Martin and Lopez-Rivas, 2008; Li et al., 2006). Epidemiologic data regarding the use of statins as singular agents in breast cancer are mixed (Beck et al., 2003; Cauley et al., 2003; Kwan et al., 2008). The apparent in vitro benefit of combining statins with other therapies suggests that statins may have a greater clinical benefit when utilized as a part of combinatorial therapies (Katz, 2005). In that regard, we have shown that cholesterol depletion synergizes with gefitinib in four EGFR TKI resistant breast cancer cell lines (Fig. 7 and 8, Table 2). Specifically, co-treatment of these cell lines with lovastatin and gefitinib significantly reduces cell proliferation compared to either drug alone (Fig. 6). Also, when CI-values were determined for the combination of cholesterol inhibitors and gefitinib, all four cell lines resistant to EGFR TKI-induced growth inhibition showed synergy (Table 2, Figs. 7 and 8). Thus, in breast cancer cells resistant to EGFR TKI-induced growth inhibition, EGFR is commonly localized to lipid rafts, and our data indicate that this localization plays a functional role in such resistance.
Failure to inhibit Akt signaling, due to mutation or loss of PTEN, constitutive activation of PI3K, or overexpression of Akt, has also been shown to be a mechanism of resistance to EGFR TKI-induced growth inhibition (Cheng et al., 1992; Hollestelle et al., 2007; Lu et al., 1999). Of the cell lines that retain the requirement of EGFR protein expression for growth, but are EGFR TKI resistant, one has a PIK3CA mutation (BT20), and one has loss of PTEN expression (HCC1937) suggesting that the PI3K/Akt pathway may be important in the tumorigenicity of these cell lines (Hollestelle et al., 2007). Indeed, Akt phosphorylation persists in the absence of EGFR kinase activity in these two cell lines and lovastatin had no effect on Akt phosphorylation (data not shown). Two other EGFR TKI resistant cell lines (SUM159 and SUM229) do not contain genetic mutations in the Akt pathway, yet retain Akt phosphorylation in the presence of gefitinib (Fig. 9, lane 3). Lovastatin treatment was sufficient to abrogate this phosphorylation in the SUM159 and SUM229 cell lines, suggesting that lipid rafts play a role in the regulation of Akt phosphorylation in a subset of EGFR TKI resistant cells (Fig. 9, lanes 2 and 4). Specifically, we suggest that lipid rafts provide a platform for Akt signaling, even in the presence of an EGFR TKI. However, as EGFR signaling is mediated by many more proteins than addressed here, it is possible that other pathways may also be downstream of EGFR-kinase independent, lipid raft-dependent activation. Nevertheless, localization of EGFR to lipid rafts is an important factor in the resistance of breast cancer cells to EGFR TKI-induced growth inhibition.
Our data suggest that the synergistic mechanism between lovastatin and gefitinib in breast cancer cells is due to depletion of cholesterol and thereby depletion of lipid rafts. However, it is important to note that while statin use has been a common method to deplete cells of lipid raft structure for many years, the mechanism of action of statin drugs is not solely through the reduction of cholesterol. Statin treatment and consequent reduction of HMG-CoA reductase activity also inhibits protein prenylation. Indeed, previous studies have demonstrated that lovastatin can potentiate the effects of gefitinib (and vice versa) in squamous cell carcinoma, non-small cell lung cancer, colon carcinoma, and glioblastoma cell lines due to decreased protein prenylation (Cemeus et al., 2008; Lacroix and Leclercq, 2004; Mantha et al., 2005; Mantha et al., 2003; Park et al., 2009). Specifically, in 2003 Mantha and colleagues combined gefitinib and lovastatin in head and neck cancer cell lines and found a synergistic interaction between these drugs due, at least in part, to protein prenylation (Mantha et al., 2003). This group later showed a synergistic interaction with this drug pairing in cervical and non-small cell lung cancers in addition to recapitulating their findings in head and neck cancer. In that manuscript, the effects of lovastatin are completely attributed to protein prenylation (Mantha et al., 2005). Further, researchers have described such an interaction between lovastatin and gefitinib in glioblastoma and non-small cell lung cancer, again attributing their effect to protein prenylation (Cemeus et al., 2008; Park et al., 2009). Most recently, Zhao and colleagues have proposed that EGFR dimerization is inhibited by treatment with lovastatin, an effect dependent on aberrant prenylation of RhoA (Lacroix and Leclercq, 2004). While all of these groups show a functional interaction between lovastatin and gefitinib, they do not eliminate the possibility that EGFR localization to lipid rafts is a potential mechanism of this effect. We have shown clearly that the synergistic interaction between lovastatin and gefitinib in breast cancer is due to cholesterol inhibition, as both lovastatin and the squalene monooxygenase inhibitor NB-598 were sufficient to sensitize EGFR TKI resistant breast cancer cells to gefitinib (Figs. 7 and 8). Taken together, these results suggest that the effects of lovastatin treatment in our study are due to cholesterol modulation and subsequent lipid raft impairment rather than decreased protein prenylation.
We have demonstrated that EGFR localizes to lipid rafts in EGFR expressing, EGFR TKI resistant, breast cancer cell lines. We have provided evidence that reducing cholesterol biosynthesis sensitizes these EGFR TKI resistant cells to the EGFR TKI gefitinib. We have shown that cholesterol reducing drugs and gefitinib act synergistically to decrease cell viability in breast cancer cells that are resistant to EGFR TKI-induced growth inhibition. We have confirmed that cholesterol depletion, rather than protein prenylation, results in a synergistic effect with gefitinib in these cells. Mechanistically while gefitinib effectively reduced MAPK phosphorylation in EGFR TKI resistant cell lines, Akt phosphorylation persisted. Lovastatin was sufficient to abrogate this phosphorylation of Akt in two of the EGFR TKI resistant cell lines. As EGFR kinase activity is completely inhibited by gefitinib treatment in these cells (Mueller et al., 2008), we hypothesize that lipid rafts provide a platform by which EGFR interacts with other proteins to phosphorylate EGFR in the presence of EGFR TKIs and activate signaling pathways including the Akt pathway. Thus, as both statin drugs and gefitinib are well tolerated and approved for use in patients, the work herein provides rationale for further exploration of the combination of these drugs in breast cancers that are resistant to EGFR TKI-induced growth inhibition.
Supplementary Material
Supplemental Figure 1: Sensitivity of breast cancer cell lines to EGFR TKI-induced growth inhibition. Cellular proliferation assays were performed in the presence of increasing doses of gefitinib for eight days. Cell counts were taken on days 1, 4, and 8. Error bars represent the standard error of the mean. Proliferation assays were performed in triplicate and repeated at least three times. (A) Sensitive cells (B) Resistant cells.
Supplemental Figure 2: Resistance of breast cancer cell lines to CI-1033, an irreversible EGFR inhibitor. Cellular proliferation assays were performed in the presence of increasing doses of CI-1033 for eight days. Cell counts were taken on days 1, 4, and 8. Error bars represent the standard error of the mean. Proliferation assays were performed in triplicate and repeated at least three times.
Acknowledgments
*We are grateful to Thomas Kocarek, PhD (Wayne State University, Detroit, MI) for technical assistance regarding NB-598. We thank the Microscopy & Imaging Resources Laboratory Shared Resource facility for providing services. The Microscopy and Imaging Resources Laboratory is supported, in part, by NIH Center grants P30ES06639 to the Institute of Environmental Health Sciences, P30CA22453 to The Karmanos Cancer Institute, Wayne State University, and U54RR020843 to The Center for Proteolytic Pathways, the Burnham Institute, and the Perinatology Research Branch of the National Institutes of Child Health and Development, Wayne State University. We would also like to thank the Translational Research Core Laboratory at the Karmanos Cancer Institute for their help with the processing of cholesterol assays. This work was supported by NCI T32-CA009531 (MEI) and Susan G. Komen for the Cure Career Catalyst Grant (KG081416; JLB).
Contract grant sponsor: NCI; Contract grant number: T32-CA009531 (MEI)
Contract grant sponsor: Susan G. Komen for the Cure; Contract grant number KG081416 (JLB).
The abbreviations used are
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinase
- FAS
fatty acid synthase
- MAPK
mitogen-activated protein kinase
- MBCD
methyl-beta cyclodextrin
- MOI
multiplicity of infection
- TKI
tyrosine kinase inhibitor
References
- Baker M. EGFR inhibitors square off at ASCO. Nat Biotechnol. 2004;22(6):641. doi: 10.1038/nbt0604-641. [DOI] [PubMed] [Google Scholar]
- Barenholz Y. Cholesterol and other membrane active sterols: from membrane evolution to “rafts”. Prog Lipid Res. 2002;41(1):1–5. doi: 10.1016/s0163-7827(01)00016-9. [DOI] [PubMed] [Google Scholar]
- Beck P, Wysowski DK, Downey W, Butler-Jones D. Statin use and the risk of breast cancer. J Clin Epidemiol. 2003;56(3):280–285. doi: 10.1016/s0895-4356(02)00614-5. [DOI] [PubMed] [Google Scholar]
- Bianco R, Troiani T, Tortora G, Ciardiello F. Intrinsic and acquired resistance to EGFR inhibitors in human cancer therapy. Endocr Relat Cancer. 2005;12 1:S159–171. doi: 10.1677/erc.1.00999. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Darzynkiewicz Z. Why Iressa failed: toward novel use of kinase inhibitors (outlook) Cancer Biol Ther. 2003;2(2):137–140. doi: 10.4161/cbt.2.2.286. [DOI] [PubMed] [Google Scholar]
- Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24(16):7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla M, Chedin M, Souvignet C, Marron J, Arnould C, Chambaz E. Estimation of epidermal growth factor receptor in 177 breast cancers: correlation with prognostic factors. Breast Cancer Res Treat. 1990;16(2):97–102. doi: 10.1007/BF01809293. [DOI] [PubMed] [Google Scholar]
- Cauley JA, Zmuda JM, Lui LY, Hillier TA, Ness RB, Stone KL, Cummings SR, Bauer DC. Lipid-lowering drug use and breast cancer in older women: a prospective study. J Womens Health (Larchmt) 2003;12(8):749–756. doi: 10.1089/154099903322447710. [DOI] [PubMed] [Google Scholar]
- Cemeus C, Zhao TT, Barrett GM, Lorimer IA, Dimitroulakos J. Lovastatin enhances gefitinib activity in glioblastoma cells irrespective of EGFRvIII and PTEN status. J Neurooncol. 2008;90(1):9–17. doi: 10.1007/s11060-008-9627-0. [DOI] [PubMed] [Google Scholar]
- Chen X, Resh MD. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J Biol Chem. 2002;277(51):49631–49637. doi: 10.1074/jbc.M208327200. [DOI] [PubMed] [Google Scholar]
- Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, Tsichlis PN, Testa JR. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci U S A. 1992;89(19):9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MH, Johnson JR, Chen YF, Sridhara R, Pazdur R. FDA drug approval summary: erlotinib (Tarceva) tablets. Oncologist. 2005;10(7):461–466. doi: 10.1634/theoncologist.10-7-461. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Valero V, Mangalik A, Royce M, Rabinowitz I, Arena FP, Kroener JF, Curcio E, Watkins C, Bacus S, Cora EM, Anderson E, Magill PJ. Phase II, randomized trial to compare anastrozole combined with gefitinib or placebo in postmenopausal women with hormone receptor-positive metastatic breast cancer. Clin Cancer Res. 2010;16(6):1904–1914. doi: 10.1158/1078-0432.CCR-09-2282. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehehalt R, Krautter M, Zorn M, Sparla R, Fullekrug J, Kulaksiz H, Stremmel W. Increased basolateral sorting of carcinoembryonic antigen in a polarized colon carcinoma cell line after cholesterol depletion-Implications for treatment of inflammatory bowel disease. World J Gastroenterol. 2008;14(10):1528–1533. doi: 10.3748/wjg.14.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Razani B, Lisanti MP. Emerging themes in lipid rafts and caveolae. Cell. 2001;106(4):403–411. doi: 10.1016/s0092-8674(01)00472-x. [DOI] [PubMed] [Google Scholar]
- Giusti RM, Shastri K, Pilaro AM, Fuchs C, Cordoba-Rodriguez R, Koti K, Rothmann M, Men AY, Zhao H, Hughes M, Keegan P, Weiss KD, Pazdur R. U.S. Food and Drug Administration approval: panitumumab for epidermal growth factor receptor-expressing metastatic colorectal carcinoma with progression following fluoropyrimidine-, oxaliplatin-, and irinotecan-containing chemotherapy regimens. Clin Cancer Res. 2008;14(5):1296–1302. doi: 10.1158/1078-0432.CCR-07-1354. [DOI] [PubMed] [Google Scholar]
- Glynn SA, O'Sullivan D, Eustace AJ, Clynes M, O'Donovan N. The 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors, simvastatin, lovastatin and mevastatin inhibit proliferation and invasion of melanoma cells. BMC Cancer. 2008;8:9. doi: 10.1186/1471-2407-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Opekarova M, Novakova L, Stolz J, Tanner W. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5(6):945–953. doi: 10.1128/EC.00206-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix M, Granja Nde M, Meszoely I, Adkins TB, Wieman BM, Frierson KE, Sanchez V, Sanders ME, Grau AM, Mayer IA, Pestano G, Shyr Y, Muthuswamy S, Calvo B, Krontiras H, Krop IE, Kelley MC, Arteaga CL. Short preoperative treatment with erlotinib inhibits tumor cell proliferation in hormone receptor-positive breast cancers. J Clin Oncol. 2008;26(6):897–906. doi: 10.1200/JCO.2007.13.5939. [DOI] [PubMed] [Google Scholar]
- Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141(4):929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazarika P, McCarty MF, Prieto VG, George S, Babu D, Koul D, Bar-Eli M, Duvic M. Up-regulation of Flotillin-2 is associated with melanoma progression and modulates expression of the thrombin receptor protease activated receptor 1. Cancer Res. 2004;64(20):7361–7369. doi: 10.1158/0008-5472.CAN-04-0823. [DOI] [PubMed] [Google Scholar]
- Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, Coldren C, Baron A, Zeng C, Franklin WA, Hirsch FR, Gazdar A, Minna J, Bunn PA., Jr Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12(23):7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- Herrero-Martin G, Lopez-Rivas A. Statins activate a mitochondria-operated pathway of apoptosis in breast tumor cells by a mechanism regulated by ErbB2 and dependent on the prenylation of proteins. FEBS Lett. 2008;582(17):2589–2594. doi: 10.1016/j.febslet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- Hollestelle A, Elstrodt F, Nagel JH, Kallemeijn WW, Schutte M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res. 2007;5(2):195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- Horie M, Tsuchiya Y, Hayashi M, Iida Y, Iwasawa Y, Nagata Y, Sawasaki Y, Fukuzumi H, Kitani K, Kamei T. NB-598: a potent competitive inhibitor of squalene epoxidase. J Biol Chem. 1990;265(30):18075–18078. [PubMed] [Google Scholar]
- Katz MS. Therapy insight: Potential of statins for cancer chemoprevention and therapy. Nat Clin Pract Oncol. 2005;2(2):82–89. doi: 10.1038/ncponc0097. [DOI] [PubMed] [Google Scholar]
- Khazaie K, Schirrmacher V, Lichtner RB. EGF receptor in neoplasia and metastasis. Cancer Metastasis Rev. 1993;12(3-4):255–274. doi: 10.1007/BF00665957. [DOI] [PubMed] [Google Scholar]
- Kwan ML, Habel LA, Flick ED, Quesenberry CP, Caan B. Post-diagnosis statin use and breast cancer recurrence in a prospective cohort study of early stage breast cancer survivors. Breast Cancer Res Treat. 2008;109(3):573–579. doi: 10.1007/s10549-007-9683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. Relevance of Breast Cancer Cell Lines as Models for Breast Tumours: An Update. Breast Cancer Research and Treatment. 2004;83(3):249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- Li YC, Park MJ, Ye SK, Kim CW, Kim YN. Elevated levels of cholesterol-rich lipid rafts in cancer cells are correlated with apoptosis sensitivity induced by cholesterol-depleting agents. Am J Pathol. 2006;168(4):1107–1118. doi: 10.2353/ajpath.2006.050959. quiz 1404-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3(9):802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun R, Wan W, Wang J, Oppenheim JJ, Chen L, Zhang N. The involvement of lipid rafts in epidermal growth factor-induced chemotaxis of breast cancer cells. Mol Membr Biol. 2007;24(2):91–101. doi: 10.1080/10929080600990500. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang X, Yu SX, Davies MA, Khan H, Furui T, Mao M, Zinner R, Hung MC, Steck P, Siminovitch K, Mills GB. The PTEN/MMAC1/TEP tumor suppressor gene decreases cell growth and induces apoptosis and anoikis in breast cancer cells. Oncogene. 1999;18(50):7034–7045. doi: 10.1038/sj.onc.1203183. [DOI] [PubMed] [Google Scholar]
- Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc Natl Acad Sci U S A. 1995;92(15):6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JL, Pike LJ. A simplified method for the preparation of detergent-free lipid rafts. J Lipid Res. 2005;46(5):1061–1067. doi: 10.1194/jlr.D400041-JLR200. [DOI] [PubMed] [Google Scholar]
- Mantha AJ, Hanson JE, Goss G, Lagarde AE, Lorimer IA, Dimitroulakos J. Targeting the mevalonate pathway inhibits the function of the epidermal growth factor receptor. Clin Cancer Res. 2005;11(6):2398–2407. doi: 10.1158/1078-0432.CCR-04-1951. [DOI] [PubMed] [Google Scholar]
- Mantha AJ, McFee KE, Niknejad N, Goss G, Lorimer IA, Dimitroulakos J. Epidermal growth factor receptor-targeted therapy potentiates lovastatin-induced apoptosis in head and neck squamous cell carcinoma cells. J Cancer Res Clin Oncol. 2003;129(11):631–641. doi: 10.1007/s00432-003-0490-2. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Vellon L, Lupu R. Targeting fatty acid synthase-driven lipid rafts: a novel strategy to overcome trastuzumab resistance in breast cancer cells. Med Hypotheses. 2005;64(5):997–1001. doi: 10.1016/j.mehy.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Mineo C, Gill GN, Anderson RG. Regulated migration of epidermal growth factor receptor from caveolae. J Biol Chem. 1999;274(43):30636–30643. doi: 10.1074/jbc.274.43.30636. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68(9):3314–3322. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro R, Gemma A, Kosaihira S, Kokubo Y, Chen M, Seike M, Kataoka K, Matsuda K, Okano T, Minegishi Y, Yoshimura A, Kudoh S. Gefitinib (IRESSA) sensitive lung cancer cell lines show phosphorylation of Akt without ligand stimulation. BMC Cancer. 2006;6:277. doi: 10.1186/1471-2407-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, Fujii T, Kuwano M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3(4):465–472. [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Park I, Kim J, Jung J, Han JY. Lovastatin overcomes gefitinib resistance in human non-small cell lung cancer cells with K-Ras mutations. Investigational New Drugs. 2009:1–9. doi: 10.1007/s10637-009-9319-4. [DOI] [PubMed] [Google Scholar]
- Peres C, Yart A, Perret B, Salles JP, Raynal P. Modulation of phosphoinositide 3-kinase activation by cholesterol level suggests a novel positive role for lipid rafts in lysophosphatidic acid signalling. FEBS Lett. 2003;534(1-3):164–168. doi: 10.1016/s0014-5793(02)03832-2. [DOI] [PubMed] [Google Scholar]
- Polychronis A, Sinnett HD, Hadjiminas D, Singhal H, Mansi JL, Shivapatham D, Shousha S, Jiang J, Peston D, Barrett N, Vigushin D, Morrison K, Beresford E, Ali S, Slade MJ, Coombes RC. Preoperative gefitinib versus gefitinib and anastrozole in postmenopausal patients with oestrogen-receptor positive and epidermal-growth-factor-receptor-positive primary breast cancer: a double-blind placebo-controlled phase II randomised trial. Lancet Oncol. 2005;6(6):383–391. doi: 10.1016/S1470-2045(05)70176-5. [DOI] [PubMed] [Google Scholar]
- Roepstorff K, Thomsen P, Sandvig K, van Deurs B. Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibits ligand binding. J Biol Chem. 2002;277(21):18954–18960. doi: 10.1074/jbc.M201422200. [DOI] [PubMed] [Google Scholar]
- Sadowski L, Pilecka I, Miaczynska M. Signaling from endosomes: location makes a difference. Exp Cell Res. 2009;315(9):1601–1609. doi: 10.1016/j.yexcr.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Sainsbury JR, Farndon JR, Needham GK, Malcolm AJ, Harris AL. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet. 1987;1(8547):1398–1402. doi: 10.1016/s0140-6736(87)90593-9. [DOI] [PubMed] [Google Scholar]
- Sethy-Coraci I, Crock LW, Silverstein SC. PAF-receptor antagonists, lovastatin, and the PTK inhibitor genistein inhibit H2O2 secretion by macrophages cultured on oxidized-LDL matrices. J Leukoc Biol. 2005;78(5):1166–1174. doi: 10.1189/jlb.0205101. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387(6633):569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, Vega-Villegas ME, Eng C, Steinhauer EU, Prausova J, Lenz HJ, Borg C, Middleton G, Kroning H, Luppi G, Kisker O, Zubel A, Langer C, Kopit J, Burris HA., 3rd EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(14):2311–2319. doi: 10.1200/JCO.2007.13.1193. [DOI] [PubMed] [Google Scholar]
- Toi M, Osaki A, Yamada H, Toge T. Epidermal growth factor receptor expression as a prognostic indicator in breast cancer. Eur J Cancer. 1991;27(8):977–980. doi: 10.1016/0277-5379(91)90262-c. [DOI] [PubMed] [Google Scholar]
- Twombly R. Failing survival advantage in crucial trial, future of Iressa is in jeopardy. J Natl Cancer Inst. 2005;97(4):249–250. doi: 10.1093/jnci/97.4.249. [DOI] [PubMed] [Google Scholar]
- Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA., Jr Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66(2):944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89(7):2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M, Anderson RG. Second cysteine-rich region of epidermal growth factor receptor contains targeting information for caveolae/rafts. J Biol Chem. 2002;277(28):24843–24846. doi: 10.1074/jbc.C200277200. [DOI] [PubMed] [Google Scholar]
- Zhuang L, Lin J, Lu ML, Solomon KR, Freeman MR. Cholesterol-rich lipid rafts mediate akt-regulated survival in prostate cancer cells. Cancer Res. 2002;62(8):2227–2231. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Sensitivity of breast cancer cell lines to EGFR TKI-induced growth inhibition. Cellular proliferation assays were performed in the presence of increasing doses of gefitinib for eight days. Cell counts were taken on days 1, 4, and 8. Error bars represent the standard error of the mean. Proliferation assays were performed in triplicate and repeated at least three times. (A) Sensitive cells (B) Resistant cells.
Supplemental Figure 2: Resistance of breast cancer cell lines to CI-1033, an irreversible EGFR inhibitor. Cellular proliferation assays were performed in the presence of increasing doses of CI-1033 for eight days. Cell counts were taken on days 1, 4, and 8. Error bars represent the standard error of the mean. Proliferation assays were performed in triplicate and repeated at least three times.