SUMMARY
Interactions with fibronectin are important in the virulence strategies of a range of disease-related bacteria. The periodontitis-associated oral spirochaete Treponema denticola expresses at least two fibronectin-binding proteins, designated Msp (major surface protein) and OppA (oligopeptide-binding protein homologue). To identify other T. denticola outer membrane fibronectin-binding proteins, the amino acid sequence of the Treponema pallidum fibronectin-binding protein Tp0155 was used to survey the T. denticola genome. Seven T. denticola genes encoding orthologous proteins were identified. All but two were expressed in Escherichia coli and purified recombinant proteins bound fibronectin. Using antibodies to the N-terminal region of Tp0155, it was demonstrated that T. denticola TDE2318, with highest homology to Tp0155, was cell surface localized. Like Tp0155, the seven T. denticola proteins contained an M23 peptidase domain and four (TDE2318, TDE2753, TDE1738, TDE1297) contained one or two LysM domains. M23 peptidases can degrade peptidoglycan whereas LysM domains recognize carbohydrate polymers. In addition, TDE1738 may act as a bacteriocin based on homology with other bacterial lysins and the presence of an adjacent gene encoding a putative immunity factor. Collectively, these results suggest that T. denticola expresses fibronectin-binding proteins associated with the cell surface that may also have cell wall modifying or lytic functions.
Keywords: fibronectin, LysM, M23 peptidase, periodontal disease, Treponema
INTRODUCTION
Periodontal disease is prevalent within the human population and results from complex communities of microorganisms, combined with the host inflammatory response, causing damage to gingival and supporting tissues. Periodontal pockets, which are indicative of periodontitis, enable the persistence of anaerobic conditions, which facilitate growth of organisms including Fusobacterium, Porphyromonas, Prevotella and Treponema species. Recent advances in understanding the growth requirements of oral Treponema have allowed the isolation and characterization of a number of different species associated with periodontal disease (Choi et al., 1994). However, there is evidence that many not yet cultivable treponemes are also present at diseased sites (Choi et al., 1994; Dewhirst et al., 2000). Possibly the best characterized of the disease-associated oral spirochaetes is Treponema denticola. There is strong evidence linking this pathogenic treponeme with disease initiation and progression. Gingivitis was more likely to develop from healthy sites where T. denticola was detected, than when it was absent (Riviere et al., 1998). Treponema denticola is most regularly detected in deep periodontal pockets and evidence suggests that it persists predominantly towards the leading edge of subgingival plaque providing direct contact with periodontal tissues (Kigure et al., 1995).
Damage and inflammation within infected periodontal pockets leads to host macromolecules, such as extracellular matrix and plasma components such as fibronectin, becoming accessible to colonizing bacteria. Fibronectin is a vital matrix component, existing either as an insoluble glycoprotein, which polymerizes to form part of the extracellular matrix thereby adding structural support to host tissues (matrix fibronectin), or as a soluble disulphide-linked plasma protein with a role in wound healing (Couchman et al., 1990). Both matrix and plasma fibronectin are found in abundance during blood clotting, tissue repair, cell migration and embryogenesis, as occurs within periodontal pockets.
Adhesion to, or degradation of, fibronectin is important in the virulence strategies of a number of pathogenic organisms. Adhesion promotes colonization (Pasula et al., 2002; Pracht et al., 2005), whereas proteolysis provides a source of peptides (Fenno et al., 2000; Samen et al., 2004) or may hamper or obstruct wound-healing processes (Heilmann et al., 2004). The capacity to avoid host immune defence mechanisms by penetrating host tissues is also enhanced by interactions with fibronectin (Dziewanowska et al., 1999; Sinha et al., 1999). Adherence to either matrix or plasma fibronectin could therefore be an essential resource in the pathogenesis of T. denticola.
Fibronectin-binding by T. denticola was first described by Dawson & Ellen (1990) and was later quantified (Haapasalo et al., 1991). Fluid-phase, but not immobilized, forms of fibronectin were bound by an OppA (oligopeptide-binding protein) homologue, involved in internalization of oligopeptides for nutrition (Fenno et al., 2000). Immunoblot overlays (Umemoto et al., 1993) suggested that T. denticola expressed three outer membrane fibronectin-binding proteins. The same technique was used by Haapasalo et al. (1992) in characterization of the 53-kDa major surface protein (Msp) of T. denticola ATCC 35405. Further analysis of Msp functions (Fenno et al., 1996; Edwards et al., 2005) suggested involvement of this surface-associated protein in fibronectin interactions.
The causative agent of syphilis, Treponema pallidum, interacts with fibronectin (Peterson et al., 1983; Thomas et al., 1986) but the molecular mechanisms involved are not well understood. Two fibronectin-binding proteins were identified in T. pallidum using a bioinformatics approach. Ten genes encoding potential outer membrane proteins were cloned and expressed and the recombinant proteins were purified (Cameron, 2003) and characterized for their abilities to bind plasma or matrix forms of fibronectin (Cameron et al., 2004). Two proteins, designated Tp0155 and Tp0483, bound fibronectin although Tp0155 bound preferentially to matrix fibronectin whereas Tp0483 showed greater attachment to the plasma form.
Comparisons between the T. pallidum Nichols and T. denticola ATCC 35405 genomes have become possible because they have both been fully sequenced and annotated. Analysis of open reading frames suggests that T. pallidum and T. denticola are derived ancestrally. The T. denticola genome contains 2786 open reading frames and T. pallidum carries 1039 open reading frames. This suggests that gene elimination may have occurred in T. pallidum, whereas duplication and expansion occurred in the T. denticola genome (Seshadri et al., 2004). It is also interesting to note that the T. denticola genome is predicted to encode 156 lipoproteins but T. pallidum encodes only 16. This may reflect the fact that T. denticola has adapted to grow and survive in an oral environment where there is strong competition with other microorganisms.
It has previously been shown that Msp has a role in mediating T. denticola binding to fibronectin (Haapasalo et al., 1992; Fenno et al., 1996; Edwards et al., 2005). However, an isogenic Msp-deficient strain (Fenno et al., 1998) was only reduced by ~ 30% in adherence to fibronectin (C.V. Bamford, unpublished data), suggesting that other factors were present that bound fibronectin. As the OppA homologue is reported to bind only fluid-phase fibronectin, additional fibronectin-binding proteins may be expressed on the surface of T. denticola. In this article we describe a family of T. denticola proteins with homology to a T. pallidum fibronectin-binding protein, Tp0155, (Cameron et al., 2004) was identified. These proteins are able to bind both matrix and plasma fibronectin, and protein sequence characteristics suggest a possible multifunctional role in T. denticola pathogenesis.
METHODS
Bacterial strains and growth conditions
Treponema denticola ATCC 35405 was maintained in pre-reduced New Oral Spirochete medium (Bamford et al., 2007) at 37°C in an anaerobic atmosphere (80% N2: 10% H2: 10% CO2). Cultures were grown for 3–4 days to late exponential phase, corresponding to an optical density (OD600) of 0.4–0.6, and were harvested by centrifugation (10,000 g, 10 min). Escherichia coli XL-1 Blue was used as host to prepare plasmid DNA, and E. coli M15 was used for production of recombinant polypeptides from pQE30 plasmids (Table 1). The E. coli was cultured on Luria Bertani (LB) agar (Sambrook et al., 1989) or in LB broth, containing appropriate antibiotics (ampicillin, 100μg ml−1; kanamycin 25μg ml−1).
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant characteristics | Source |
|---|---|---|
| Treponema denticola | ||
| ATCC 35405 | Type strain | (Chan et al., 1993) |
| Escherichia coli | ||
| XL-1 Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac [F ‘proAB laclqZΔM15 Tn10 (TetR)] | Stratagene |
| M15 | (pREP-4 KnR) nals strs rifs thi− lac− ara+ gal+ mtl−F− recA+ uvr+ lon+ | Qiagen |
| Plasmids | ||
| pQE30 | bla ColE1ori (ApR) | Qiagen |
| pQE30-TDE2318 | bla ColE1ori | This study |
| pQE30-TDE2753 | bla ColE1ori | This study |
| pQE30-TDE1738 | bla ColE1ori | This study |
| pQE30-TDE0110 | bla ColE1ori | This study |
| pQE30-TDE2189 | bla ColE1ori | This study |
| pQE30-TDE1136 | bla ColE1ori | This study |
| pQE30-TDE1297 | bla ColE1ori | This study |
Bioinformatics analyses
Treponema denticola ATCC 35405 and T. pallidum Nichols peptide sequences were acquired from the TIGR Comprehensive Microbial Resource (http://cmr.tigr.org.tigr-scripts/cmr/CmrHomePage.cgi) and analysed using a number of in silico investigations. BLAST analyses to identify homologous peptide sequences were achieved using appropriate tools at http://cmr.tigr.org.tigr-scripts/cmr/CmrHomePage.cgi. Properties of individual peptide sequences were studied to assess peptide mass (www.us.expasy.org/cgi-bin/peptide-mass.html) and presence of signal sequence (http://bioinformatcs.leeds.ac.uk/prot_analysis/signal.html) and to examine other structural or functional domains (www.ebi.ac.uk/InterProScan/) within the peptide sequence.
Expression and purification of recombinant Tp0155 orthologues
Coding sequences of T. denticola DNA that corresponded to potential orthologues of T. pallidum Tp0155 were identified from predicted open reading frames of the T. denticola genome (Seshadri et al., 2004) using BLAST similarity analyses (Altschul et al., 1990). Chromosomal DNA of T. denticola was extracted using a method adapted from Nelson & Selander (1994) and a polymerase chain reaction (PCR) was carried out with a Platinum Pfx PCR system (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The PCR amplimers were generated using specific forward and reverse primers for each gene (see Supporting information, Table S1) and, where relevant, without inclusion of sequences encoding leader peptides that may result in products toxic to E. coli (Fenno et al., 1996; Edwards et al., 2005). Included within the forward primer sequences was an SphI restriction site, and within the reverse primer a SalI site to ensure in-frame insertion of coding sequences within the His6-tag expression vector pQE-30 (Qiagen). The PCR products were purified using a PCR cleanup kit (Qiagen), then digested and ligated into SphI/SalI-digested and dephosphorylated pQE-30 vector. Eschierchia coli XL-1 Blue was transformed with each ligation mix and colonies were screened first by PCR, then potential recombinant plasmids were purified (Qiagen miniprep) and sequenced to confirm in-frame fusion and sequence identity (Seqlon, Goettingen, Germany). The expression host E. coli M15 was then transformed with appropriate constructs and recombinant His6-tagged proteins were expressed after addition of 1 mM isopropyl-β-D-thiogalactopyranoside. Purification was achieved under denaturing conditions (8 M urea) using nickel-nitrilotriacetic acid resin (Qiagen) as described previously (Edwards et al., 2005). Recombinant proteins were dialysed against 4 M and 2 M urea and finally against distilled H2O, and then freeze-dried for storage.
Extraction of T. denticola outer sheath proteins
Proteins of T. denticola ATCC 35405 outer membranes were isolated as previously described (Bamford et al., 2007). Briefly, exponential phase T. denticola cells were harvested and washed twice in TE buffer (10 mM Tris–HCl, 1 mM ethylenediaminetetraacetic acid, pH 8) in the presence of 0.05 mM phenylmethylsulphonyl fluoride (PMSF). Approximately 3.6 × 109 cells ml−1 were incubated for 16 h at 4°C with TE buffer containing 1% Triton-X 114 and 0.05 mM PMSF. Outer membrane proteins were separated from non-periplasmic material by centrifugation at 21,000 g for 1 h at 4°C and membrane proteins in supernatants were analysed by sodium dodecyl sulphate–polyacrylamide gel eletrophoresis (SDS–PAGE).
SDS–PAGE and Western blot analysis
SDS–PAGE was carried out as described by Laemmli (1970). Rehydrated purified recombinant His6-tagged T. denticola polypeptides or T. denticola outer sheath proteins were mixed with sample buffer containing 2% SDS and 5 mMβ-mercaptoethanol and incubated at 20°C or 100°C before electrophoresis within a gel containing 10% acrylamide. Proteins were visualized by silver nitrate staining or electroblotted onto nitrocellulose membrane (Hybond ECL, Amersham, Amersham, UK). The membrane was incubated for 16 h at 4°C in Tris-buffered saline (TBS) containing 5% [weight/volume (w/v)] Fraction V bovine serum albumin (BSA) or 10% (w/v) skimmed milk powder and washed with TBS-0.1% Tween 20 (TBST). Membranes were then incubated with either anti-tetra His antibodies (Qiagen) diluted 1: 2000, or antibodies raised in chickens to the N terminus (amino acids 46–187) of Tp0155 diluted 1: 50 in TBST-0.1% BSA (TBSTB) or 1% skimmed milk powder (TBSTM) for 2 h at 20°C. The membranes were washed twice with TBSTB or TBSTM as appropriate and incubated with either horseradish peroxidase (HRP) -conjugated secondary anti-mouse antibodies (1: 1000) or HRP-Protein A for 2 h at 20°C. Visualization of proteins was achieved using 4-chloro-1-naphthol solution or enhanced chemiluminescence (Amersham).
Fibronectin adherence assay
Human plasma (Roche, Basel, Switzerland) or fibroblast (Calbiochem, San Diego, CA) fibronectin (5μg) in carbonate coating buffer (0.02 M NaHCO3, 0.02 M Na2CO3, pH 9.3) were adsorbed on to the surface of 96-well plastic plates (Greiner, Frickenhausen, Germany) for 16 h at 4°C (confirmed using anti-fibronectin antibodies, data not shown). Wells were blocked for 16 h at 4°C with 5% (w/v) BSA in phosphate-buffered saline (PBS). After removal of the blocking agent, 50μl His6-tagged purified recombinant T. denticola peptides (0–200μg/ml) were added to the wells, and incubated for 2 h at 20°C. Wells were washed twice with PBS containing 0.1% Tween 20 and 0.1% (w/v) BSA (PBSTB), and once with PBS for 10 min at 20°C. Recombinant peptides were detected using anti-tetra His antibodies diluted 1: 1000 in PBSTB for 2 h at 20°C. Wells were washed twice with PBSTB and once with PBS for 10 min each and HRP-conjugated secondary antibodies diluted 1: 1000 in PBSTB were added. After 2 h incubation at 20°C the wells were washed twice with PBSTB and once with PBS for 10 min. Antibody binding to recombinant protein was detected by incubating with o-phenylenediamine colour reagent and developing with 0.65 M H2SO4 (Edwards et al., 2005). Experiments were carried out in triplicate, and results are expressed as means with standard deviations from the mean.
Immunofluorescence microscopy
Stationary-phase T. denticola cells were pelleted by centrifugation (10,000 g for 10 min) and washed with PBS. Sterile glass coverslips (13 mm) were coated with filter-sterilized 0.1 mg ml−1 poly-lysine (Sigma, St Louis, MO) in borate buffer (0.1 M, pH 8.5) for 30 min at 20°C, washed thoroughly in sterile distilled H2O and dried. Approximately 4.8 × 107 cells were added to the poly-lysine coated coverslips in a 24-well plate and this was centrifuged at 3000 g for 5 min. Coverslips were gently washed twice with PBS, blocked with 5% BSA in PBS (1 h at 20°C) and washed with 0.1% Tween 20 in PBS. Antibodies to Tp0155 and to Tp0249, a T. pallidum flagellar sheath protein (Cameron et al., 2008) were pre-absorbed with Treponema phagedenis (Kazan). Antibodies and corresponding preimmune sera controls in PBSTB were added at suitable dilutions and incubated for 1 h at 20°C. Excess antibodies were removed by washing twice with PBSTB and once with PBS and then fluorescein isothiocyanate-conjugated immunoglobulin G was added for 1 h at 20°C. Coverslips were washed as above, placed onto slides with a drop of PermaFluor (Thermo Electron), and imaged using phase and fluorescence microscopy.
Statistics
To establish statistical significance, Student’s t-test was used on paired samples.
RESULTS
Identification of T. denticola proteins with homology to Tp0155 of T. pallidum
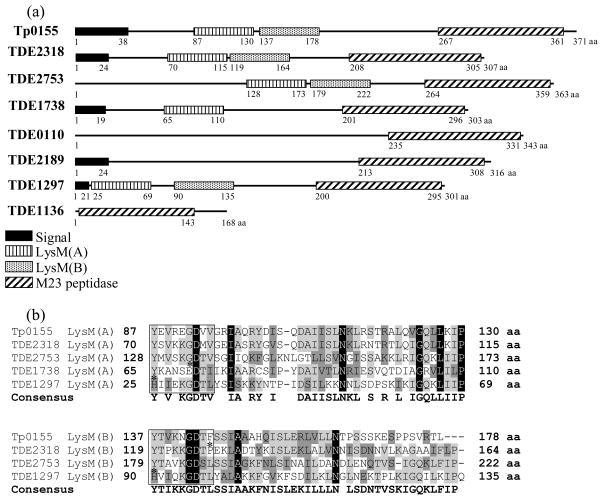
The amino acid (aa) sequence of T. pallidum Tp0155 fibronectin-binding protein (371 aa residues) was compared with the T. denticola genome (Seshadri et al., 2004) using BLAST similarity analyses. Significant similarities between Tp0155 and seven T. denticola predicted proteins were identified (Table 2), with TDE2318 showing the highest overall identity (43% identity) and TDE1136 being the least similar (14% identity). The aa sequences of the T. pallidum and T. denticola proteins were analysed to identify potential structural or functional regions. A number of distinct domains were evident in Tp0155 and in the seven T. denticola proteins (Fig. 1A). Tp0155 was composed of a leader peptide (38 aa residues), two N-terminal LysM domains, labelled for clarity LysM(A) (44 aa residues) and LysM(B) (42 aa residues), and an M23 peptidase sequence (95 aa residues) at the C-terminal end. The C-terminal M23 peptidase domain was present in all of the T. denticola polypeptide sequences and showed the highest levels of homology (Table 2) with each other and with the M23 sequence of Tp0155. Three of the seven T. denticola proteins contained two predicted N-terminal LysM domains (TDE2318, TDE2753, TDE1297), whereas TDE1738 polypeptide had only one LysM domain (Fig. 1A). TDE0110 and TDE2189 contained no potential LysM domains but TDE1136 appeared to consist solely of an extended M23 peptidase sequence of 144 aa residues (Fig. 1A). Alignment of the seven proteins with Tp0155 and the positions of the various domains within the sequences are shown in supplementary material (see Supporting information, Fig. S1 and Fig. S2, respectively).
Table 2.
Comparative bioinformatic analyses of Treponema pallidum (Tp) fibronectin-binding protein Tp0155 and Treponema denticola (TDE) homologues
| ORF | aa residues | Predicted molecular mass (Da) | % identity to Tp0155
|
|||
|---|---|---|---|---|---|---|
| Complete | LysM(A)1 | LysM(B)2 | M233 | |||
| Tp0155 | 371 | 40,664 | 100 | 100 | 100 | 100 |
| TDE 2318 | 307 | 33,607 | 42.8 | 73.3 | 31.4 | 61.1 |
| TDE 2753 | 363 | 39,731 | 29.7 | 34.8 | 26.9 | 52.6 |
| TDE 1738 | 303 | 33,766 | 26.7 | 33.3 | –4 | 43.4 |
| TDE 0110 | 343 | 38,172 | 23.6 | – | – | 36 |
| TDE 2189 | 316 | 36,052 | 20.8 | – | – | 34.3 |
| TDE 1297 | 301 | 32,820 | 16.8 | 28.9 | 18 | 23.6 |
| TDE 1136 | 168 | 18,642 | 13.9 | – | – | 20.4 |
The LysM domain located closest to the N terminus (Fig. 1) has been designated LysM(A).
Where a second LysM domain sequence was identified (Fig. 1), it was assigned the title LysM(B). % identity of T. denticola LysM sequences were calculated through comparison with the most similar domains in Tp0155.
The eight polypeptides all carry an M23 peptidase domain, see Fig. 1A.
Indicates no LysM domain present
Figure 1.
(A) Predicted identifiable peptide motifs (www.ebi.ac.uk/InterProScan/) of Treponema pallidum fibronectin-binding protein Tp0155 and potential orthologues identified from the Treponema denticola genome using BLAST similarity searches. LysM domains are shown with vertical bars (A) or dots (B). Treponema pallidum and T. denticola M23 peptidase domains (diagonal bars) retain 37% identity. Potential signal sequences (solid black) were identified using http://bioinformatics.leeds.ac.uk/prot_analysis/signal.html. (B) Alignments of LysM(A) and LysM(B) domain sequences from T. pallidum Tp0155 and potential T. denticola orthologues showing high degree of consensus across the different peptide sequences. Degree of identity is shown from highest (e.g.
 ) to lowest (e.g. H). The LysM motif YxxxxGxx(Hyd) (Turner et al., 2004) is boxed. Asterisk indicates residues that do not fit the specified motif.
) to lowest (e.g. H). The LysM motif YxxxxGxx(Hyd) (Turner et al., 2004) is boxed. Asterisk indicates residues that do not fit the specified motif.
Other T. pallidum fibronectin-binding proteins have been described including Tp0483 (Cameron et al., 2004) and Tp0136 (Brinkman et al., 2008). An orthologous protein to Tp0483 was identified in T. denticola (TDE1984). However, the low sequence identity (21%) between these proteins, by comparison to the Tp0155 orthologues, led to no further functional investigations at this time. No orthologues to Tp0136 were found.
Generation of recombinant T. denticola proteins
To determine the fibronectin-interacting activities of T. denticola proteins, genes were cloned and proteins were expressed in E. coli. The appropriate gene sequences were amplified by PCR with proof-reading enzymes (see Supporting information, Fig. S3) and primers based on the sequence of the T. denticola ATCC 35405 genome on the Comprehensive Microbial Resource (http://cmr.tigr.org.tigr-scripts/cmr/CmrHomePage.cgi). Forward primers were constructed such that start codons, and where appropriate signal sequences, were absent from the expressed proteins to counteract possible expression problems in E. coli. Previous work suggests that some leader sequences of T. denticola polypeptides may be toxic to E. coli (Fenno et al., 1996; Edwards et al., 2005). Termination of translation was effected by including the stop codon of each gene within the reverse PCR primer (see Supporting information, Table S1). In silico restriction analyses were carried out (www.restrictionmapper.org) to identify enzymes that did not cut any of the T. denticola sequences, so allowing a common strategy to be used for cloning and expression of the genes.
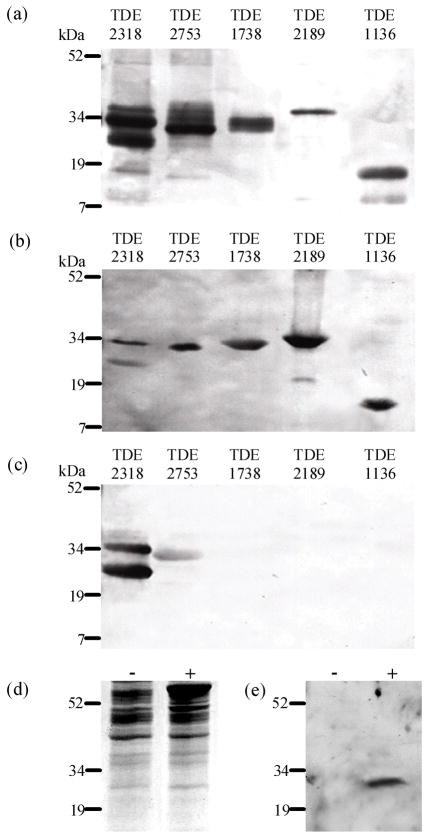
Recombinant His6-tagged T. denticola proteins were expressed and purified from E. coli for five proteins, as cloning was unsuccessful for TDE0110 and TDE1297 (Fig. 2A). All polypeptides migrated closely to their predicted molecular mass and were detected using HRP-conjugated anti-tetra His antibodies and appropriate HRP-conjugated secondary antibodies (Fig. 2B). A peptide doublet was observed for rTDE2318, which may suggest degradation at the C terminus because the 6×His Tag was at the N terminus.
Figure 2.
Recombinant Treponema denticola His-tagged proteins (5μg), which show homology to fibronectin-binding protein Tp0155 of Treponema pallidum, purified using nickel affinity chromatography, analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) stained with silver nitrate (A), and by Western blot, using anti-tetra His antibodies to detect each protein (B) or antibodies to the T. pallidum protein Tp0155, which had first been pre-adsorbed with T. phagedenis (Kazan) (C). Treponema denticola ATCC35405 outer membrane proteins extracted with 1% Triton X-114 incubated at 20°C or 100°C before separation by SDS–PAGE analyses (d) exposed to antibodies to T. pallidum Tp0155 (E). Molecular mass standards (kDa) are indicated to the side of the figure.
Adherence to fibronectin
Differential adherence of T. pallidum protein Tp0155 to matrix or plasma fibronectin has already been characterized (Cameron et al., 2004), with matrix fibronectin being preferentially bound. Quantitative assessment of binding to fibronectin immobilized to wells of a plastic plate by the recombinant T. denticola proteins indicated that all, except rTDE2753, were better able to adhere to matrix fibronectin (Fig. 3). Significant differences (P < 0.05) in binding matrix versus plasma fibronectin were observed for rTDE1738, rTDE2189 and rTDE1136 (Fig. 3). This figure was based on dose-dependent (Fig. S4) adherence to matrix or plasma fibronectin (p < 0.05) that was shown for all proteins tested, compared with binding to blocked wells (controls). The recombinant T. denticola proteins were also immobilized by Western blot on nitrocellulose and assessed for their ability to bind fluid-phase fibronectin detected using anti-fibronectin antibodies. None of the recombinant proteins bound fibronectin under these conditions (data not shown), suggesting that affinity towards fluid-phase fibronectin was much lower than to immobilized fibronectin. These results identify five novel fibronectin-binding proteins from T. denticola with greater ability to bind immobilized forms of fibronectin.
Figure 3.
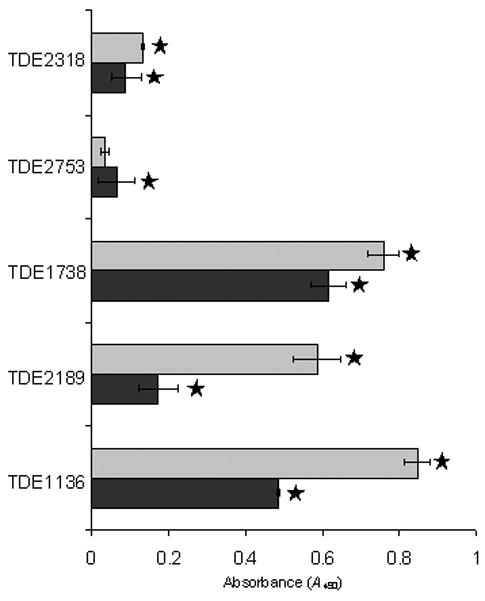
Adhesion of recombinant Treponema denticola proteins to matrix (grey bars) or plasma fibronectin (black bars). Recombinant protein (5 μg) was added to each well, except for TDE2753 and TDE1738, which appeared to bind non-specifically to the substrates when more than 1.25μg was added. Stars denote significant difference (P < 0.05) between adhesion to either matrix or plasma fibronectin and bovine serum albumin-blocked wells. Error bars indicate ± SD of the mean from triplicates of a representative experiment.
Surface localization of Tp0155 orthologues in T. denticola
Sequence analyses of the T. denticola proteins indicated that TDE2318, TDE1738, TDE2189 and TDE1297 had leader peptides (Fig. 1) suggesting cell surface localization or secretion. To investigate surface exposure of TDE2318, which has the highest homology to T. pallidum fibronectin-binding protein Tp0155 on the basis of aa sequence (Table 1), antibodies specific to Tp0155 were used to probe T. denticola outer membrane proteins in Western blot analyses and immunofluorescence microscopy investigations of T. denticola cells. These antibodies were shown to detect both TDE2318 and TDE2753 in Western blot analyses (Fig. 2C) but lack of predicted leader sequence in TDE2753 indicates that this protein might not be secreted. Polyclonal antibodies to Tp0155 were first pre-adsorbed to a closely related organism T. phagedenis (Kazan) to reduce non-specific binding. Preimmune sera were similarly preadsorbed against T. phagedenis.
A single peptide of ~ 33 kDa was visualized in T. denticola outer sheath proteins, although only in a sample heated to 100°C before separation. This peptide showed similar molecular mass to that predicted for TDE2318. This suggests that this peptide is excreted to the T. denticola outer membrane but may be a multimeric complex in its native form.
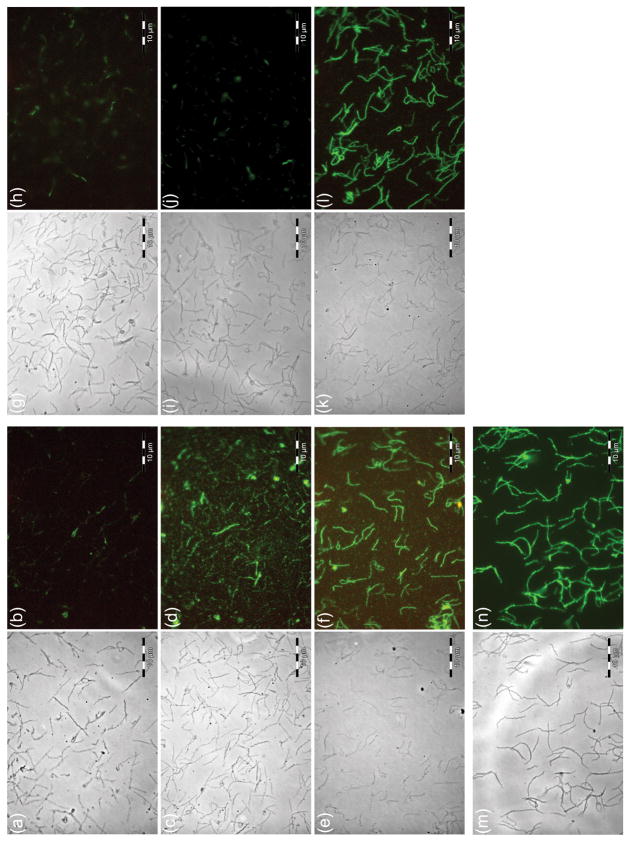
Immunofluorescence analyses demonstrated control (preimmune) antibodies did not react with T. denticola cells in analyses (Fig. 4A,B). However, antibodies to Tp0155 reacted with intact T. denticola cells (Fig. 4C,D). Based on fluorescence distribution, it appeared that the orthologous proteins may not be expressed by all cells of the population. In addition, proteins may not be expressed uniformly across the cell surface (Fig. 4D). When cells were permeabilized with Triton X-114, all cells present reacted with Tp0155 antibody (Fig. 4E,F). To demonstrate that T. denticola cells were all intact in Fig. 4(C,D), the T. denticola cells were reacted with anti-Tp0249 (FlaA) antibodies generated to the flagella sheath proteins (Cameron et al., 2008) that are beneath the outer membrane, and so not exposed in intact cells. Neither preimmune antibody (Fig. 4G,H) nor anti-FlaA antibody (Fig. 4I,J) reacted with T. denticola cells, but cells permeabilized with Triton X-114 were all reactive (Fig. 4K,L). These were as reactive as whole cell antibodies to intact cells (Fig. 4M,N). The results show that TDE2318 is surface exposed in T. denticola.
Figure 4.
Detection of proteins on the surface of Treponema denticola ATCC 35405 using antibodies to the Treponema pallidum protein Tp0155 which had first been pre-adsorbed against Treponema phagedenis (Kazan). (A, C, E, G, I, K and M) Phase-contrast images; (B, D, F, H, J, L and N) corresponding immunofluorescence. (A, B) Preimmune serum for Tp0155-specific antibodies; (C, D), Tp0155-specific antibodies, non-detergent-treated T. denticola; (E, F), Tp0155-specific antibodies, Triton X-114-treated T. denticola; (G, H), preimmune serum for Tp0249-specific antibodies; (I, J), Tp0249-specific antibodies, non-detergent-treated T. denticola (negative control); (K, L), Tp0249-specific antibodies, Triton X-114-treated T. denticola; (M, N), anti-T. denticola antibodies (control) (Edwards et al., 2005).
DISCUSSION
Normal epithelial shedding or tissue damage, caused by trauma or infection of mucosal surfaces as witnessed deep within the periodontal pocket, will expose plasma and extracellular matrix proteins including fibronectin. It has been known for some time that T. denticola can bind both matrix fibronectin (Ellen et al., 1994) and plasma fibronectin (Haapasalo et al., 1991; Fenno et al., 1996; Edwards et al., 2005). The major surface protein Msp has been shown to bind fibronectin (Haapasalo et al., 1992; Edwards et al., 2005) but an Msp mutant of T. denticola (Fenno et al., 1998) still retains the ability to adhere to fibronectin (C.V. Bamford, unpublished data) suggesting that other factors are involved. To identify these molecules we used comparative sequence analyses, based on the T. pallidum fibronectin-binding protein Tp0155 we identified seven potential fibronectin-binding proteins from the T. denticola genome. Five of these were produced as recombinant polypeptides and shown to adhere to matrix or plasma fibronectin. Protein TDE2318, the closest homologue to Tp0155, was shown to be cell surface exposed and others with signal sequences, but not detected here, may be similarly localized to the cell membrane. A role for these proteins in colonization of damaged host tissues by T. denticola is suggested. The ability of these proteins to also bind plasma fibronectin may interfere with tissue regeneration and disrupt healing processes.
Distinct domains are present in all of the seven T. denticola proteins described in this study. However, it is not clear at this stage which of these domains might be involved in adherence to fibronectin. None of the protein sequences contain the consensus bacterial fibronectin-binding protein domain motif (Schwarz-Linek et al., 2004) that is often a component of amino acid sequence repeats. Four of the polypeptides contain a LysM domain, and all of them contain the M23 peptidase domain (Fig. 1).
LysM domains consist of about 40 aa residues, containing a lysin motif sequence YxxxxGxx(Hyd) (Turner et al., 2004). The LysM domains of Tp0155 and the T. denticola proteins all showed high homology and carried the lysin motif (Fig. 1B). LysM domains are commonly found in proteins with modular functions including autolysis (MurA, Listeria monocytogenes, Carroll et al., 2003). A surface protein (Sep) of Lactobacillus fermentum BR11 (Turner et al., 2004) and the autolysin, Aae, of Staphylococcus epidermidis (Heilmann et al., 2003) use the peptidoglycan-binding LysM domains as cell wall anchors. LysM domains of proteins with accompanying lytic domains may act to target activity to a specific substrate (Steen et al., 2003).
The LysM domains of autolysins Aae (Staphylococcus epidermidis) and Aaa (Staphylococcus aureus), were shown to bind vitronectin, the Aα and Bβ chains of fibrinogen and the 29-kDa fragment of fibronectin (Heilmann et al., 2003, 2005). This may indicate a similar role for the LysM domains within the T. denticola polypeptide. However, ability to bind either matrix or plasma fibronectin did not correlate with the numbers of LysM domains present. For example, TDE2318 and TDE2753 both have two LysM domains, but show a much lower adherence level to fibronectin compared with TDE1136. The latter consists almost entirely of an M23 peptidase domain with no LysM sequence and is able to bind fibronectin to a higher level than TDE2318 or TDE2753. A development of this work will be to produce truncated recombinant polypeptides of TDE1738 and TDE1136 to try to delineate more precisely the fibronectin-binding domains.
Interactions of LysM sequences with peptidoglycan, or peptidoglycan precursors, appear to be one property, so potentially targeting LysM-containing proteins to the cell surface. In T. denticola it has been shown that binding to immobilized fibronectin occurs through interactions with the bacterial cell tip (Dawson & Ellen, 1990). Binding levels were reduced ~50% at 4°C compared with 37°C, suggesting that metabolic activity was important (Dawson & Ellen, 1990). Recent work shows that the cell tips of T. denticola and T. pallidum have complex morphologies with T. pallidum having higher structural complexity (Izard et al., 2008, 2009). Peptidoglycan forms a thin layer between the cytoplasmic membrane and outer membrane, and is thicker at the cell ends (Izard et al., 2009). It is possible therefore that the LysM domain proteins described in this article may be involved in peptidoglycan modelling as well as fibronectin adherence.
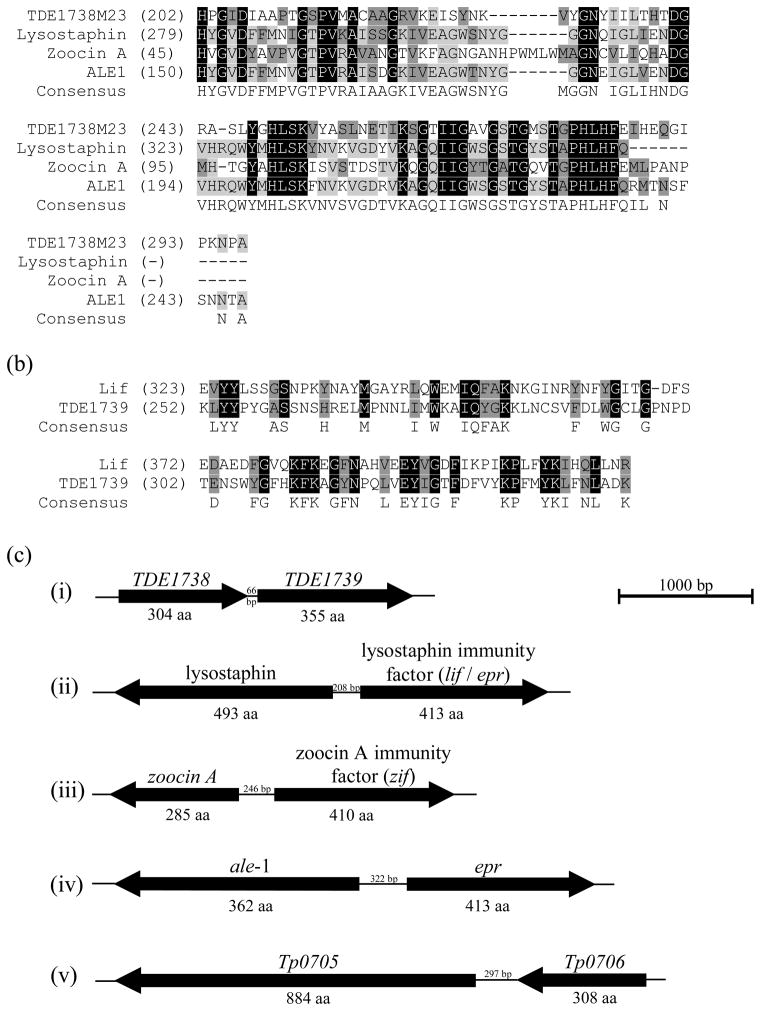
All the T. denticola polypeptides contained M23 peptidase domains which are zinc metalloproteases for which the active sites are HxH or HxxxD. Analysis of Tp0155 and the seven T. denticola polypeptides with M23 peptidase domains indicated that all except TDE1297 had conserved HxH (HLH or HVH) and HxGxD sequences (Fig. S2). Characterization of other M23 peptidases has suggested activity against glycine–glycine bonds (Schindler & Schuhardt, 1965). Many proteins containing the M23 domain have specific hydrolytic activity on peptidoglycan, resulting in bacteriolytic capabilities (Schindler & Schuhardt, 1964; Thumm & Gotz, 1997). Perhaps the best characterized of these lytic M23 peptidase-containing proteins is lysostaphin produced by Staphylococcus simulans biovar staphylolyticus (Schindler & Schuhardt, 1964; Naverre & Schneewind, 1999). It specifically cleaves between the third and fourth Gly residueS of the pentaglycine cross bridges which are unique to the peptidoglycan of staphylococcal species (Thumm & Gotz, 1997; Recsei et al., 1987). Other known lytic proteins with similar peptidoglycanolytic activities are Zoocin A (Streptococcus equi subsp. zooepidemicus 4881, Simmonds et al., 1996, 1997) and ALE-1 (Staphylococcus capitis EPK1, Sugai et al., 1997a). These proteins are all conserved for the M23 protease motifs (Fig. 5A). It is possible therefore that the T. denticola proteins have lytic activity against other closely related or closely co-habiting bacteria. We have tested the recombinant polypeptides for their ability to bind to, or degrade, cells walls from S. aureus or T. denticola, but have been unable to demonstrate any clumping or lytic activities (data not shown).
Figure 5.
(A) Alignments of M23 peptidase domains from TDE1738 of Treponema denticola and homologous regions of peptidoglycan-degrading enzymes lysostaphin from Staphylococcus simulans biovar staphylolyticus, zoocin A from Streptococcus equi subsp zooepidemicus and ALE-1 from Staphylococcus capitis. The M23 peptidase domain of TDE1738 retains 43% identity to lysostaphin, 43% identity to zoocin A and 40% identity to ALE-1 with conservation across the HxGxD and HLH active sites. (B) Alignments of a 92 amino acid (aa) residue portion regions of TDE1739 of T. denticola and lysostaphin immunity factor from S. simulans biovar staphylolyticus, demonstrating homology (29% identity) between these peptides. Sequence consensus is shown as is degree of identity from highest (e.g.
 ) to lowest (e.g. A). (C) Diagram depicting genes encoding potential and known peptidoglycan degradation enzymes and adjacent immunity factors which provide protection from autolysis by the peptidoglycanolytic enzyme. (i) The chromosomal gene TDE1738 from T. denticola encodes for a potential peptidoglycan-degrading enzyme, and TDE1739 for a potential immunity factor. (ii) Lysostaphin and adjacent lysostaphin immunity factor (Lif/Epr) are encoded on the plasmid pACK1 of S. simulans biovar staphylolyticus. (iii) The gene encoding zoocin A and corresponding zoocin A immunity factor (Zif) is on the chromosome of Streptococcus equi subsp zooepidemicus, while (iv) ale-1 and adjacent endopeptidase resistance (epr) genes are located on a plasmid in Staphylococcus capitis. (v) Tp0706 from the Treponema pallidum chromosome, with homology to TDE1738 may also be an enzyme with peptidoglycan-degrading activity and the adjacent gene Tp0705 is annotated as a pencillin-binding protein in the same manner as zoocin A immunity factor (Zif).
) to lowest (e.g. A). (C) Diagram depicting genes encoding potential and known peptidoglycan degradation enzymes and adjacent immunity factors which provide protection from autolysis by the peptidoglycanolytic enzyme. (i) The chromosomal gene TDE1738 from T. denticola encodes for a potential peptidoglycan-degrading enzyme, and TDE1739 for a potential immunity factor. (ii) Lysostaphin and adjacent lysostaphin immunity factor (Lif/Epr) are encoded on the plasmid pACK1 of S. simulans biovar staphylolyticus. (iii) The gene encoding zoocin A and corresponding zoocin A immunity factor (Zif) is on the chromosome of Streptococcus equi subsp zooepidemicus, while (iv) ale-1 and adjacent endopeptidase resistance (epr) genes are located on a plasmid in Staphylococcus capitis. (v) Tp0706 from the Treponema pallidum chromosome, with homology to TDE1738 may also be an enzyme with peptidoglycan-degrading activity and the adjacent gene Tp0705 is annotated as a pencillin-binding protein in the same manner as zoocin A immunity factor (Zif).
Many bacteriolysins are targeted to bind peptidoglycan via a substrate-binding domain (e.g. LysM) at the C-terminal end of the protein with the M23 peptidase domain at the N terminus (Baba & Schneewind, 1996; Simmonds et al., 1996; Nilsen et al., 2003). In four of the T. denticola proteins there is opposite orientation, with LysM at the N-terminal and M23 peptidase domain at the C-terminal ends. We speculate that the LysM domain is responsible for anchoring the protein to the cell surface environment of T. denticola through binding peptidoglycan-like material associated with the outer membranes. Such a function for LysM sequences has been shown for surface protein Sep of Lactobacillus fermentum BR11 (Turner et al., 2004) and the Aae autolysin of Staphylococcus epidermidis (Heilmann et al., 2003).
Bacteria that produce M23 peptidase domain bacteriolytic proteins all appear to produce immunity factors. These factors protect the producer organism from self-peptidoglycanolysis. Some immunity factors (Sugai et al., 1997b; Beatson et al., 1998) are enzymic and replace peptidoglycan glycines with serines (DeHart et al., 1995; Heath Farris et al., 2003), which prevents successful targeting of the substrate-binding domain and inhibiting hydrolysis (Lai et al., 2002). The genes encoding for immunity factors are commonly adjacent to the bacteriocin peptidase gene (Fig. 5B) although they may not always be in the same orientation (Recsei et al., 1987; Heath et al., 1989, 2004; Sugai et al., 1997b). Analysis of the T. denticola genome shows that immediately downstream of TDE1738, the gene TDE1739 encodes a protein with similarity to lysostaphin immunity factor (Fig. 5B). The TDE1739 gene is annotated within the genome as encoding a FemAB homologue. FemAB imparts resistance to methicillin by altering the glycine ratio of peptidoglycan in a similar manner to other immunity factors (Maidhof et al., 1991; Heath et al., 2005). This strongly suggests that TDE1738 could be a bacteriocin, with the gene product of TDE1739 necessary to protect T. denticola from autolysis. The periodontal pocket is a highly competitive environment for bacteria and T. denticola bacteriocins could lyse organisms to release otherwise unavailable nutrients. It has been shown that Prevotella intermedia, Eubacterium nodatum, Veillonella parvula and Fusobacterium nucleatum all stimulate growth of T. denticola (ter Steeg & van der Hoeven, 1990). The possibility that T. denticola has the ability to lyse anaerobic bacteria is currently being investigated along with attempted genetic knockouts of the relevant genes to further develop functional analyses.
The comparative analyses carried out in this article have identified seven T. denticola proteins that show various degrees of homology to Tp0155. Further investigation of the T. pallidum genome reveals that six of these T. denticola proteins have corresponding homologous proteins in T. pallidum (Table S2). Close phylogenetic relationships between individual T. pallidum and T. denticola proteins is retained, with each containing similar structural arrangements of LysM and M23 peptidase domains (Table S2). These observations suggest that this gene set is functionally conserved within Treponema. The T. pallidum genome also contains a gene, Tp0806, that encodes a protein with homology to lysostaphin immunity factor. The ZoocinA immunity factor (Zif) acts as a penicillin-binding protein (Heath et al., 2004) capable of peptidoglycan reorganization, and downstream and in the same orientation as the T. pallidum M23 peptidase gene Tp0706 (homologous to TDE1738) is a gene, Tp0705 (Fig. 5C), annotated as a penicillin-binding protein, suggesting a role in protection from autolysis.
In summary, this paper describes a family of seven conserved T. denticola genes. The products of five of these genes bind fibronectin and TDE2318, the orthologue of T. pallidum protein Tp0155, is cell-surface exposed. Six of the T. denticola genes have orthologues in T. pallidum. Based on sequence similarities, and genetic organization of adjacent genes, one of these polypeptides (TDE1738), and the orthologue T. pallidum (Tp0706), may be a bacteriocin. The other proteins may have autolytic or peptidoglycan remodelling activities in addition to recognizing fibronectin. It will be important to determine that these molecules are expressed in vivo and if their expression correlates with virulence or disease severity.
Supplementary Material
Figure S1 Amino acid sequence alignments of seven Treponema denticola proteins with Treponema pallidum Tp0155 fibronectin-binding protein showing sequence consensus and degree of identity from highest (e.g. A) to lowest (e.g. A).
Figure S2 Alignments of homologous Treponema pallidum and Treponema denticola proteins. Predicted signal sequences are highlighted in green, LysM domains in blue with the LysM motif in pink, M23 peptidase sequences in red, and active sites of the M23 peptidase in yellow.
Figure S3 Polymerase chain reaction amplification of DNA corresponding to genes of Treponema denticola that were homologous to known Treponema pallidum fibronectin-binding proteins. Positions of base-pair markers are indicated at the side of the figure.
Acknowledgments
We thank M. Freeman, J. L. Brittan, and L.C. Dutton, and for expert technical assistance, J. Chris Fenno for providing T. denticola strains, and A. M. Edwards and J. Spencer for helpful discussions.
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–97. [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford CV, Fenno JC, Jenkinson HF, Dymock D. The chymotrypsin-like protease complex of Treponema denticola ATCC 35405 mediates fibrinogen adherence and degradation. Infect Immun. 2007;75:4364–72. doi: 10.1128/IAI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatson SA, Sloan GL, Simmonds RS. Zoocin A immunity factor: a femA-like gene found in group C streptococcus. FEMS Microbiol Lett. 1998;163:73–7. doi: 10.1111/j.1574-6968.1998.tb13028.x. [DOI] [PubMed] [Google Scholar]
- 5.Brinkman MB, McGill MA, Pettersson J, et al. A novel Treponema pallidum antigen, Tp0136, is an outer membrane protein that binds human fibronectin. Infect Immun. 2008;76:1848–57. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron CE. Identification of a Treponema pallidum laminin binding protein. Infect Immun. 2003;71:2525–33. doi: 10.1128/IAI.71.5.2525-2533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron CE, Brown EL, Kuroiwa JM, Schnapp LM, Brouwer NL. Treponema pallidum fibronectin-binding proteins. J Bacteriol. 2004;186:7019–22. doi: 10.1128/JB.186.20.7019-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron CE, Kuroiwa JMY, Yamada M, Francescutti T, Chi B, Kuramitsu HK. Heterologous expression of the Treponema pallidum laminin-binding adhesin Tp0751 in the culturable spirochete Treponema phagedenis. J Bacteriol. 2008;190:2565–71. doi: 10.1128/JB.01537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll SA, Hain T, Technow U, et al. Identification and characterisation of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muraminidase needed for cell separation. J Bacteriol. 2003;185:6801–8. doi: 10.1128/JB.185.23.6801-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan ECS, Siboo R, Keng T, et al. Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int J Sys Bacteriol. 1993;43:196–203. doi: 10.1099/00207713-43-2-196. [DOI] [PubMed] [Google Scholar]
- 11.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–95. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couchman JR, Austria MR, Woods A. Fibronectin-cell interactions. J Invest Dermatol. 1990;94:7S–14S. doi: 10.1111/1523-1747.ep12874973. [DOI] [PubMed] [Google Scholar]
- 13.Dawson JR, Ellen RP. Tip-oriented adherence of Treponema denticola to fibronectin. Infect Immun. 1990;58:3924–8. doi: 10.1128/iai.58.12.3924-3928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeHart HP, Heath HE, Heath LS, LeBlanc PA, Sloan GL. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Env Microbiol. 1995;61:1475–9. doi: 10.1128/aem.61.4.1475-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewhirst FE, Tamer MA, Ericson RE, et al. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol. 2000;15:196–202. doi: 10.1034/j.1399-302x.2000.150308.x. [DOI] [PubMed] [Google Scholar]
- 16.Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, Bohach GA. Fibronectin binding protein and host cell tyrosine kinase are required for internalisation of Staphylococcus aureus by epithelial cells. Infect Immun. 1999;67:4673–8. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73:2891–8. doi: 10.1128/IAI.73.5.2891-2898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellen RP, Song M, McCulloch CA. Degradation of endogenous plasma membrane fibronectin concomitant with Treponema denticola 35405 adhesion to gingival fibroblasts. Infect Immun. 1994;62:3033–3037. doi: 10.1128/iai.62.7.3033-3037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenno JC, Muller KH, McBride BC. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–97. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenno JC, Tamura M, Hannam PM, Wong GW, Chan RA, McBride BC. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect Immun. 2000;68:1884–92. doi: 10.1128/iai.68.4.1884-1892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenno JC, Wong GW, Hannam PM, McBride BC. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett. 1998;163:209–15. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 22.Haapasalo M, Muller KH, Uitto VJ, Leung WK, McBride BC. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60:2058–5. doi: 10.1128/iai.60.5.2058-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haapasalo M, Singh U, McBride BC, Uitto VJ. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect Immun. 1991;59:423–7. doi: 10.1128/iai.59.11.4230-4237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath Farris M, Heath LS, Heath HE, LeBlanc PA, Simmonds RS, Sloan GL. Expression of the genes for lysostaphin and lysostaphin resistance in streptococci. FEMS Microbiol Lett. 2003;288:115–19. doi: 10.1016/S0378-1097(03)00743-2. [DOI] [PubMed] [Google Scholar]
- 25.Heath HE, Heath LS, Nitterauer JD, Rose KE, Sloan GL. Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem Biophys Res Comm. 1989;15:1106–9. doi: 10.1016/s0006-291x(89)80117-2. [DOI] [PubMed] [Google Scholar]
- 26.Heath LS, Gargis SR, Smithberg SR, et al. Plasmid-specified FemBX-like immunity factor in Staphylococcus sciuri DD4747. FEMS Microbiol Lett. 2005;249:227–31. doi: 10.1016/j.femsle.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Heath LS, Heath HE, LeBlanc PA, et al. The streptococcolytic enzyme ZoocinA is a penicillin-binding protein. FEMS Microbiol Lett. 2004;236:205–11. doi: 10.1016/j.femsle.2004.05.038. [DOI] [PubMed] [Google Scholar]
- 28.Heilmann C, Hartleib J, Hussain MS, Peters G. The multifunctional Staphylococcus aureus autolysin, Aaa, mediates adherence to immobilised fibrinogen and fibronectin. Infect Immun. 2005;73:4793–802. doi: 10.1128/IAI.73.8.4793-4802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heilmann C, Niemann S, Sinha B, Herrmann M, Kehrel BE, Peters G. Staphylococcus aureus fibronectin-binding protein (FnBP)-mediated adherence to platelets, and aggregation of platelets induced by FnBPA but not by FnBPB. J Infect Dis. 2004;190:321–9. doi: 10.1086/421914. [DOI] [PubMed] [Google Scholar]
- 30.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterisation of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149:2769–78. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 31.Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izard J, Renken C, Hsieh CE, et al. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191:7566–80. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods. J Periodontal Res. 1995;30:332–41. doi: 10.1111/j.1600-0765.1995.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Lai AC, Tran S, Simmonds RS. Functional characterisation of domains found within a lytic enzyme produced by Streptococcus equi subsp. zooepidemicus. FEMS Microbiol Lett. 2002;215:133–8. doi: 10.1111/j.1574-6968.2002.tb11382.x. [DOI] [PubMed] [Google Scholar]
- 36.Maidhof H, Reinicke B, Blumel P, Berger-Bachi B, Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. 1991;173:3507–13. doi: 10.1128/jb.173.11.3507-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naverre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson K, Selander RK. Analysis of genetic variation by polymerase chain reaction-based nucleotide sequencing. Methods Enzymol. 1994;235:174–83. doi: 10.1016/0076-6879(94)35139-2. [DOI] [PubMed] [Google Scholar]
- 39.Pasula R, Wisniowski P, Martin WJ., 2nd Fibronectin facilitates Mycobacterium tuberculosis attachment to murine alveolar macrophages. Infect Immun. 2002;70:1287–92. doi: 10.1128/IAI.70.3.1287-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson KM, Baseman JB, Alderete JF. Treponema pallidum receptor binding proteins interact with fibronectin. J Exp Medicine. 1983;157:1958–70. doi: 10.1084/jem.157.6.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pracht D, Elm C, Gerber J, et al. PavA of Streptococcus pneumoniae modulates adherence invasion and meningeal inflammation. Infect Immun. 2005;73:2680–9. doi: 10.1128/IAI.73.5.2680-2689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recsei PA, Gruss AD, Novick RP. Cloning, sequence and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–31. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riviere GR, DeRouen TA. Association of oral spirochetes from periodontally healthy sites with development of gingivitis. J Periodontol. 1998;69:496–501. doi: 10.1902/jop.1998.69.4.496. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Samen U, Gottschalk B, Eikmanns BJ, Reinscheid DJ. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J Bacteriol. 2004;186:1398–408. doi: 10.1128/JB.186.5.1398-1408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindler CA, Schuhardt VT. Lysostaphin: a new bacteriolytic agent for the staphylococcus. Proc Natl Acad Sci USA. 1964;51:414–21. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindler CA, Schuhardt VT. Purification and properties of lysostaphin – a lytic agent of Staphylococcus aureus. Biochem Biophys Acta. 1965;97:242–50. doi: 10.1016/0304-4165(65)90088-7. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz-Linek U, Hook M, Potts JR. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol Microbiol. 2004;52:631–41. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 49.Seshadri R, Myers GS, Tettelin H, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA. 2004;101:5646–51. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmonds RS, Simpson WJ, Tagg JR. Cloning and sequence analysis of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene. 1997;189:255–61. doi: 10.1016/s0378-1119(96)00859-1. [DOI] [PubMed] [Google Scholar]
- 51.Simmonds RS, Pearson L, Kennedy RC, Tagg JR. Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl Env Microbiol. 1996;62:4536–41. doi: 10.1128/aem.62.12.4536-4541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha B, Francois PP, Nusse O, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1999;1:101–17. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 53.Steen A, Buist G, Leenhouts KJ, et al. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem. 2003;278:23874–81. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- 54.Sugai M, Fujiwara T, Akiyama T, et al. Purification and molecular characterisation of glycineglycine endopeptidase produced by Staphylococcus capitis EPK1. J Bacteriol. 1997a;179:1193–202. doi: 10.1128/jb.179.4.1193-1202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugai M, Fujiwara T, Ohta K, Komatsuzawa H, Ohara M, Suginaka H. epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J Bacteriol. 1997b;179:4311–18. doi: 10.1128/jb.179.13.4311-4318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.ter Steeg PF, van der Hoeven JS. Growth stimulation of Treponema denticola by periodontal microorganisms. Antonie Van Leeuwenhoek. 1990;57:63–70. doi: 10.1007/BF00403156. [DOI] [PubMed] [Google Scholar]
- 57.Thomas DD, Baseman JB, Alderete JF. Enhanced levels of attachment of fibronectin-primed Treponema pallidum to extracellular matrix. Infect Immun. 1986;52:736–41. doi: 10.1128/iai.52.3.736-741.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thumm G, Gotz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol. 1997;23:1251–65. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- 59.Turner MS, Hafner LM, Walsh T, Giffard PM. Identification and characterisation of the Novel LysM domain-containing surface protein Sep from Lactobacillus fermentum BR11 and its use as a peptide fusion partner in Lactobacillus and Lactococcus. Appl Env Microbiol. 2004;70:3673–80. doi: 10.1128/AEM.70.6.3673-3680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Umemoto T, Nakatani Y, Nakamura Y, Namikawa I. Fibronectin-binding proteins of a human oral spirochete Treponema denticola. Microbiol Immunol. 1993;37:75–8. doi: 10.1111/j.1348-0421.1993.tb03182.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Amino acid sequence alignments of seven Treponema denticola proteins with Treponema pallidum Tp0155 fibronectin-binding protein showing sequence consensus and degree of identity from highest (e.g. A) to lowest (e.g. A).
Figure S2 Alignments of homologous Treponema pallidum and Treponema denticola proteins. Predicted signal sequences are highlighted in green, LysM domains in blue with the LysM motif in pink, M23 peptidase sequences in red, and active sites of the M23 peptidase in yellow.
Figure S3 Polymerase chain reaction amplification of DNA corresponding to genes of Treponema denticola that were homologous to known Treponema pallidum fibronectin-binding proteins. Positions of base-pair markers are indicated at the side of the figure.