Abstract
BACKGROUND
If liquid-chromatography–multiple-reaction–monitoring mass spectrometry (LC-MRM/MS) could be used in the large-scale preclinical verification of putative biomarkers, it would obviate the need for the development of expensive immunoassays. In addition, the translation of novel biomarkers to clinical use would be accelerated if the assays used in preclinical studies were the same as those used in the clinical laboratory. To validate this approach, we developed a multiplexed assay for the quantification of 2 clinically well-known biomarkers in human plasma, apolipoprotein A-I and apolipoprotein B (apoA-I and apoB).
METHODS
We used PeptideAtlas to identify candidate peptides. Human samples were denatured with urea or trifluoroethanol, reduced and alkylated, and digested with trypsin. We compared reversed-phase chromatographic separation of peptides with normal flow and microflow, and we normalized endogenous peptide peak areas to internal standard peptides. We evaluated different methods of calibration and compared the final method with a nephelometric immunoassay.
RESULTS
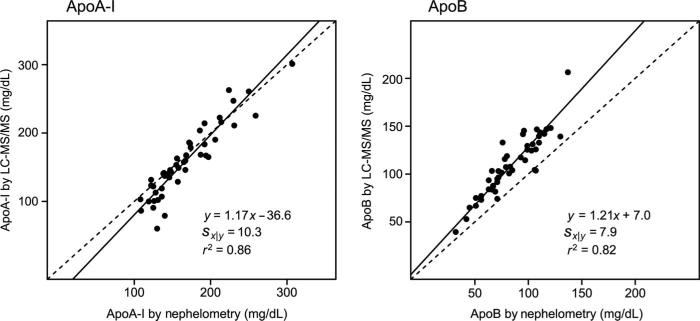
We developed a final method using trifluoroethanol denaturation, 21-h digestion, normal flow chromatography-electrospray ionization, and calibration with a single normal human plasma sample. For samples injected in duplicate, the method had intraassay CVs <6% and interassay CVs <12% for both proteins, and compared well with immunoassay (n = 47; Deming regression, LC-MRM/MS = 1.17 × immunoassay – 36.6; Sx|y = 10.3 for apoA-I and LC-MRM/MS = 1.21 × immunoassay + 7.0; Sx|y = 7.9 for apoB).
CONCLUSIONS
Multiplexed quantification of proteins in human plasma/serum by LC-MRM/MS is possible and compares well with clinically useful immunoassays. The potential application of single-point calibration to large clinical studies could simplify efforts to reduce day-to-day digestion variability.
Liquid chromatography–tandem mass spectrometry (LC-MS/MS)2 is a powerful technique for the absolute quantification of small molecules. It facilitates high-throughput, low limit of quantification, and specific detection of analytes in complex matrices, including whole blood and plasma. Currently, however, most clinical laboratories use immunoassays, which can suffer from limitations including interfering antibodies, poor interplatform concordance, cross-reactivity, and high-dose hook effects (1).
Substantial progress has been made toward the quantification of proteins using isotope dilution LC-MS/MS (2–9). The most robust applications, considered by some to be the gold standard proteomic methods (10), rely on the denaturation of all proteins in a sample and the proteolytic digestion of proteins into peptides. The resulting peptides act as surrogates for the intact protein and are quantified by selected reaction monitoring and stable isotope–labeled internal standard peptides, which normalize for ion suppression and variable mass spectrometer performance (11, 12). There is intense interest in using liquid-chromatography–multiple-reaction–monitoring mass spectrometry (LC-MRM/MS) to measure putative biomarkers in large verification studies (13–15), but formal validation of multiplexed LC-MRM/MS assays with existing clinical immunoassays for known biomarkers has not yet been described.
Apolipoproteins have become important screening biomarkers for predicting cardiovascular risk in the general population (16, 17). Several studies suggest that they may be better predictors than traditional serum lipid measurements (18, 19). Apolipoprotein B (apoB) is the main structural protein in atherogenic particles, and its quantification in plasma estimates the number of atherogenic particles. In contrast, measurement of the concentration of apolipoprotein A-I (apoA-I) in plasma estimates the number of HDL particles, which are atheroprotective by facilitating reverse cholesterol transport and inhibiting inflammation (20, 21). The ratio of the 2 proteins may have improved diagnostic sensitivity and specificity in assessing cardiovascular disease risk compared to either protein alone (22). Many other proteins in HDL and LDL are pivotal in lipid metabolism and in the pro- and anti-atherogenic functions of the lipoproteins, but clinical immunoassays are not yet available for the majority of them (23).
Previous studies have suggested that LC-MRM/MS may be useful in the quantification of apolipoproteins in human plasma (7, 24). We investigated whether LC-MRM/MS methodologies could be applied to the absolute quantification in plasma and serum of apolipoproteins A-I and B, which are quite disparate in size, tertiary structure, and carbohydrate content. Our results demonstrate that multiple proteins can be reliably quantified by using LC-MRM/MS and by using appropriately designed tryptic digestion conditions.
Materials and Methods
CHEMICALS AND HUMAN SAMPLES
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. The use of human specimens was approved by the Human Subjects Division at the University of Washington.
ANALYSIS OF APOLIPOPROTEINS BY NEPHELOMETRIC IMMUNOASSAY
ApoA-I and apoB were measured on a BNII nephelometer (Dade Behring/Siemens) according to the manufacturer's instructions. The assays had interassay CVs of 4.63% at 79.6 mg/dL (796 mg/L) for apoA-I and 8.83% at 35.6 mg/dL (356 mg/L) for apoB. The limits of quantification were 5 mg/dL (50 mg/L) and 24 mg/dL (240 mg/L) for each assay, respectively.
INTERNAL STANDARDS
We prepared internal standard peptides V121QPYLDDFQK*130 for apoA-I and V1968SALLTPAEQTGTWK*1982 for apoB by standard solid-phase synthesis. We purchased stable isotope–labeled lysine (K*, 13C6 15N2) from Cambridge Isotopes and incorporated it at the C-terminus of each peptide. The peptides were >90% pure by HPLC. Because we did not use internal standard peptides as a concentration reference, we did not perform amino acid analysis.
CALIBRATION MIXTURES
External calibrator mixtures for the LC-MRM/MS assays contained purified, delipidated apolipoproteins added to normal serum matrix and were prepared with the same WHO/IFCC traceable preparation (traceable to reference materials BCR-393 and SP3–07) that was used for quality control in the nephelometric assay (Dade Behring/Siemens). However, it was reconstituted with one-half of the recommended volume of water [apoA-I, 249 mg/dL (2490 mg/L); apoB, 142 mg/dL (1420 mg/L)], and serial dilutions were made with water. Final concentrations of apoA-I were 249, 124.5, 62, and 31 mg/dL, and of apoB, 142, 71, 35, and 17.5 mg/dL. Each calibration mixture dilution was divided into aliquots and frozen at –80 °C until used.
DENATURATION AND DIGESTION OF CALIBRATION MIXTURE AND SAMPLES
For denaturation with urea, we diluted 10 μL serum or plasma with 30 μL of 8 mol/L urea (made fresh). For denaturation with trifluoroethanol (TFE), we diluted 10 μL serum or plasma with 25 μL of 0.5 mol/L aqueous ammonium bicarbonate, and then added 35 μL of trifluoroethanol to the diluted sample. For both urea and TFE, we heated samples for 1 h with agitation (37 °C or 65 °C for urea or TFE denaturation, respectively) before adding 1 μL of 0.5 mol/L aqueous dithiothreitol (final concentration 7 mmol/L). We again heated the samples for 1 h at 37 °C or 65 °C with agitation. The samples were then alkylated for 30 min with aqueous iodoacetamide (final concentration 38 mmol/L) in the dark at room temperature. Excess alkylating agent was quenched with 1 μL of 0.5 mol/L dithiothreitol. Samples were diluted with 0.7 mL of 0.14 mol/L aqueous ammonium bicarbonate and digested with acetylated, nonsequencing-grade trypsin (40 μg trypsin/10 μL sample) for 2 h at 37 °C with agitation. We added more trypsin (final concentration 80 μg trypsin/10 μL sample), and the samples were digested again at 37 °C with agitation (total digestion time ≤21 h). Samples were acidified with 1 μL formic acid and purified by using 1 mL, 30 mg Oasis HLB solid phase extraction columns (Waters). Columns were wetted with 1 mL of methanol and equilibrated with 1 mL of 0.1% formic acid in water before the addition of the samples. We washed the columns with 1 mL of 0.1% formic acid in water and eluted the peptides in 1 mL methanol. The eluant was evaporated in a centrifugal vacuum concentrator to dryness and reconstituted in 300 μL of 0.1% formic acid, 5% acetonitrile in water. Before analysis by normal-flow LC-MRM/MS, we added stable isotope–labeled internal standard (2 μL of 50 μmol/L each peptide) to 20 μL sample. For microflow LC-MRM/MS, we added 2 μL internal standard to 4 μL sample and diluted it with 14 μL of 0.2% formic acid in water.
TIME-COURSE EXPERIMENT
To evaluate the kinetics of protein digestion, proteins were denatured, reduced, and alkylated as usual. However, portions (100 μL) were withdrawn and acidified at 0, 1, 2, 4, 6, 8, and 21 h of total trypsin digestion. All samples other than the time-course experiment were digested for a total of 21 h.
MICROFLOW LIQUID CHROMATOGRAPHY
Peptides (1 μL mix containing internal standard) were desalted on a C18 trapping column (Dionex, Acclaim Pepmap 100 Å, 5 μm, 5 by 1.0 mm), by use of a Tempo 1D Plus autosampler-liquid chromatography system (AB Sciex), eluted onto a capillary C18 analytical column (Michrom, Magic 200 Å, 5 μm, 150 by 0.15 mm), and separated over 8 min with a linear gradient of acetonitrile (5%–40%) in 0.2% formic acid in water at a flow rate of 1 μL/min. Peptides were ionized with a Microionspray II ion source (AB Sciex). Pertinent settings for the ion source and interface: curtain gas parameter, 20; nebulizer gas parameter; 27; and ion-spray voltage, 3100 V.
NORMAL-FLOW LIQUID CHROMATOGRAPHY
Peptides (4 μL mix containing internal standard) were resolved by standard C18 reversed-phase chromatography (Restek, Pinnacle Aqueous, 140 Å, 3 μm, 100 by 3.2 mm) on a Shimadzu LC-20AD chromatographic system with a linear gradient over 10 min from 20% to 95% acetonitrile in 0.2% formic acid in water at 400 μL/min flow rate. Peptides were ionized with a turbo ion spray interface (AB Sciex). Pertinent settings for the ion source and interface: curtain gas parameter, 20; nebulizer gas parameter, 40; auxiliary desolvation gas parameter, 90; ion-spray voltage, 2500 V; and auxiliary desolvation gas temperature, 400 °C.
TANDEM MASS SPECTROMETRY AND PEAK INTEGRATION
The peptides analyzed by MRM, their position in the primary sequence of the proteins, and the transitions used to monitor them are listed in Table 1. Mass analysis was performed with an API 4000 QTRAP triple quadrupole mass spectrometer (AB Sciex). Pertinent settings for the mass spectrometer: collision cell gas pressure, 12 U; entrance potential, 10 V; declustering potential, 80–90 V; collision cell exit potential, 7–15 V; and dwell time, 25 ms. Collision energies used for each transition are also listed in Table 1. We determined peak areas for the endogenous and internal standard peptides in Analyst 1.4.2. The 2 transitions per peptide or internal standard were summed. We calculated the response of each peptide as the ratio of the peak area of each endogenous peptide to the peak area of an internal standard. Each sample was injected in duplicate, and the results were averaged. We normalized the peak areas of the apoA-I and apoB peptides to those of the stable isotope–labeled peptides VQPYLDDFQK* and VSALLT PAEQTGTWK*, respectively.
Table 1.
Peptides studied for the quantification of apoA-I and apoB.a
| Protein and peptide | Position in sequenceb | m/z | Fragment 1 | Fragment 2 | CE, eV |
|---|---|---|---|---|---|
| ApoA-I | |||||
| VQPYLDDFQK | 121–130 | 626.8 | 422.2 (y3) | 1025.5 (y8) | 32/23 |
| DYVSQFEGSALGK | 52–64 | 700.8 | 1023.5 (y10) | 808.4 (y8) | 38 |
| LLDNWDSVTSTFSK | 70–83 | 806.9 | 1386.6 (y12) | 971.5 (y9) | 43 |
| ATEHLSTLSEK | 220–230 | 405.9 | 572.8 (y10+2) | 522.3 (y9+2) | 19 |
| ApoB | |||||
| VSALLTPAEQTGTWK | 1968–1982 | 801.4 | 1017.5 (y9) | 1118.6 (y10) | 34 |
| FPEVDVLTK | 3791–3799 | 524.3 | 450.8 (y8+2) | 803.5 (y7) | 28 |
| NLQNNAEWVYQGAIR | 4107–4121 | 888.4 | 707.4 (y6) | 992.5 (y8) | 46 |
MRM transitions were assembled into a single analysis program. The first quadrupole was set to the m/z listed, and after collision-induced dissociation at the collision energy (CE) listed, either fragment 1 or 2 was monitored for 25 ms.
Results
PEPTIDE SELECTION
To develop a quantitative method for the quantification of apoA-I and apoB in human serum and plasma, 10 potentially useful tryptic peptides and fragment ions for each protein were selected by using PeptideAtlas (25), which is a database of tandem mass spectra from proteomic analyses of many different sample types, including human plasma. The peptides chosen were the most commonly observed peptides across human plasma experiments in the database, and the fragment ions selected were the most abundant peaks in the consensus spectra. Peptides containing cysteines or methionines and products of incomplete digestion were excluded. In preliminary experiments, 10 human samples were digested and analyzed by microflow LC-MRM/MS for the 20 potentially useful peptides. Those peptides that appeared to correlate well with nephelometry in this initial experiment (r > 0.70; data not shown) were pursued as viable peptides for the assay and are located throughout the linear sequences of the proteins (Table 1).
EVALUATION OF TRIFLUOROETHANOL AND UREA AS DENATURANTS
To completely denature the proteins in plasma, we compared a protocol that used trifluoroethanol (26) with one that used a more traditional approach using urea. The commercially available calibration mixtures and human samples digested in TFE exhibited a 2- to 6-fold increase in peak area in samples processed with TFE compared with samples processed with urea (P < 0.001). On the basis of the improved signal, we used TFE as the denaturing agent for the remaining serum or plasma samples.
EVALUATION OF NORMAL-FLOW LIQUID CHROMATOGRAPHY
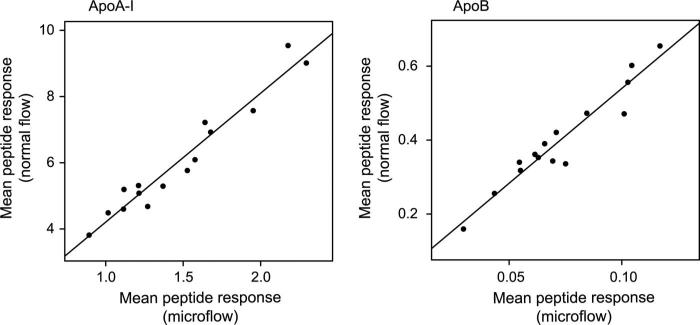
Most proteomics work flows rely on nano- or micro-flow liquid chromatography (≤1 μL/min) for improved limits of detection. Unfortunately, the systems used to ionize molecules at these low flow rates are not as robust as normal-flow systems. In addition, previous experiments have led to questions about the ability of normal-flow systems to reliably detect certain peptides (24). To evaluate whether normal flow rates could be used to detect peptides from the apolipoproteins, we analyzed serum digests from 15 patients by using both normal-flow and microflow systems. The Pearson correlation coefficients (r2) were 0.94 for apoA-I and 0.92 for apoB (Fig. 1). In addition, when compared with nephelometry, the normal-flow system was comparable to the microflow system for apoA-I (r2 = 0.91 vs 0.94; data not shown) and apoB (r2 = 0.97 vs 0.93; data not shown). These results suggested that the more robust normal-flow system would be capable of reliably detecting target analytes in tryptic digests of plasma.
Fig. 1. Comparison of normal-flow and microflow liquid chromatography.
Mean peptide responses for apoA-I and apoB are shown for normal flow (0.4 mL/min) and microflow (1 μL/min) chromatographic separation and electrospray ionization for the same samples digested on different days and supplemented with different amounts of internal standard.
TIME-COURSE EXPERIMENTS
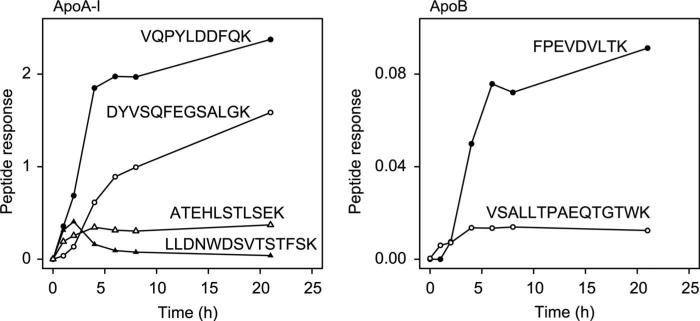
It was unclear whether a single digestion time would be appropriate for all proteins in complex mixtures. Given the large difference in size and extent of glycosylation of apoA-I vs apoB, we examined the efficiency of digestion of the 2 proteins in plasma over time by sampling tryptic digests of a human plasma sample at 0, 1, 2, 4, 6, 8, and 21 h (Fig. 2). Interestingly, the observed signals from 2 apoA-I peptides continued to increase over the time course with the greatest increase by 4–6 h (VQPYLDDFQK and DYVSQFEGSALGK). In striking contrast, the observed signal for 2 other apoA-I peptides reached a maximum in 2–4 h and then plateaued or decreased during the remainder of the time course (ATEHLSTLSEK and LLDNWDSVTSTFSK). The signal for 2 peptides in apoB behaved similarly to peptides in apoA-I, with one reaching a maximum after 4 h and the other increasing over the course of the digestion (VSALLTPAEQTGTWK and FPEVDVLTK, respectively).
Fig. 2. Time course of digestion.
Responses for peptides in apoA-I and apoB are plotted against the length of digestion time of human plasma.
COMPARISON OF PEPTIDES FROM THE SAME PROTEIN
As it was apparent that peptides were generated at varying rates during trypsin digestion, we next investigated if peptides were liberated from proteins consistently from patient sample to patient sample. Fourteen patient samples were digested for 21 h, and we examined the correlation of the response of peptides to one another (Fig. 3). Two pairs of apoA-I peptides (VQPYLDDFQK and DYVSQFEGSALGK; ATEHLSTLSEK and DYVSQFEGSALGK) correlated well with one another (r2 = 0.96 and 0.82, respectively). For apoB, peptides VSALLTPAEQTGTWK and FPEVDVLTK correlated much better (r2 = 0.89) than VSALLTPAEQTGTWK and NLQNNAEWVYQGAIR (r2 = 0.01). The stronger correlations between certain peptides suggested that they are released similarly from sample to sample, which supports the proposal that ratios of peptides from the same protein might be used in the future to detect interferences in the assay (2, 27).
Fig. 3. Peptide comparisons.
To evaluate the consistency of peptide generation among samples, responses for peptides from the same proteins are plotted against one another for apoA-I and apoB.
SELECTION OF EXTERNAL CALIBRATION MATERIALS
To calibrate peptide responses in the assay, we first used an external calibration curve that was generated from the same human serum–based preparation used in the nephelometric immunoassay. This calibration mixture was synthesized by spiking purified/delipi-dated apolipoproteins into a human serum matrix. The results of a comparison of the calibrated LC-MRM/MS assay by using the TFE denaturation step with the nephelometric assay were surprising. Whereas the correlations were acceptable (r > 0.90 for apoA-I and apoB; n = 7), the slope of the Deming regression was 2.10 for apoA-I and 1.26 for apoB (Table 2). To test whether the denaturation step was behaving differently in the calibrators derived from purified proteins vs native human serum, we compared a calibrated LC-MRM/MS assay that used the urea denaturation step with the nephelometric immunoassay; a Deming regression slope of 0.89 was observed for apoA-I, but the slope for apoB increased to 5.37 (Table 2). As an alternative strategy, we used a single patient plasma sample with apoA-I and apoB concentrations defined by the nephelometric immunoassay to calibrate the LC-MRM/MS assay. The calibrator was run in triplicate, and the responses were averaged. By use of this approach, the slopes of the Deming regression for the TFE and urea denaturation steps were markedly improved for apoA-I (1.06 and 0.94, respectively) and apoB (1.05 and 1.13, respectively), suggesting that a single-point calibration could be used with either denaturation step to adequately calibrate the response in the LC-MRM/MS assay. Further, it appears that a calibration curve made from purified proteins spiked into a standard matrix does not adequately reflect the digestion efficiency of native proteins in actual human samples.
Table 2.
Method comparisons between nephelometry and LC-MRM/MS for the quantification of apoA-I and apoB.a
| Protein and calibration | Denaturant | Slope | Intercept |
|---|---|---|---|
| ApoA-I | |||
| Calibration curve | TFE | 2.10 | –78.1 |
| Urea | 0.89 | –3.4 | |
| Single calibrator | TFE | 1.06 | 1.68 |
| Urea | 0.94 | 10.9 | |
| ApoB | |||
| Calibration curve | TFE | 1.26 | –5.2 |
| Urea | 5.37 | –132.9 | |
| Single calibrator | TFE | 1.05 | –8.8 |
| Urea | 1.13 | –2.1 |
Deming regression coefficients are presented for the method comparison of immunoassays and LC-MRM/MS assays, which were performed with 2 different denaturation reagents and 2 different approaches to calibration.
METHOD EVALUATION
We next determined how well the new single-point-calibrated assay, with a TFE denaturation step, a 21-h digestion time, and analysis by normal flow LC-MRM/MS, behaved in a clinical work flow. First, we measured 47 different patient specimens on 3 days spread over 3 weeks (approximately 15 specimens per day) and compared the results to nephelometric immunoassay (Fig. 4). The Deming regression equations were y = 1.17x – 36.6 (r = 0.93; range, 108–307 mg/dL) for ApoA-I and y = 1.21x + 7.0 (r = 0.91; range, 32–137 mg/dL) for apoB. These correlations over multiple days of analysis indicated that a single-point calibration curve using native, intact lipoproteins in a matrix identical to samples was effective at correcting for between-day variability due to digestion and liquid chromatograph/mass spectrometer performance. It is interesting to note that the individual peptides behaved similarly for apoA-I but differently for apoB (see Supplemental Fig. 1, which accompanies the online version of this article at http://www.clinchem.org/content/vol56/issue12). Next, we measured 14 samples over 7 days and estimated the intraassay and interassay imprecision (28). Intraassay imprecision (CV) was 6.3% for apoA-I (156 mg/dL) and 5.8% for apoB (106 mg/dL), and the interassay imprecision was 11.2% for apoA-I and 11.4% for apoB. We assessed linearity by using a patient sample with apoA-I concentration 172 mg/dL and apoB concentration 117 mg/dL, which was serially diluted with water. This approach generated a dilution curve with varying amounts of protein and lipid, which could potentially affect the adequacy of denaturation and digestion. Nonetheless, by using this approach, the assay was linear for apoA-I concentrations between 9 and 172 mg/dL and apoB concentrations between 4 and 117 mg/dL (see online Supplemental Fig. 2), demonstrating the robustness of the TFE denaturation and 21 h digestion steps. Additional experiments demonstrated no significant interference from increased endogenous triglycerides (400–717 mg/dL) for either protein and no effect of multiple freeze-thaw cycles for apoA-I, but a modest reduction in measured apoB concentration was observed after 4 or more freeze-thaw cycles (see online Supplemental Tables 1 and 2).
Fig. 4. Method comparison of LC-MRM/MS assay with immunoassay.
Concentrations of apoA-I and apoB measured by LC-MRM/MS assay calibrated with a single human plasma calibrator were compared with concentrations of the proteins measured by nephelometric immunoassay. The results of Deming regression are shown with the standard deviation of the residuals (Sx|y) and the Pearson correlation coefficient (r2). Deming regression was performed in R (35) with the MethComp package and 1000 bootstrap iterations and variance ratio (LC-MS/MS:immunoassay = 4).
Discussion
We developed a multiplexed LC-MRM/MS assay for the absolute quantification of 2 clinically important apolipoproteins in human serum and plasma. The proteins differ in size (approximately 30 kDa for apoA-I vs approximately 500 kDa for apoB-100), percent carbohydrate (0% vs 9% by mass, respectively (29), and disulfide bridging (0 vs 8, respectively). As a result, the selection of the proper digestion conditions and calibration method was critical for a favorable comparison with a clinically used nephelometric immunoassay platform.
There are several approaches to the quantification of proteins using LC-MRM/MS in clinical samples. For abundant proteins in human plasma, such as those studied here (2–30 μmol/L), plasma or serum can be digested and peptides quantified directly from the digest (3). For lower-abundance proteins, some sample manipulation before analysis is necessary. For example, target proteins can be enriched by an immunoaffinity step before analysis by mass spectrometry (30), or abundant, nontargeted proteins can be depleted by using immunoaffinity (6). For even lower-abundance proteins, an interesting approach, termed SISCAPA (stable isotope standards and capture by anti-peptide antibodies (31), has now been successful in a few different clinical/pharmaceutical situations and uses immunoaffinity enrichment of peptides after digestion of serum or plasma (4, 5, 8).
By use of these approaches, progress has been made in the quantification of proteins in human serum or plasma by LC-MRM/MS. In 1 study, the putative biomarker for prostate cancer, Zn-α2-glycoprotein, was quantified in digested plasma (3). This assay was not compared against an immunoassay. Other assays have required extensive sample preparation before analysis. For example, Kuhn et al. (6) quantified C-reactive protein by first depleting high-abundance proteins and then further reducing sample complexity by size-exclusion chromatography. A limited method comparison demonstrated that LC-MRM/MS and immunoassay gave results at a similar order of magnitude. In more recent experiments, the low-abundance clinical targets thyroglobulin and troponin I have been quantified by using immunoaffinity enrichment of peptides from digests, and comparisons with immunoassay have been very promising (r2 > 0.80) (4, 5).
An important study that evaluated the interlaboratory reproducibility of LC-MRM/MS assays of peptides spiked into digests of human plasma suggested that trypsin digestion can be quite variable from laboratory to laboratory, but that the quantification of peptides could be quite reliable (2, 32). It is possible that the reproducibility of digestion after TFE denaturation would help reduce interlaboratory digestion variability, but this needs to be tested in future experiments.
The interassay CVs of our apoA-I and apoB assays were 11.2% and 11.4%, respectively. Of note, the internal standard peptides were added after digestion and SPE purification of tryptic peptides. It is possible that incorporation of stable isotope–labeled internal standard peptides or proteins earlier in the process could help reduce the variability of LC-MRM/MS assays (33), which needs to be explored further.
The calibration of proteomic LC-MRM/MS assays is an important problem that has not yet drawn much attention. Two important steps in the assay influence the observed response: the digestion step and the LC-MRM/MS analysis. One approach to calibration makes the assumptions that (1) protein digestion goes to completion, (2) peptides are detected to the same extent from sample to sample, and (3) the response (peak area of endogenous peptide divided by peak area of the internal standard) of the peptide is linear across the measurement range. If all 3 of these assumptions hold true, then the response for each peptide can be used to calculate the concentration of endogenous peptide, because the amount of internal standard is known. Unfortunately, all 3 of these assumptions are frequently false (2, 4, 34). Another attractive approach uses an external calibration curve made with peptides spiked into a matrix of digested plasma (7). Although this approach helps control for LC-MS/MS analytical variability, it does nothing to control for the variability of digestion from day to day.
In contrast with these approaches, it was our initial hope that commercially available calibration mixtures, which were made from delipidated apolipoproteins spiked into a human serum-based matrix and digested in parallel with unknown samples, could control for the variability of digestion. Unfortunately, purified proteins spiked into serum do not appear to digest with efficiencies similar to those of native human lipoproteins in plasma, and different denaturation reagents led to very different results when this approach was used (Table 2). By using an actual human plasma specimen as a single-point external calibration mixture, with apolipoprotein concentrations defined by immunoassay, we were able to effectively calibrate the LC-MRM/MS assay over multiple days (Fig. 4). Thus, a truly matrix-matched human plasma calibration mixture containing native proteins can control for the variability in digestion completeness observed from day to day. With this approach, the correlation with immunoassay did not rely as heavily on the denaturation reagents used for the digestion (Table 2).
It has always been assumed that the quantification of different peptides from the same protein will lead to similar quantitative results. Even for the use of TFE (the use of which led to greater signals for the apoB peptides), the results of time-course experiments were different for different peptides (Fig. 2), and peptides responses correlated variably with one another (Fig. 3) and with immunoassay results. Thus, each peptide in a protein could lead to a different absolute quantification of patient samples. For the 2 proteins presented here, there were at least 2 peptides from each protein with responses that correlated well with each other as well as with immunoassay results. The mechanisms to explain the different behavior of peptides from the same protein are unclear, but could involve peptide degradation due to contaminating enzymes in the nonsequencing-grade trypsin used, peptide aggregation or nonspecific association with the plastic tube, or peptide oxidation. Further experiments are needed to elucidate the important mechanisms that lead to the observed discrepancies.
Two previous studies provided preliminary evidence that LC-MRM/MS could be useful in the quantification of apoA-I and apoB in human serum and plasma. In the first, Kay et al. (24) demonstrated that ultrahigh-pressure liquid chromatography could be used to resolve peptides from apoA-I and apoB from tryptic digests of human serum. The authors used external calibration with a commercially available serum-based calibration mixture and a stable isotope–labeled internal standard peptide to quantify apoA-I with reasonable intraassay precision and accuracy compared with a clinically used immunoassay. However, no effort was made to simultaneously quantify apoB or measure interassay precision. In the second study, Kuzyk et al. (7) used external calibration with a peptide-based calibration mixture and demonstrated reasonable precision and accuracy. The authors relied on capillary flow rates to detect and quantify 45 serum proteins in their assay, and their results were not compared directly with immunoassays for apoA-I and apoB. The current study complements these 2 important investigations and further demonstrates the potential clinical utility of LC-MRM/MS quantification of proteins.
The throughput of the new LC-MRM/MS assay is not as high as that of traditional automated immunoassays. However, the normal flow-chromatographic methods described herein can accommodate approximately 15 samples per day (in addition to calibration mixtures), which is significantly better than nontargeted proteomic methods, which are generally limited to 3 or fewer samples per day (11, 12). With improvements in the work flow using automation and shorter chromatographic run times, multiplexed LC-MRM/ MS protein assays may be translated to large clinical studies and eventually to regular clinical laboratory operations. The assay becomes even more attractive in these settings as other protein analytes are added.
Supplementary Material
Acknowledgments
The authors thank Christine Miller and Jessica Becker for helpful discussions and Geoff Baird for critically reading the manuscript.
Research Funding: A.N. Hoofnagle, Waters, Inc., Bruker Daltonics, Inc., the Nutrition and Obesity Research Center (NIH P30DK035816), and the Diabetes and Endocrinology Research Center (NIH P30DK017047). This work was supported by the Clinical Mass Spectrometry Facility at the University of Washington.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: LC-MS/MS, liquid chromatography–tandem mass spectrometry; MRM, multiple reaction monitoring; apoB, apolipoprotein B; apoA-I, apolipoprotein A-I; TFE, trifluoroethanol; SISCAPA, stable isotope standards and capture by anti-peptide antibodies.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: A.N. Hoofnagle, Association for Mass Spectrometry: Applications to the Clinical Laboratory, Inc.
Consultant or Advisory Role: A.N. Hoofnagle, Thermo-Fisher.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondar OP, Barnidge DR, Klee EW, Davis BJ, Klee GG. LC-MS/MS quantification of Zn-alpha2 glycoprotein: a potential serum biomarker for prostate cancer. Clin Chem. 2007;53:673–8. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- 4.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54:1796–804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55:1108–17. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–86. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 7.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, et al. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8:1860–77. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neubert H, Gale J, Muirhead D. Online high-flow peptide immunoaffinity enrichment and nanoflow liquid chromatography/tandem mass spectrometry: assay development for total salivary pepsin/pepsinogen. Clin Chem. 2010;56:1413–23. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 9.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein bio-markers. Mol Cell Proteomics. 2010;9:184–96. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–62. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 11.Brun V, Masselon C, Garin J, Dupuis A. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics. 2009;72:740–9. doi: 10.1016/j.jprot.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44:1637–60. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 13.Anderson NL, Anderson NG, Pearson TW, Borchers CH, Paulovich AG, Patterson SD, et al. A human proteome detection and quantitation project. Mol Cell Proteomics. 2009;8:883–6. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr SA, Anderson L. Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem. 2008;54:1749–52. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteom Clin Appl. 2008;2:1386–402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contois JH, McConnell JP, Sethi AA, Csako G, Devaraj S, Hoefner DM, Warnick GR. Apolipoprotein B and cardiovascular disease risk: position statement from the AACC Lipoproteins and Vascular Diseases Division Working Group on Best Practices. Clin Chem. 2009;55:407–19. doi: 10.1373/clinchem.2008.118356. [DOI] [PubMed] [Google Scholar]
- 17.Davidson MH. Apolipoprotein measurements: is more widespread use clinically indicated? Clin Cardiol. 2009;32:482–6. doi: 10.1002/clc.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holme I, Aastveit AH, Hammar N, Jungner I, Walldius G. Lipoprotein components and risk of congestive heart failure in 84,740 men and women in the Apolipoprotein MOrtality RISk study (AMORIS). Eur J Heart Fail. 2009;11:1036–42. doi: 10.1093/eurjhf/hfp129. [DOI] [PubMed] [Google Scholar]
- 19.Lamarche B, Moorjani S, Lupien PJ, Cantin B, Bernard PM, Dagenais GR, Despres JP. Apolipoprotein A-I and B levels and the risk of ischemic heart disease during a five-year follow-up of men in the Quebec cardiovascular study. Circulation. 1996;94:273–8. doi: 10.1161/01.cir.94.3.273. [DOI] [PubMed] [Google Scholar]
- 20.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res. 2004;95:764–72. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 21.Movva R, Rader DJ. Laboratory assessment of HDL heterogeneity and function. Clin Chem. 2008;54:788–800. doi: 10.1373/clinchem.2007.101923. [DOI] [PubMed] [Google Scholar]
- 22.Walldius G, Jungner I. The apoB/apoA-I ratio: a strong, new risk factor for cardiovascular disease and a target for lipid-lowering therapy—a review of the evidence. J Intern Med. 2006;259:493–519. doi: 10.1111/j.1365-2796.2006.01643.x. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle AN, Heinecke JW. Lipoproteomics: using mass spectrometry-based proteomics to explore the assembly, structure, and function of lipoproteins. J Lipid Res. 2009;50:1967–75. doi: 10.1194/jlr.R900015-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay RG, Gregory B, Grace PB, Pleasance S. The application of ultra-performance liquid chromatography/tandem mass spectrometry to the detection and quantitation of apolipoproteins in human serum. Rapid Commun Mass Spectrom. 2007;21:2585–93. doi: 10.1002/rcm.3130. [DOI] [PubMed] [Google Scholar]
- 25.Deutsch EW, Eng JK, Zhang H, King NL, Nesvizhskii AI, Lin B, et al. Human plasma PeptideAtlas. Proteomics. 2005;5:3497–500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 26.Meza JE, Miller CA, Fischer SM. [August 2010];Improved tryptic digestion of proteins using 2,2,2-trifluoroethanol (TFE) http://www.chem.agilent.com/Library/posters/Public/5989-1781EN.pdf.
- 27.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 2010;56:291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodbard D. Statistical quality control and routine data processing for radioimmunoassays and immunoradiometric assays. Clin Chem. 1974;20:1255–70. [PubMed] [Google Scholar]
- 29.La Belle M, McCall MR, Krauss RM, Forte TM. Unique structural properties of apolipoprotein B in low-density lipoproteins produced by several human hepatoma-derived cell lines. Biochim Biophys Acta. 1990;1046:288–93. doi: 10.1016/0005-2760(90)90243-q. [DOI] [PubMed] [Google Scholar]
- 30.Lopez MF, Rezai T, Sarracino DA, Prakash A, Krastins B, Athanas M, et al. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin Chem. 2010;56:281–90. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 31.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J Proteome Res. 2004;3:235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 32.Hoofnagle AN. Quantitative clinical proteomics by liquid chromatography-tandem mass spectrometry: assessing the platform. Clin Chem. 2010;56:161–4. doi: 10.1373/clinchem.2009.134049. [DOI] [PubMed] [Google Scholar]
- 33.Hoofnagle AN. Peptide lost and found: internal standards and the mass spectrometric quantification of peptides. Clin Chem. 2010;56:1515–17. doi: 10.1373/clinchem.2010.152181. [DOI] [PubMed] [Google Scholar]
- 34.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, et al. Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–82. [PubMed] [Google Scholar]
- 35.R Development Core Team . R: A language and environment for statistical computing [computer program] R Foundation for Statistical Computing; Vienna (Austria): 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.