Abstract
Alcoholism is a major health problem in the United States and worldwide, and alcohol remains the single most significant cause of liver-related diseases and deaths. Alcohol is known to influence nutritional status at many levels including nutrient intake, absorption, utilization, and excretion, and can lead to many nutritional disturbances and deficiencies. Nutrients can dramatically affect gene expression and alcohol-induced nutrient imbalance may be a major contributor to pathogenic gene expression in alcohol-induced liver disease (ALD). There is growing interest regarding epigenetic changes, including histone modifications that regulate gene expression during disease pathogenesis. Notably, modifications of core histones in the nucleosome regulate chromatin structure and DNA methylation, and control gene transcription. This review highlights the role of nutrient disturbances brought about during alcohol metabolism and their impact on epigenetic histone modifications that may contribute to ALD. The review is focused on four critical metabolites, namely, acetate, S-adenosylmethionine, nicotinamide adenine dinucleotide and zinc that are particularly relevant to alcohol metabolism and ALD.
Keywords: Alcohol, Liver disease, Nutrients, Metabolism, Histone, Epigenetic modifications, S-adenosylmethionine, Acetate, Zinc, Nicotinamide adenine dinucleotide
INTRODUCTION
Alcoholism is a growing health problem worldwide. In the United States, alcoholism is a major cause of liver-related disease and deaths. Recent statistics reveal 52% of US adults to be “current regular” drinkers (Summary Health Statistics for U.S. Adults: National Health Interview Survey, 2009) and the death rate for alcohol-induced causes to be on the rise (National Vital Statistics Reports, May 2010). As a result, alcohol-induced liver disease (ALD) continues to be studied, with the objectives of elucidating the underlying mechanisms and discovering potential therapeutic targets.
ALD consists of the spectrum of pathological changes including fatty liver, alcoholic hepatitis and alcoholic cirrhosis. The clinical manifestations of these changes and the pathogenesis of this disease have been extensively studied and described. There is a growing body of evidence supporting the involvement of epigenetic mechanisms in response to environmental inputs in the development of human disease. Accordingly, in recent years, there has been increasing interest in understanding the role of epigenetic mechanisms in the initiation and/or progression of ALD.
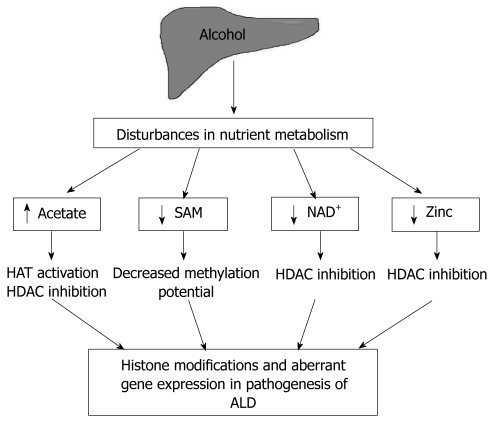
Nutrient fluctuations can impact transcriptional activity and expression of selective genes by modulating epigenetic parameters including histone modifications, DNA methylation, and nucleosome positioning. In ALD, especially in chronic alcoholics, the combined effect of alcohol metabolism and compromised nutrition causes major nutrient disturbances that are likely to influence epigenetic mechanisms, gene expression and disease pathogenesis. Covalent modifications of the amino termini of the core histones in nucleosomes play a key role in regulating chromatin structure as well as DNA methylation status. This review interrelates alcohol-mediated nutrient disturbances and consequent histone modifications that may have a contributory role in ALD (Figure 1). Specifically, the review is focused on fluctuations in four critical metabolites, namely, acetate, S-adenosylmethionine (SAM, also known as SAMe or AdoMet), nicotinamide adenine dinucleotide (NAD+) and zinc that are relevant to alcohol metabolism and ALD.
Figure 1.
Nutrient disturbances, a potential link between alcohol metabolism and histone modifications in alcohol-induced liver disease. SAM: S-adenosylmethionine; NAD+: Nicotinamide adenine dinucleotide; HAT: Histone acetyltransferase; HDAC: Histone deacetylase; ALD: Alcohol-induced liver disease.
ALCOHOL AND ACETATE
The principal route of ethanol oxidation is through the enzyme liver alcohol dehydrogenase, which converts alcohol to aldehyde with the reduction of NAD+ to NADH[1]. Acetaldehyde is then further oxidized by acetaldehyde dehydrogenase to acetate. The other major route of oxidation is through the microsomal ethanol oxidizing system (MEOS), in which the chief enzyme catalyzing alcohol oxidation is the cytochrome P450 mixed-function oxidase isoenzyme CYP2E1[1]. This route is engaged when alcohol is ingested in large quantities or in chronic alcoholics, who upregulate CYP2E1 expression. Thus, the end-product of both pathways of ethanol metabolism in the liver is free acetate[1]. This free acetate is then incorporated into acetyl-coenzyme A (acetyl-coA), by the catalytic action of the cytosolic and mitochondrial enzymes acetyl-coA synthetases[2]. Acetyl-coA is the substrate for histone acetylation, in addition to being utilized in the Krebs cycle, fatty acid synthesis and acetylation of other proteins[3].
Alcohol increases acetate production
Ethanol consumption has been shown to increase blood acetate levels significantly in several studies, dating back to the 1980s. Short-term ethanol administration in humans led to a sustained steady state concentration of 0.4 to 0.6 mmol acetate within 2 to 5 h following ingestion in a study consisting of healthy male and female volunteers[4]. Although this phenomenon was also seen in alcoholics, there were variations in the kinetics of acetate production, with chronic alcoholics eliminating alcohol faster and producing more acetate[5,6]. Acetate levels were significantly higher both in the hepatic vein (1.79 and 1.15 mmol) and peripherally (0.91 and 0.52 mmol) in alcoholics than in non-alcoholics respectively[6]. In another study employing an enzymic acetate detection method, acetate concentration in peripheral human blood increased to about 20 times the normal level with ethanol consumption, whereas neither fasting nor the intake of a fatty meal significantly influenced acetate concentration[7].
Acetate and histone modifications
Amongst the various modifications documented at the tails of histone proteins in humans, acetylation and methylation are the best characterized[8]. Generally, acetylation at the lysine residues of histones depicts a transcriptionally permissive state, allowing opening up of the chromatin structure and access to transcriptional machinery[8]. Hyperacetylation of histones has been observed after alcohol administration in both, cell culture and animal studies[9-13]. In a 2003 study by Park et al[12], ethanol increased acetylation at lysine 9 on histone 3 (H3K9Ac) in a dose- and time-dependent manner in isolated rat hepatocytes. There was a remarkable 8-fold increase in the amount of H3K9Ac at 24 h by 100 mmol ethanol without an increase in histone 3 protein expression[12]. Also, inhibition of the metabolism of ethanol to acetate largely abolished this effect, suggesting that production of the ethanol metabolite, acetate, was critical to the process of acetylation[12]. Similar experiments in rat hepatic stellate cells (HSCs) demonstrated an increase in H3K9Ac levels with no changes at lysines 14 or 18[9]. An in vivo study of the effect of acute ethanol (binge drinking) in rats concluded that the ethanol-induced increase in H3K9Ac was largely restricted to the liver, lung and spleen, with the liver showing a maximal increase of ~6-fold in a 12 h period[10].
An increase in the acetylation of histones in response to ethanol may be brought about by an orchestration of events that (1) increase the substrate for the reaction, acetyl-coA, (2) and/or modulate the enzymes controlling histone acetylation (HATs, HDACs). A study by Shukla’s group in 2005 demonstrated that ethanol increased histone 3 acetylation at lysine 9 by specifically modulating HAT(s) targeting lysine 9 in rat hepatocytes[13]. However, it was not determined whether ethanol induced this effect by increasing transcriptional expression of HAT(s) or by specifically augmenting their activity. Accordingly, H3K9Ac and HAT activity was also increased by acetate in these cells, again indicating it as the likely mediator of ethanol-induced histone acetylation[13]. Interestingly, signaling pathway analysis showed that mitogen-activated protein kinase kinase (MEK) and c-Jun N-terminal kinase (JNK) inhibitors reduced ethanol-induced acetylation without affecting ethanol-induced HAT activity. This suggests a role for MEK and JNK in histone 3 acetylation induced by ethanol; however, the mitogen-activated protein kinase (MAPK) cascades may influence histone 3 acetylation without involving HAT activity. In similar experiments, acetate-induced acetylation was not affected by MEK or JNK inhibitors further indicating that the MAPK pathway was not downstream of acetate in the process of acetylation. The precise role of MAPKs in ethanol-induced histone acetylation needs further investigation[13]. Another study in which rats were fed ethanol intragastrically demonstrated that levels of P300, a histone acetyltransferase, increased corresponding to the peaks in urinary alcohol levels, and this correlated with an increase in histone 3 acetylation at lysine 9[14].
A recent study by Day’s group very elegantly demonstrated that the formation of acetate from alcohol is key to the process of alcohol-induced inflammatory gene expression by promoter histone acetylation in acute alcoholic hepatitis[15]. Treatment of Monomac6 cells (human macrophage cell line modeling Kupffer cell responses in ethanol) with ethanol increased global H3 and H4 acetylation and reduced HDAC activity significantly. Ethanol also induced the expression of acetyl-coA synthetases (ACSS1 and 2), the enzymes required for conversion of acetate to acetyl-coA, the substrate for acetylation reactions. Corresponding to this effect, increased acetylation was observed at the promoters of inflammatory cytokines IL-6 and tumor necrosis factor (TNF)-α, with an increase in their mRNA expression. Notably, when these experiments were performed using acetate, these effects could be reproduced. What underscores the critical role of acetate in these ethanol-induced effects is that inhibition of ethanol metabolism to acetate using 4-methylpyrazole (4-MP) completely abrogated the effects and histone acetylation remained at baseline[15]. This confirms that acetate is indeed, the mediator of alcohol-induced histone acetylation.
Although there are no published reports regarding the exact mechanisms by which acetate formed by ethanol metabolism may affect histone acetylation, some hypotheses have been suggested. Acetate may increase HAT activity simply by increasing substrate availability for the reaction. Since acetate is also the product of deacetylation reaction, free acetate may cause feedback inhibition of HDACs[15]. It should also be noted that most of the studies done with regard to ethanol-induced acetylation measure global acetylation and studies focusing on specific genes affected by alcohol metabolism are only beginning to be performed. One such gene examined in hepatocytes is ADH1 (Class 1 alcohol dehydrogenase)[13], while TNFα and IL-6 have been examined in Monomac6 cells[15] in response to alcohol. Also of interest are the findings that alcohol seems to have an effect on the acetylation at certain lysine residue, such as lysine 9 on H3 (H3K9), but not others (H3K14, H3K18)[9,12]. Further research is required to explore the significance and relevance of these findings.
ALCOHOL AND SAM
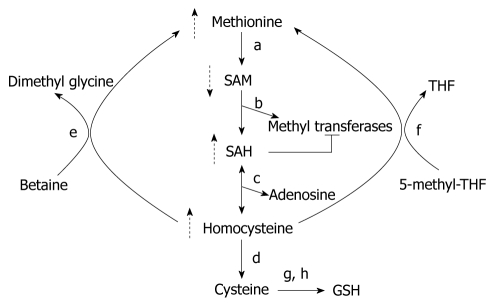
SAM is the one of the most widely used cofactors in nature, probably second only to ATP[16]. The liver is the main source of SAM in humans, and is also largely the site where SAM is metabolized by methyltransferases to S-adenosylhomocysteine (SAH)[17,18] (Figure 2). SAH is a potent inhibitor of all methyltransferases, and needs to be promptly eliminated by the body by a reaction catalyzed by SAH hydrolase[19]. SAM is an essential molecule that is vital to numerous cellular processes and is the principal biological methyl donor required for methylation of histones; as also other proteins, DNA, RNA, biogenic amines and phospholipids. It gives away its high energy methyl group to methyltransferases in transmethylation reactions, and thus, plays a central role in the epigenetic regulation of genes that are controlled by histone or DNA methylation[19]. The ratio of SAM to SAH is a critical determinant of the efficiency of transmethylation reactions and hence this ratio is referred to as the cellular methylation potential[20]. SAM dependent methyltransferases is a broad class of enzymes that contains over a hundred genes[21,22]. Besides, SAM also contributes to gene regulation by methylation of non-histone proteins such as tumor suppressor p53, transcriptional factor TAF10 and the receptor for angiogenic factor VEGF, VEGFR1[23].
Figure 2.
S-adenosylmethionine metabolism in alcohol-induced liver disease. Effects of alcohol are indicated by dotted arrows; a: Methionine adenosyltransferase; b: Enzymes involved in transmethylation reactions; c: S-adenosylhomocysteine (SAH) hydrolase; d: Cystathionine-β-synthase; e: Betaine homecysteine methyltransferase; f: Methionine synthase; g: glutamate-cysteine synthetase; h: Glutathione (GSH) synthetase. SAM: S-adenosylmethionine; THF: Tetrahydrofolate.
Alcohol causes SAM deficiency
SAM deficiency in alcohol-induced liver disease was first described in the early 1980s[24]. Hepatic MAT activity was found to be subnormal in alcohol-dependent individuals, blocking the conversion of methionine to SAM and resulting in hypermethioninemia[25-27]. Alcohol-dependent individuals often display glutathione (GSH) deficiency because GSH synthesis requires SAM[27,28]. Hepatic SAM depletion in response to chronic alcohol consumption has been studied both in humans and in animal models. SAM deficiency has been associated with hepatitis in humans[29], and different stages of alcohol-induced liver injury in rats[30], baboons[31] and micropigs[32].
Ethanol may deplete hepatic SAM by more than one mechanism (Figure 2). Ethanol administration reduces hepatic MAT activity by the oxidation or nitrosylation of the cysteine residue at position 121 and this may be affected by the reactive oxygen and nitrogen species generated during ethanol metabolism[33,34]. MAT activity has also been shown to be reduced due to decreased gene expression of MAT1 (liver specific MAT) in alcoholic hepatitis patients[29] and ethanol-fed micropigs[35]. In another study in rats, chronic ethanol administration decreased SAM levels and glutathione concentration without affecting MAT and it was proposed that SAM consumption was increased to fuel glutathione synthesis[36]. In addition to increased consumption, SAM deficiency may occur because its synthesis may be inhibited by the unavailability of methionine, the endogenous precursor of SAM. Chronic ethanol administration can influence methionine synthesis by decreasing methionine synthase (MS) activity, and the hepatic levels of folate and betaine[35,37-39]. Thus, alcohol consumption can affect aspects of SAM production and metabolism at multiple levels to result in hepatic SAM deficiency in ALD.
SAM and histone modifications
Unlike histone acetylation, which is generally a transcriptionally permissive modification, histone methylation is known to exhibit differential effects that depend on the position of the particular residue that is modified. Methylation of the lysine at position 9 on H3 (H3K9) is a silencing event[40,41], and opposes the transcription-activating acetylation (H3K9Ac) of the same residue. On the other hand, methylation of lysine 4 on H3 (H3K4) activates transcription, and trimethylation at this residue (H3K4Me3) is strongly correlated with active transcription[42]. Also, in contrast to histone acetylation, histone methylation appears to be a more permanent mark and is relatively irreversible[43,44].
The effect of ethanol on histone and DNA methylation has been studied in cell culture and animal studies[9,10,45,46]. However, in comparison to histone acetylation, studies involving methylation changes with ethanol are only beginning to be documented. Pal-Bhadra et al[45] examined the effect of ethanol on H3 methylation in primary rat hepatocytes, and reported contrasting methylation patterns at H3K9 and H3K4 following ethanol treatment. H3K9 dimethylation was decreased whereas H3K4 dimethylation was increased. Further analysis showed that K9 methylation was associated with the promoters of ethanol-downregulated genes [L-serine dehydratase (Lsdh) and Cytochrome P450 2C11 (CYP2C11)] and K4 methylation with those of ethanol-upregulated genes [Alcohol dehydrogenase-1 (Adh-1) and Glutathione S-transferase Yc2 (GST-Yc2)] in these cells[45]. In earlier studies in rat liver and rat hepatic stellate cells, ethanol had been shown to increase acetylation at H3K9 with little, if any, change in methylation at the same residue[9,10]. In another recent study using a chronic rat model for ethanol (intragastric feeding model), a significant increase was noted in dimethyl histone 3 lysine 4 (H3K4Me2). Trimethylation of histone 3 lysine 27 (H3K27Me3), a transcriptionally silencing modification, also increased significantly after chronic ethanol feeding[46]. Similarly, the effect of SAM treatment on histone methylation[47,48] and gene expression[49] has been documented in some studies. When SAM was administered along with ethanol intragastrically to rats for a month (chronic model), SAM remarkably attenuated the ethanol-induced liver injury[48]. Histone 3 trimethylation at lysine 27 (H3K27Me3) was significantly increased with SAM treatment, irrespective of ethanol feeding. SAM also prevented most of the changes in gene expression caused by ethanol feeding. Since H3K27 trimethylation correlates with gene repression, it was postulated that SAM stabilized global gene expression and prevented the blood alcohol level (BAL) cycle through this epigenetic modification[48]. Other experiments in RAW and Kupffer cells show that SAM can inhibit the LPS-stimulated expression of pro-inflammatory genes such as TNF-α and i-NOS at the transcriptional level. In these studies, it was found that LPS increased the trimethylation of H3K4 at the promoters of these genes, and treatment with SAM reversed this effect[47]. Overall, research thus far indicates that SAM deficiency may be an important mediator of histone modifications in ethanol-induced liver disease.
ALCOHOL AND NAD+
The ubiquitous biological molecule, NAD+, was first discovered in 1906, and since then, has been widely studied for its ever-expanding repertoire of cellular functions. NAD+ is best known for its role in oxidation-reduction reactions in cell metabolism[50]. NAD+ is also the precursor of the important second messenger cyclic ADP-ribose[51], and more recently, has been found to be absolutely essential for the protein deacetylase activity of sirtuins[52,53]. Sirtuins or Sir2 proteins are a class of histone deacetylases and in addition to their role in gene transcription, are also involved in the regulation of ageing and metabolic processes[54]. Thus, from being a coenzyme in redox reactions, NAD+ has come a long way, now being called a critical metabolic regulator of transcription, longevity, calorie-restriction mediated life-span extension and several age-associated diseases, including diabetes, cancer and Alzheimer’s disease[55].
Alcohol depletes NAD+
Alcohol metabolism utilizes NAD+ at the very initial steps of breakdown[1]. NAD+ is used up both when alcohol dehydrogenase converts alcohol to acetaldehyde and when acetaldehyde dehydrogenase further converts it to acetate. In both these reactions, NAD+ is reduced to NADH. Several acute effects of ethanol are caused by the reduction of the NAD+/NADH ratio in the liver, as a consequence of ethanol metabolism. An increased NAD+/NADH ratio in the liver disrupts fatty acid oxidation and can induce ketogenesis, lactic acidosis, hyperuricemia and hypoglycemia[56]. Even after conversion of ethanol to acetate, NAD+ continues to be consumed. Acetate forms acetyl-coA, which reduces NAD+ to NADH once it enters the Krebs cycle. Chronic ethanol consumption also recruits the CYP2E1 pathway for metabolism, which adds to the imbalance in the hepatic redox state by reducing NAD+ and increasing hydroxyl radicals[57]. The oxidative stress and reactive oxygen species (ROS) caused by ethanol consumption are only aggravated by the poor nutritional status in chronic alcoholics, and play a major part in depleting NAD+ and advancing liver injury[58].
NAD+ and histone modifications
The NAD+/NADH ratio in the liver likely plays a major role in the regulation of histone modifications and thus, gene transcription/silencing[59]. Histone deacetylases are categorized into three main classes, and class III HDACs or sirtuins (SIRT), are activated only in the presence of NAD+. In sirtuin-mediated deacetylase reactions, NAD+ is hydrolyzed into nicotinamide and accepts the acetate moiety, O-acetyl-ribose. The nicotinamide formed is a potent inhibitor of sirtuin HDAC activity. Thus, the histone deacetylase activity of sirtuins is intricately controlled by NAD+ metabolism, requiring NAD+ for catalysis and being inhibited by nicotinamide. Ethanol, as discussed earlier, increases histone acetylation in the liver. The mechanisms underlying this effect of ethanol are not well characterized; however, there are reports of increased HAT activity[10] and/or decreased HDAC activity[11]. It may then be postulated that ethanol-induced inhibition of HDACs is due to the depletion of NAD+ caused during its metabolism. Ethanol has already been shown to inhibit sirtuin expression and activity in other studies in the liver[60,61]. Chronic administration of ethanol in mice reduced hepatic SIRT1 protein levels and significantly inhibited its deacetylase activity in a recent study by You et al[61]. In an earlier report, ethanol was shown to increase SREBP-1c (Sterol regulatory binding protein-1c) lysine acetylation and transcriptional activity in rat H4IIEC3 cells, and this effect was at least in part, mediated by SIRT1 inhibition[62]. Results from another study reaffirm the link between ethanol consumption and SIRT1 reduction, and in this study ethanol reduced hepatic SIRT1 mRNA expression to half its baseline levels[60]. In addition, independent studies have shown that oxidative stress can inhibit HDACs[63-66].
In light of these findings, it can be hypothesized that the interplay between NAD+ levels, reactive intermediates and oxidative stress will impact histone modifications and gene expression in ALD.
ALCOHOL AND ZINC
Zinc is an essential trace element and is vital in carbohydrate and protein metabolism, glucose control, wound healing, the immune system, digestion, fertility, and growth[67-69]. Zinc plays an important role in controlling gene expression, antioxidant defense and DNA repair. Abundant changes in gene expression in dietary zinc deficiency have been profiled in several tissues, including the liver[70,71], although the mechanisms have not been investigated. Zinc is an essential cofactor for over 300 enzymes; of particular relevance to this review are the zinc-requiring enzymes, namely (1) histone deacetylases (HDACs) that catalyze the removal of an acetyl group from lysines on histones protein tails, thereby increasing the accessibility of nucleosomal DNA and resulting in transcriptionally active chromatin; and (2) histone demethylases that demethylate Lys residues on histones and modulate gene expression.
Alcohol causes zinc deficiency
Zinc deficiency is often observed in chronic alcoholics and approximately 30%-50% of alcoholics are thought to have low zinc status[67,69], possibly due to decreases in intestinal absorption, increased urinary excretion and/or inadequate dietary zinc intake. Clinical studies have demonstrated that zinc concentrations in both serum and liver were significantly reduced in patients with alcoholic steatosis, hepatitis, and cirrhosis[72-75]. Indeed, zinc insufficiency is one of the most commonly observed nutritional manifestations of alcoholic liver disease[76]. Zinc depletion in the liver has also been documented in animal models of ethanol-induced liver injury[77-82]. Investigations of zinc metabolism in alcoholics has demonstrated that ethanol consumption leads to increased Zn excretion in urine[83] and decreased Zn absorption from intestine[84,85]; the latter is also seen in a chronic ethanol feeding animal model[86].
Zinc and histone modifications
While class III HDACs require the cofactor NAD+ for their deacetylase function, the class I, II and IV HDACs are structurally distinct and require zinc for their deacetylase function[87]. A suboptimal zinc concentration, as is observed in alcoholic individuals, is highly likely to significantly decrease the activity of HDACs, leading to altered gene expression. Indeed, the alcohol-induced decrease in HDAC activity observed by others[88] and us [unpublished results] may be attributed, at least in part, to lowered zinc levels. A direct effect of zinc on epigenetic histone changes was demonstrated in a recent study using chromatin immunoprecipitation assays, wherein zinc treatment rapidly decreased Lys4-trimethylated and Lys9-acetylated histone H3 in the metallothionein1 (MT1) promoter and decreased total histone H3. Also, the micrococcal nuclease sensitivity of the MT1 promoter was also increased by zinc, suggesting that the chromatin structure in the MT1 promoter may be disrupted by zinc-induced nucleosome removal[89]. Another in vitro study showed that p21 transcription was downregulated by lowered zinc in HepG2 cells[90]. Zinc deficiency led to a reduction in acetylated histone-H4 on the p21 promoter resulting in reduced p21 promoter accessibility, which contributed to the decrease in p21 promoter activity and the downregulation of p21 mRNA and protein expression in zinc-depleted HepG2cells. Specifically, the amounts of acetylated histone-4 associated with the proximal and distal p21 promoter regions were decreased in severe zinc-deficient (73% and 64%, respectively) and mild zinc deficient (82% and 77%, respectively) cells compared with zinc-normal (100% and 100%, respectively)[90].
A recently discovered histone lysine demethylase, LSD2, was shown to not only contain a CW-type zinc finger motif, but also to bind zinc with 3:1 molar ratio (zinc:protein), suggesting that zinc maybe important for its activity[91]. Also, a zinc finger motif located at the end of the conventionally defined JmjC domain of histone demethylases, such as jumonji-type JMJD2A, is thought to be essential for enzymatic function[92]. Changes in zinc status as seen with chronic alcohol consumption are expected to cause dramatic alterations in gene expression, leading to phenotypic changes. In addition to direct epigenetic effects, zinc deficiency is also known to reduce the utilization of methyl groups from SAM in rat liver, resulting in genomic DNA hypomethylation and histone hypomethylation[93,94]. This occurs due to the fact that the enzyme betaine-homocysteine S-methyltransferase (BMHT) is a zinc metalloenzyme[95]. Overall, we believe that alcohol consumption-induced zinc deficiency may greatly impact gene expression via direct and indirect epigenetic histone modifications and modulation of chromatin structure and gene expression.
In addition to the nutritional alterations mentioned in this review, alcohol is known to cause disturbances in other nutrients, which may also play a role in the epigenetic changes effected by alcohol. Alcohol has also been shown to influence other histone modifications, such as glycation[96] and phosphorylation[97]. The significance of these modifications in relation to gene expression is, however, not clear.
CONCLUSION
Alcohol metabolism is inextricably connected to the regulation of key nutrient metabolites in the liver. There is a growing body of literature suggesting a role for the complex interplay between alcohol-induced nutrient changes, histone modifications and gene expression. It is becoming widely accepted that specific aberrant patterns of histone modifications play a fundamental role in chromatin structure and function contributing to the development of disease processes. Epigenetic histone modifications provide a plausible link between alcohol-mediated nutrient alterations and pathogenic gene expression. However, in ALD, the precise contribution of histone modifications in the alteration of expression of specific genes remains largely unknown. Clearly more advances are needed and will be witnessed in this area that will enhance our knowledge about the epigenetic mechanisms underpinning ALD pathogenesis and lead to the development of relevant therapeutic strategies.
Footnotes
Supported by The National Institute of Alcohol Abuse and Alcoholism grants AA014371 (to Joshi-Barve S), AA015970 (to McClain CJ), and Office of Dietary Supplements, NIH
Peer reviewer: Dr. Milan Jirsa, Laboratory of Experimental Medicine-building Z1, Institute for Clinical and Experimental Medicine, Videnska 1958/9, Praha 4, 14000, Czech Republic
S- Editor Tian L L- Editor O’Neill M E- Editor Ma WH
References
- 1.Lieber CS. Metabolism of alcohol. Clin Liver Dis. 2005;9:1–35. doi: 10.1016/j.cld.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Fujino TIY, Osborne TF, Takahashi S, Yamamoto TT, Sakai J. Sources of acetyl-coA: acetyl-coA synthetase 1 and 2. Curr Med Chem Immunol Endocr Metabol Agents. 2003;3:207–210. [Google Scholar]
- 3.Yamashita H, Kaneyuki T, Tagawa K. Production of acetate in the liver and its utilization in peripheral tissues. Biochim Biophys Acta. 2001;1532:79–87. doi: 10.1016/s1388-1981(01)00117-2. [DOI] [PubMed] [Google Scholar]
- 4.Hannak D, Bartelt U, Kattermann R. Acetate formation after short-term ethanol administration in man. Biol Chem Hoppe Seyler. 1985;366:749–753. doi: 10.1515/bchm3.1985.366.2.749. [DOI] [PubMed] [Google Scholar]
- 5.Bruno R, Iliadis A, Treffot MJ, Mariotti B, Cano JP, Jullien G. Evolution of plasma acetate concentration during ethanol metabolism in man. Forensic Sci Int. 1983;21:215–221. doi: 10.1016/0379-0738(83)90125-1. [DOI] [PubMed] [Google Scholar]
- 6.Nuutinen H, Lindros K, Hekali P, Salaspuro M. Elevated blood acetate as indicator of fast ethanol elimination in chronic alcoholics. Alcohol. 1985;2:623–626. doi: 10.1016/0741-8329(85)90090-4. [DOI] [PubMed] [Google Scholar]
- 7.Lundquist F, Tygstrup N, Winkler K, Mellemgaard K, Munck-petersen S. Ethanol metabolism and production of free acetate in the human liver. J Clin Invest. 1962;41:955–961. doi: 10.1172/JCI104574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 9.Kim JS, Shukla SD. Histone h3 modifications in rat hepatic stellate cells by ethanol. Alcohol Alcohol. 2005;40:367–372. doi: 10.1093/alcalc/agh170. [DOI] [PubMed] [Google Scholar]
- 10.Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- 11.Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin Exp Res. 2008;32:829–839. doi: 10.1111/j.1530-0277.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- 12.Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys Res Commun. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- 13.Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1124–G1136. doi: 10.1152/ajpgi.00091.2005. [DOI] [PubMed] [Google Scholar]
- 14.Bardag-Gorce F, French BA, Joyce M, Baires M, Montgomery RO, Li J, French S. Histone acetyltransferase p300 modulates gene expression in an epigenetic manner at high blood alcohol levels. Exp Mol Pathol. 2007;82:197–202. doi: 10.1016/j.yexmp.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendrick SF, O’Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, Day CP. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51:1988–1997. doi: 10.1002/hep.23572. [DOI] [PubMed] [Google Scholar]
- 16.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Mato JM, Corrales FJ, Lu SC, Avila MA. S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 2002;16:15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 18.Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 19.Lu SC, Mato JM. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J Gastroenterol Hepatol. 2008;23 Suppl 1:S73–S77. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 21.Huang S. Histone methyltransferases, diet nutrients and tumour suppressors. Nat Rev Cancer. 2002;2:469–476. doi: 10.1038/nrc819. [DOI] [PubMed] [Google Scholar]
- 22.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Horowitz JH, Rypins EB, Henderson JM, Heymsfield SB, Moffitt SD, Bain RP, Chawla RK, Bleier JC, Rudman D. Evidence for impairment of transsulfuration pathway in cirrhosis. Gastroenterology. 1981;81:668–675. [PubMed] [Google Scholar]
- 25.Cabrero C, Duce AM, Ortiz P, Alemany S, Mato JM. Specific loss of the high-molecular-weight form of S-adenosyl-L-methionine synthetase in human liver cirrhosis. Hepatology. 1988;8:1530–1534. doi: 10.1002/hep.1840080610. [DOI] [PubMed] [Google Scholar]
- 26.Duce AM, Ortíz P, Cabrero C, Mato JM. S-adenosyl-L-methionine synthetase and phospholipid methyltransferase are inhibited in human cirrhosis. Hepatology. 1988;8:65–68. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 27.Mato JM, Alvarez L, Ortiz P, Pajares MA. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 28.Chawla RK, Lewis FW, Kutner MH, Bate DM, Roy RG, Rudman D. Plasma cysteine, cystine, and glutathione in cirrhosis. Gastroenterology. 1984;87:770–776. [PubMed] [Google Scholar]
- 29.Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2004;28:173–181. doi: 10.1097/01.ALC.0000108654.77178.03. [DOI] [PubMed] [Google Scholar]
- 30.Barak AJ, Beckenhauer HC, Tuma DJ. S-adenosylmethionine generation and prevention of alcoholic fatty liver by betaine. Alcohol. 1994;11:501–503. doi: 10.1016/0741-8329(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 31.Lieber CS, Casini A, DeCarli LM, Kim CI, Lowe N, Sasaki R, Leo MA. S-adenosyl-L-methionine attenuates alcohol-induced liver injury in the baboon. Hepatology. 1990;11:165–172. doi: 10.1002/hep.1840110203. [DOI] [PubMed] [Google Scholar]
- 32.Halsted CH, Villanueva JA, Devlin AM, Niemelä O, Parkkila S, Garrow TA, Wallock LM, Shigenaga MK, Melnyk S, James SJ. Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci USA. 2002;99:10072–10077. doi: 10.1073/pnas.112336399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukamoto H, Lu SC. Current concepts in the pathogenesis of alcoholic liver injury. FASEB J. 2001;15:1335–1349. doi: 10.1096/fj.00-0650rev. [DOI] [PubMed] [Google Scholar]
- 34.Caballero F, Fernández A, Matías N, Martínez L, Fucho R, Elena M, Caballeria J, Morales A, Fernández-Checa JC, García-Ruiz C. Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: impact on mitochondrial S-adenosyl-L-methionine and glutathione. J Biol Chem. 2010;285:18528–18536. doi: 10.1074/jbc.M109.099333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva JA, Halsted CH. Hepatic transmethylation reactions in micropigs with alcoholic liver disease. Hepatology. 2004;39:1303–1310. doi: 10.1002/hep.20168. [DOI] [PubMed] [Google Scholar]
- 36.Aleynik SI, Lieber CS. Polyenylphosphatidylcholine corrects the alcohol-induced hepatic oxidative stress by restoring s-adenosylmethionine. Alcohol Alcohol. 2003;38:208–212. doi: 10.1093/alcalc/agg066. [DOI] [PubMed] [Google Scholar]
- 37.de la Vega MJ, Santolaria F, González-Reimers E, Alemán MR, Milena A, Martínez-Riera A, González-García C. High prevalence of hyperhomocysteinemia in chronic alcoholism: the importance of the thermolabile form of the enzyme methylenetetrahydrofolate reductase (MTHFR) Alcohol. 2001;25:59–67. doi: 10.1016/s0741-8329(01)00167-7. [DOI] [PubMed] [Google Scholar]
- 38.Barak AJ, Beckenhauer HC, Junnila M, Tuma DJ. Dietary betaine promotes generation of hepatic S-adenosylmethionine and protects the liver from ethanol-induced fatty infiltration. Alcohol Clin Exp Res. 1993;17:552–555. doi: 10.1111/j.1530-0277.1993.tb00798.x. [DOI] [PubMed] [Google Scholar]
- 39.Kharbanda KK, Rogers DD, Mailliard ME, Siford GL, Barak AJ, Beckenhauer HC, Sorrell MF, Tuma DJ. A comparison of the effects of betaine and S-adenosylmethionine on ethanol-induced changes in methionine metabolism and steatosis in rat hepatocytes. J Nutr. 2005;135:519–524. doi: 10.1093/jn/135.3.519. [DOI] [PubMed] [Google Scholar]
- 40.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 41.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 42.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 43.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 44.Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148:558–567. doi: 10.1016/0003-9861(72)90174-9. [DOI] [PubMed] [Google Scholar]
- 45.Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81:979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardag-Gorce F, Oliva J, Dedes J, Li J, French BA, French SW. Chronic ethanol feeding alters hepatocyte memory which is not altered by acute feeding. Alcohol Clin Exp Res. 2009;33:684–692. doi: 10.1111/j.1530-0277.2008.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ara AI, Xia M, Ramani K, Mato JM, Lu SC. S-adenosylmethionine inhibits lipopolysaccharide-induced gene expression via modulation of histone methylation. Hepatology. 2008;47:1655–1666. doi: 10.1002/hep.22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bardag-Gorce F, Li J, Oliva J, Lu SC, French BA, French SW. The cyclic pattern of blood alcohol levels during continuous ethanol feeding in rats: the effect of feeding S-adenosylmethionine. Exp Mol Pathol. 2010;88:380–387. doi: 10.1016/j.yexmp.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Bardag-Gorce F, Oliva J, Dedes J, French BA, French SW. Gene expression modifications in the liver caused by binge drinking and S-adenosylmethionine feeding. The role of epigenetic changes. Genes Nutr. 2009:Epub ahead of print. doi: 10.1007/s12263-009-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–690. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 51.Guse AH. Cyclic ADP-ribose. J Mol Med. 2000;78:26–35. doi: 10.1007/s001090000076. [DOI] [PubMed] [Google Scholar]
- 52.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 53.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 54.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 56.Lieber CS. ALCOHOL: its metabolism and interaction with nutrients. Annu Rev Nutr. 2000;20:395–430. doi: 10.1146/annurev.nutr.20.1.395. [DOI] [PubMed] [Google Scholar]
- 57.Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- 58.Lieber CS. Alcoholic liver disease: new insights in pathogenesis lead to new treatments. J Hepatol. 2000;32:113–128. doi: 10.1016/s0168-8278(00)80420-1. [DOI] [PubMed] [Google Scholar]
- 59.Luo J, Kuo MH. Linking nutrient metabolism to epigenetics. Cell Sci Rev. 2009;6:49–54. [Google Scholar]
- 60.Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–48. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 61.You M, Liang X, Ajmo JM, Ness GC. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 62.You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP) J Biol Chem. 2002;277:29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- 63.Barnes PJ, Ito K, Adcock IM. Corticosteroid resistance in chronic obstructive pulmonary disease: inactivation of histone deacetylase. Lancet. 2004;363:731–733. doi: 10.1016/S0140-6736(04)15650-X. [DOI] [PubMed] [Google Scholar]
- 64.Furukawa A, Tada-Oikawa S, Kawanishi S, Oikawa S. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD+ depletion. Cell Physiol Biochem. 2007;20:45–54. doi: 10.1159/000104152. [DOI] [PubMed] [Google Scholar]
- 65.Wu A, Ying Z, Gomez-Pinilla F. Oxidative stress modulates Sir2alpha in rat hippocampus and cerebral cortex. Eur J Neurosci. 2006;23:2573–2580. doi: 10.1111/j.1460-9568.2006.04807.x. [DOI] [PubMed] [Google Scholar]
- 66.Wu Z, Lauer TW, Sick A, Hackett SF, Campochiaro PA. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282:22414–22425. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]
- 67.Prasad AS. Zinc: an overview. Nutrition. 1995;11:93–99. [PubMed] [Google Scholar]
- 68.Lipscomb WN, Sträter N. Recent Advances in Zinc Enzymology. Chem Rev. 1996;96:2375–2434. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 69.King J, Cousins RJ. Zinc. In: Shils ME, Shike M, Ross AC, Caballero B, Cousins RJ, et al., editors. Modern Nutrition in Health and Disease, 10th ed. Baltimore, MD: Lippincott Williams & Wilkins; 2005. pp. 271–285. [Google Scholar]
- 70.Pfaffl MW, Gerstmayer B, Bosio A, Windisch W. Effect of zinc deficiency on the mRNA expression pattern in liver and jejunum of adult rats: monitoring gene expression using cDNA microarrays combined with real-time RT-PCR. J Nutr Biochem. 2003;14:691–702. doi: 10.1016/j.jnutbio.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 71.Sun JY, Wang JF, Zi NT, Jing MY, Weng XY. Gene expression profiles analysis of the growing rat liver in response to different zinc status by cDNA microarray analysis. Biol Trace Elem Res. 2007;115:169–185. doi: 10.1007/BF02686028. [DOI] [PubMed] [Google Scholar]
- 72.Kiilerich S, Dietrichson O, Loud FB, Naestoft J, Christoffersen P, Juhl E, Kjems G, Christiansen C. Zinc depletion in alcoholic liver diseases. Scand J Gastroenterol. 1980;15:363–367. doi: 10.3109/00365528009181484. [DOI] [PubMed] [Google Scholar]
- 73.Bode JC, Hanisch P, Henning H, Koenig W, Richter FW, Bode C. Hepatic zinc content in patients with various stages of alcoholic liver disease and in patients with chronic active and chronic persistent hepatitis. Hepatology. 1988;8:1605–1609. doi: 10.1002/hep.1840080622. [DOI] [PubMed] [Google Scholar]
- 74.Rodríguez-Moreno F, González-Reimers E, Santolaria-Fernández F, Galindo-Martín L, Hernandez-Torres O, Batista-López N, Molina-Perez M. Zinc, copper, manganese, and iron in chronic alcoholic liver disease. Alcohol. 1997;14:39–44. doi: 10.1016/s0741-8329(96)00103-6. [DOI] [PubMed] [Google Scholar]
- 75.Schölmerich J, Löhle E, Köttgen E, Gerok W. Zinc and vitamin A deficiency in liver cirrhosis. Hepatogastroenterology. 1983;30:119–125. [PubMed] [Google Scholar]
- 76.McClain CJ, Antonow DR, Cohen DA, Shedlofsky SI. Zinc metabolism in alcoholic liver disease. Alcohol Clin Exp Res. 1986;10:582–589. doi: 10.1111/j.1530-0277.1986.tb05149.x. [DOI] [PubMed] [Google Scholar]
- 77.Giménez A, Caballería J, Parés A, Alié S, Deulofeu R, Andreu H, Rodés J. Influence of dietary zinc on hepatic collagen and prolyl hydroxylase activity in alcoholic rats. Hepatology. 1992;16:815–819. doi: 10.1002/hep.1840160331. [DOI] [PubMed] [Google Scholar]
- 78.Cabré M, Folch J, Giménez A, Matas C, Parés A, Caballería J, Paternain JL, Rodés J, Joven J, Camps J. Influence of zinc intake on hepatic lipid peroxidation and metallothioneins in alcoholic rats: relationship to collagen synthesis. Int J Vitam Nutr Res. 1995;65:45–50. [PubMed] [Google Scholar]
- 79.Zhou Z, Wang L, Song Z, Saari JT, McClain CJ, Kang YJ. Zinc supplementation prevents alcoholic liver injury in mice through attenuation of oxidative stress. Am J Pathol. 2005;166:1681–1690. doi: 10.1016/S0002-9440(10)62478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang YJ, Zhou Z. Zinc prevention and treatment of alcoholic liver disease. Mol Aspects Med. 2005;26:391–404. doi: 10.1016/j.mam.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 81.Kang X, Song Z, McClain CJ, Kang YJ, Zhou Z. Zinc supplementation enhances hepatic regeneration by preserving hepatocyte nuclear factor-4alpha in mice subjected to long-term ethanol administration. Am J Pathol. 2008;172:916–925. doi: 10.2353/ajpath.2008.070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Z, Liu J, Song Z, McClain CJ, Kang YJ. Zinc supplementation inhibits hepatic apoptosis in mice subjected to a long-term ethanol exposure. Exp Biol Med (Maywood) 2008;233:540–548. doi: 10.3181/0710-RM-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sullivan JF, Blotcky AJ, Jetton MM, Hahn HK, Burch RE. Serum levels of selenium, calcium, copper magnesium, manganese and zinc in various human diseases. J Nutr. 1979;109:1432–1437. doi: 10.1093/jn/109.8.1432. [DOI] [PubMed] [Google Scholar]
- 84.Dinsmore W, Callender ME, McMaster D, Todd SJ, Love AH. Zinc absorption in alcoholics using zinc-65. Digestion. 1985;32:238–242. doi: 10.1159/000199243. [DOI] [PubMed] [Google Scholar]
- 85.Valberg LS, Flanagan PR, Ghent CN, Chamberlain MJ. Zinc absorption and leukocyte zinc in alcoholic and nonalcoholic cirrhosis. Dig Dis Sci. 1985;30:329–333. doi: 10.1007/BF01403841. [DOI] [PubMed] [Google Scholar]
- 86.Antonson DL, Vanderhoof JA. Effect of chronic ethanol ingestion on zinc absorption in rat small intestine. Dig Dis Sci. 1983;28:604–608. doi: 10.1007/BF01299920. [DOI] [PubMed] [Google Scholar]
- 87.Hernick M, Fierke CA. Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch Biochem Biophys. 2005;433:71–84. doi: 10.1016/j.abb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 88.Shepard BD, Joseph RA, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Alcohol-induced alterations in hepatic microtubule dynamics can be explained by impaired histone deacetylase 6 function. Hepatology. 2008;48:1671–1679. doi: 10.1002/hep.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okumura F, Li Y, Itoh N, Nakanishi T, Isobe M, Andrews GK, Kimura T. The zinc-sensing transcription factor MTF-1 mediates zinc-induced epigenetic changes in chromatin of the mouse metallothionein-I promoter. Biochim Biophys Acta. 2011;1809:56–62. doi: 10.1016/j.bbagrm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 90.Wong SH, Zhao Y, Schoene NW, Han CT, Shih RS, Lei KY. Zinc deficiency depresses p21 gene expression: inhibition of cell cycle progression is independent of the decrease in p21 protein level in HepG2 cells. Am J Physiol Cell Physiol. 2007;292:C2175–C2184. doi: 10.1152/ajpcell.00256.2006. [DOI] [PubMed] [Google Scholar]
- 91.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 93.Wallwork JC, Duerre JA. Effect of zinc deficiency on methionine metabolism, methylation reactions and protein synthesis in isolated perfused rat liver. J Nutr. 1985;115:252–262. doi: 10.1093/jn/115.2.252. [DOI] [PubMed] [Google Scholar]
- 94.Dreosti IE. Zinc and the gene. Mutat Res. 2001;475:161–167. doi: 10.1016/s0027-5107(01)00067-7. [DOI] [PubMed] [Google Scholar]
- 95.Breksa AP, Garrow TA. Random mutagenesis of the zinc-binding motif of betaine-homocysteine methyltransferase reveals that Gly 214 is essential. Arch Biochem Biophys. 2002;399:73–80. doi: 10.1006/abbi.2001.2751. [DOI] [PubMed] [Google Scholar]
- 96.Lakatos A, Jobst K, Juricskay Z, Kalász V. The effect of ethanol on histone glycation in diabetic rats. Alcohol Alcohol. 2000;35:145–147. doi: 10.1093/alcalc/35.2.145. [DOI] [PubMed] [Google Scholar]
- 97.Lee YJ, Shukla SD. Histone H3 phosphorylation at serine 10 and serine 28 is mediated by p38 MAPK in rat hepatocytes exposed to ethanol and acetaldehyde. Eur J Pharmacol. 2007;573:29–38. doi: 10.1016/j.ejphar.2007.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]