Abstract
Current genome-wide association studies (GWAS) often involve populations that have experienced recent genetic admixture. Genotype data generated from these studies can be used to test for association directly, as in a non-admixed population. As an alternative, these data can be used to infer chromosomal ancestry, and thus allow for admixture mapping. We quantify the contribution of allele-based and ancestry-based association testing under a family-design, and demonstrate that the two tests can provide non-redundant information. We propose a joint testing procedure, which efficiently integrates the two sources information. The efficiencies of the allele, ancestry and combined tests are compared in the context of a GWAS. We discuss the impact of population history and provide guidelines for future design and analysis of GWAS in admixed populations.
Introduction
During the past few years, hundreds of genome-wide association studies (GWAS) have been reported. It is only recently, however, that several large-scale GWAS are underway, which focus on genetically admixed populations, such as African Americans or Hispanics. Genome-wide association studies in recently admixed populations present special challenges. Under a case-control design, spurious association may arise when subjects in the case and control groups differ in allele frequencies, which can easily arise from differences in ancestry proportions. A well-known example of this sort is the putative association between a Gm haplotype and the risk of type 2 diabetes in Pima Indians. After adjusting for the European admixture of each individual, estimated by the number of European grandparents, the putative association disappears (Williams et al., 2000). In this situation, the Native American ancestry proportion acts as a confounding variable, correlated with both the disease prevalence and with the frequency of the Gm haplotype. Although various adjustment strategies have been proposed (Redden et al., 2006), the adequacy of these methods in GWAS remains uncertain. Hence family-based association methods provide a robust defense against population stratification. In its simplest form, the Transmission Disequilibrium Test (TDT) focuses on the transmission from a heterozygous parent to an affected child (Ewens and Spielman, 1995). This test was subsequently generalized to accommodate quantitative traits, as well as to allow for non-additive models (Laird and Lange, 2008; Weinberg et al., 1998; Kistner and Weinberg, 2004).
While admixed populations pose challenges for case-control association studies, these populations provide unique opportunities to detect risk variants that occur in some, but not all, of the ancestral populations, and to disentangle the genetic contribution of the ethnicity-specific patterns of disease prevalence. The prospect of admixture mapping, which exploits the extended linkage disequilibrium generated in the process of recent admixing, can be traced to Chakraborty and Weiss (1988). The rationale of admixture mapping parallels that of conventional association testing procedures: while conventional association procedures seek genomic locations in which a specific allele or genotype occurs more frequently among cases than controls, admixture mapping searches for locations in which a particular ancestry is over-represented among the cases compared to expectations (Smith and O'Brien, 2005). Under a family-based design, a TDT-like admixture mapping statistic was proposed by McKeigue (1997). This test, as well as all other admixture mapping tests that have been proposed to date, infers ancestry at each marker location from surrounding genotype data. To reduce the genotyping cost, panels of ancestry informative markers (AIMs) have been designed for admixture mapping in African Americans or Hispanics (Smith et al., 2004; Tian et al., 2006; Price et al., 2007). In the context of a GWAS, however, computational methods have been developed, which allow accurate inference of locus-specific ancestry using genotype data generated from standard dense genotyping platforms (Tang et al., 2006; Sankararaman et al., 2008; Sundquist et al., 2008; Price et al., 2009).
In a GWAS that focuses on a recently admixed population, researchers have the option to use the same genotype data to perform either or both genotype-based association mapping and admixture mapping. A natural question to ask is, do genotype and ancestry provide non-overlapping information? In this paper, we show that the answer is yes, and we propose a testing procedure that integrates the genotype and ancestry information. We evaluate the power of the genotype, ancestry and combined tests through a combination of theoretical calculation, simulation and empirical data. Our analysis also quantifies the degree to which genotype and ancestry provide non-overlapping information. The results are relevant to the design and analysis of family-based GWAS studies in recently admixed populations.
Model and Method
In this section, we first define the allelic and ancestry-TDT statistics. Under the null hypothesis of no association or no linkage, each of these test statistics has an approximate chi-square distribution and under the alternative has a noncental chi-square distribution, for which we derive the non-centrality parameter. As explained in Clarke and Whittemore (2007), the square root of the TDT coincides with a score statistic. Thus, the non-centrality parameters for both allele and ancestry tests can be derived by computing the expectation of the efficient score. We refer readers to Clarke and Whittemore (2007) or Siegmund and Yakir (2007) for a more complete discussion.
Data structure and notations
Consider a study that recruited N nuclear families, each of which consists of two parents and an affected child. The underlying population has experienced recent admixture of two ancestral populations, X and Y, with average admixture proportions of f and 1 − f. Let S and s denote two alleles at a disease predisposing locus. Let , and ps be the frequencies of S in the two ancestral populations and in the admixed population, respectively. The location of S is unknown, and the polymorphism itself may or may not be genotyped. We assume a multiplicative disease model, as it simplifies the analytic derivation (McKeigue, 1997; Zhang et al., 2004; Clarke and Whittemore, 2007). The test statistics remain valid regardless of the underlying disease model, although there may be a loss of power with incorrect specification of the mode of inheritance. Letting D denote the disease phenotype, the penetrance function at the S locus satisfies:
Let A and a denote the alleles at a diallelic genotyped marker, and let F be the frequency of A in the admixed population. We parameterize the gametic phase disequilibrium between A and S by ΠA and Πa, which denote the frequencies, in the admixed population, of S conditioning on the marker alleles being A and a, respectively.
Testing allelic association
The premise of the TDT is that, if the genotype at marker A is not associated with the disease risk, then a heterozygous parent, with genotype Aa, transmits the A or a allele to an affected child with equal probability. Let NAa be the number of heterozygous parents in the sample, of which NA transmitted the A allele while Na transmitted to an affected child the a allele. The TDT statistic, in its classic form, is
Under the null hypothesis of no association or no linkage, TG asymptotically follows a χ2 distribution with one degree of freedom. To emphasize that this test is based on comparing the transmitted and non-transmitted alleles, we refer to this test as the allelic-TDT.
In order to evaluate the power of this test under an alternative hypothesis, we compute the asymptotic non-centrality parameter of the test statistic. In so doing, we make the conventional assumption of statistical large sample theory: that N is large, while γ − 1 is small, in such a way that N1/2(γ − 1) is a finite, positive number. The numerator of TG represents the allelic transmission bias. Conditional on the individual being diseased, it has the expectation:
| (1) |
where θ is the recombination fraction between A and S. This is equivalent to equation (37) in Clarke and Whittemore (2007). For the asymptotic expected value of the denominator of TG under the assumption that γ − 1 approaches 0 and Hardy-Weinberg equilibrium, we obtain
| (2) |
By standard central limit theorem and law of large numbers arguments, the asymptotic non-centrality parameter for the allele-TDT statistic is
where r = (ΠA − Πa)[F(1 − F)]1/2/[ps(1 − ps)]1/2 is the correlation of gametic phase disequilibrium (linkage disequilibrium). See also Siegmund and Yakir (2007), pp 302–303. Note that the test will have power to reject the null hypothesis if the non-centrality parameter is non-zero, or equivalently if both θ < 1/2 and (ΠA − Πa) ≠ 0.
Testing ancestry association
For ancestry association testing, we adopt an analogous statistic, originally proposed by McKeigue (1997), which tests whether a parent heterozygous in ancestry (that is, parents with one allele from population X and the other allele from population Y) transmits the two alleles with equal probability. Here we assume that the ancestry at each marker can be inferred without error from the genotype at that marker as well as genotypes at neighboring markers. At a given locus, let ZX (respectively, ZY) denote the number of parents who are XY in ancestry and transmit to the children an allele from the X (respectively Y) chromosome. Let ZXY be the number of parents heterozygous in ancestry. The ancestry-TDT statistic takes the form:
In the same fashion as we computed the non-centrality parameter for the allelic-TDT, the non-centrality parameter of the ancestry-TDT can be derived analytically assuming a multiplicative disease model and known population admixing history:
| (3) |
Again, for the non-centrality parameter to be non-zero requires that θ < 1/2 and . The term, , in μA shows that the ancestry-TDT only has power if the underlying risk variant occurs at different frequencies between the two ancestral populations. Another way of interpreting is that the ancestry at the marker is in “linkage disequilibrium” with the causal variant (where perfect disequilibrium would be a variant that is fixed in one population is absent in the other, while pefect equilibrium occurs when the allele occurs at the same frequency in both populations).
Remarks
Although the preceding calculations are based for simplicity on the model of multiplicative penetrances, under the asymptotic scenario that γ → 1 as N → ∞, the multiplicative model is asymptotically equivalent to an additive model. The allelic-TDT test presupposes parental allelic heterozygosity to remove the effect of population stratification, which otherwise can lead to an inflated level of false positive errors. In principle, the admixture statistic need not be restricted to parents who are heterozygous in ancestry at the candidate marker. Indeed, McKeigue (1998) proposed to use a parent's genome-wide average ancestry, and he argued that excluding homozygous parents can lead to as much as a 50% loss of efficiency under some conditions. However, this approach implicitly assumes that the population ancestry proportions are constant across the genome, thus ignoring region specific variability in ancestry. We believe that the statistic that conditions on heterozygosity in ancestry is more robust. Finally, it may be worth noting that an appropriate degree of freedom can be added to these statistics if it is felt that dominance may play an important role. However, conditioning on parental heterozygosity (either for genotype or for ancestry) would require that both parents be heterozygous in order to estimate the dominance deviation. This might shrink the effective sample size too much to be useful.
There is a caveat in our calculation of the non-centrality parameter for the ancestry-based test. In computing the expected number of ancestrally heterozygous parents, we have assumed that all parents have the same ancestry proportion (from X) of f. In reality, there is often substantial variation in ancestry proportions among individuals from recently admixed populations (Parra et al., 1998; Silva-Zolezzi et al., 2009). Heterogeneity in ancestry proportion decreases the proportion of ancestrally heterozygous parents. The magnitude of the deficiency depends on the admixing history. Assuming individuals mated randomly subsequent to admixing, the variation in ancestry proportion decays by a factor of 2 in each successive generation, and the variation would be negligible in 10 ~ 20 generations. However, several studies have found complex and non-random mating patterns among some admixed populations (Krieger et al., 1965; Risch et al., 2009). Therefore, the non-centrality parameter derived above and the sample size calculation presented below are somewhat optimistic.
Joint test of association
We now consider a test statistic that combines the genotype and ancestry information. Consider the two-dimension vector that represents the bias in allele and ancestry transmission:
| (4) |
Let MGA denote the covariance between the two components in Eqn (4). As a combined test statistic we propose
| (5) |
which in large samples has approximately a χ2 distribution with two degrees of freedom under the null hypothesis.
To derive the non-centrality parameter of this combined test, note that the expectation of all terms in Eqn (5) have been derived in the preceding sections with the exception of MGA. By taking the second derivative of the log likelihood function or by direct calculation, we find the covariance between the two components is
where AXaY is the number of parents, who have a X-derived chromosome carrying the A allele and a Y-derived chromosome carrying the a allele; likewise AYaX is the number of parents, who have a Y-derived chromosome carrying the A allele and a X-derived chromosome carrying the a allele. This covariance term has an intuitive interpretation. For a parent in the AXaY group, the two components in DGA are perfectly correlated: the transmission of the X-derived chromosome always co-occurs with the transmission of the A allele, and thus contributes an increment of 1 to both components; similarly, the transmission of the Y-derived chromosome simultaneously transmit the a allele, contributing a decrement of 1 in both components. In contrast, the two components in DGA are negatively correlated for parents in the AYaX group: the transmission of the X-derived chromosome increases ZX − ZY by one, but because this chromosome carries the a allele, NA − Na is decreased by one. Parents of all other genotype-ancestry combinations contribute to at most one of the components in DGA. To compute the expectation of MGA, let and denote the allele frequency of A at the marker in ancestral populations X and Y, respectively. Under the null hypothesis, it is easy to see that:
Conditions where the combined test is more powerful than a single degree of freedom tests will be discussed in the Results section. In the special case where the marker allele A is the same as the disease allele S, there is no new information provided by the ancestry test beyond that already provided by the allelic test. To see this, observe that the noncentrality parameter of the two degrees of freedom statistic is of the form ξ′R−1ξ. Here ξ = (ξ1, ξ2)′, where ξ1 is the signed square root of ϕG, while ξ2 is the signed square root of ϕA; and R is the correlation matrix of the (signed square root of the) two standardized statistics. Let ρ denote the off diagonal element of R. Then the noncentrality parameter can be written . In the case where the allele tested is actually the disease susceptibility allele, so πA − πa = 1 (e.g. perfect LD between disease variant and candidate marker), it is easy to see from the computations reported above that ξ2 = ρξ1, so the second degree of freedom adds nothing to the non-centrality parameter of the allelic statistic.
Genome-wide significance level
The non-centrality parameters we derived in the previous sections relate directly to the statistical power of the respective tests at a specific candidate locus. In the context of a genome-wide scan, however, the significance level at each marker needs to be adjusted to account for the large number of markers tested. A common approach for this adjustment is the Bonferroni correction, which simply divides the significance level by the number of markers tested. This approach, however, can be overly conservative if the test statistics are highly correlated between linked markers. The correlation of TG along the genome depends on the linkage disequilibrium in the admixed population, while the correlation of TA depends on the population history of admixture. Incorporating known population history and the empirical linkage disequilibrium pattern from existing data, we derived approximations for the genome-wide significance levels for allelic-TDT, ancestry and combined test statistics. The derivations of these significance approximations are beyond the scope of this manuscript, and will be described elsewhere.
Alternatively, one could simulate the null distribution of the genome-wide maximum test statistics using a permutation procedure, and assess the significance of the observed test statistics based on the empirical quantiles. To illustrate this procedure, we make use of data generated in a GWAS of asthma in a Mexican population, which consists of 492 parents-child trios genotyped using Illumina 550K arrays. The recruitment and characteristics of this cohort are described in Hancock et al. (2009). The Mexican population derived ancestries from European, African and AmerIndian ancestral populations. The average ancestry proportions in our cohort was 27%, 3%, and 69%, respectively. Although the ancestry-TDT can be generalized to admixture of more than two parental populations, in this case we combined the European and African derived chromosomes and test for only AmerIndian vs non-AmerIndian ancestry. We generate permuted data by randomly assigning the transmitted and non-transimitted chromosome in each parent. For the allelic-TDT, we simply record the maximum TG achieved across the genome in each permuted set of data. For the ancestry-TDT, we inferred ancestry for each haplotype by means of a modified algorithm of Sundquist et al. (2008), which uses phased genotype data. We treat the inferred ancestry as fixed and record the genome-wide maximum TA in each permuted data set. Likewise, we compute TGA on the permuted data and record the genome-wide maximum. The critical values at a .05 genome-wide significance level for TG, TA and TGA are defined by the 95% empirical quantiles of the respective null distributions, based on 10, 000 permutations. We will not describe a full application of the proposed methods to this datase here because a much larger sample size is required to have adequate power to find effects of the modest magnitudes that are reasonable to expect. Furthermore, for the purpose of understanding the performance of the methods, the results of such an analyses will not provide much insight in the absence of a gold standard.
Results
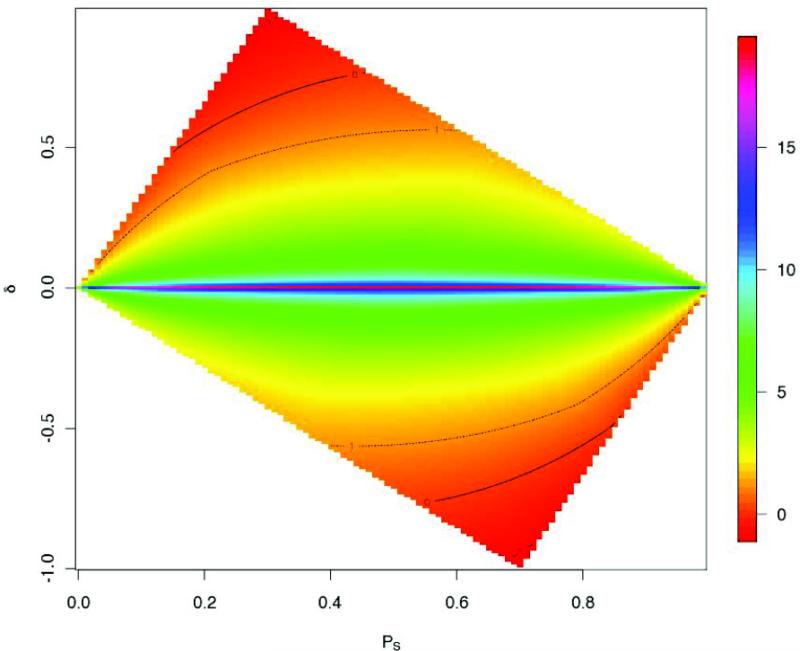
We begin by comparing the magnitude of the noncentrality parameters of the allelic and the ancestry-TDT. Consider an admixed population with proportions 0.3 and 0.7 from two ancestral populations. We take the LD between A and S to be fixed at D′ = 0.8 in each ancestral population. The frequencies of S in the two ancestral populations are allowed to vary, and the genotypic relative risk is set at γ = 1.5. The noncentrality parameter of the combined test is not directly comparable because this test has two-degrees of freedom, and hence requires a larger significance threshold. Figure (1) shows the log ratio of the noncentrality parameter of the allele-TDT, ϕG, compared to that of the ancestry-TDT, ϕA as a function of ps and . As expected, we find the ancestry test compares favorably to the allelic test near the top and bottom corners, regions of maximal allele frequency difference of S between the two ancestral populations. In contrast, in the middle belt, where S occurs at similar frequencies in the ancestral populations, the ancestry test essentially has no power. The outer solid contour lines define a region within which ϕG is at least as large as ϕA. The inner dotted contour lines define the region in which ϕG is at least twice as large as ϕA. The specific values in this heatmap depend on various model parameters; in particular, ϕG depends on the allele frequency of A. For the results presented in Figure (1), the frequency of A in each ancestral population is computed by shrinking the corresponding population-specific frequency of S towards 0.5. This and our additional analyses indicate that in a wide range of parameter settings, the allelic-TDT tends to provide more information than the ancestry-TDT.
Figure 1.
As explained above, the genome-wide significance level depends on the correlation of a test statistic at neighboring markers. We simulated the null distribution of the maximum TG, TA and TGA on a Mexican asthma GWAS study. Figure (2) displays the realizations of the test statistics along one chromosome for one permuted dataset, with the three panels corresponding to the genotype, ancestry and combined tests, respectively. We find little correlation between TG at neighboring markers; and the 95% quantile of the null distribution of the genome-wide maximum statistic was 27.32. This is only slightly lower than a Bonferroni corrected threshold for 483, 000 SNPs, which would lead to a critical value of 28.31. In contrast, we find much stronger correlation between neighboring markers for TA, whose 95% quantile of the null distribution of the genome-wide maximum statistic was 20.15, corresponding to a test of ~ 7000 independent hypotheses. Finally, we observe that the distribution of the genome-wide maximum combined test-statistic has a slightly longer tail than that of the allelic test, with a 95% quantile of 30.15. In other words, the value of the two degree of freedom statistic required to declare genome-wide significance is only marginally higher than that for the allelic test, and hence the combined statistic can be more powerful than the allelic test if the true disease allele has only a moderate frequency difference in the ancestral populations. Our analytic approximation for the genome-wide false positive rate produces critical values that are close to the simulated values: assuming a 15 generation admixing time for the Mexican population, the genome-wide critical values at α = 0.05 level, are 30.9, 27.6 and 20.7, for TGA, TG and TA respectively.
Figure 2.
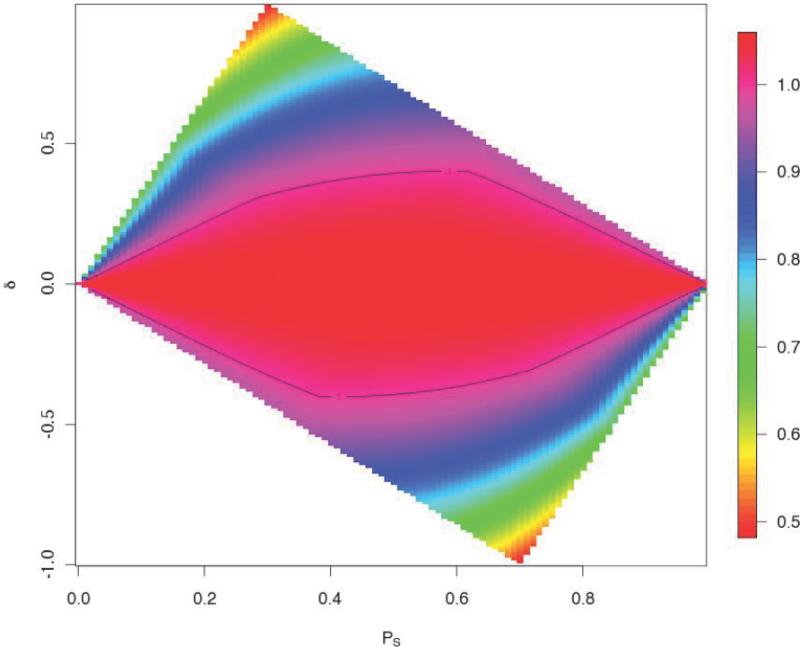
Figure (3) compares the sample size required for the two-df test and for the allelic test under the same model specification as in Figure (1), but we now account for the differences in degrees of freedom and genome-wide critical values for the two tests. The contour lines define the parameters for which the power of the combined test and allelic tests are equal. It is easy to see that taking ancestry into account can improve power in some cases. (We have made a similar comparison of the ancestry test and the allelic test. While the power of the ancestry test relative to that of the allelic test improves compared to the situation described in Figure 1, despite the much lower genome-wide critical value of the ancestry test, it still has lower power than the allelic test in most situations. The exception is when the risk variant occurs with considerable ancestry-specificity.)
Figure 3.
The preceding analyses explore the relative efficiencies of the various tests primarily as a function of the frequencies of the true risk variant. However, the power of the tests depends on numerous aspects of the model, which makes a completely satisfactory comparison difficult. To incorporate a more realistic pattern of LD and of diversity in allele frequencies, we performed a simulation using the data generated from the Phase 3 data from the HapMap project (http://www.hapmap.org). We extracted all SNPs that are typed on the Illumina 550K platform, and consider an admixed population that is 30% European (CEU) and 70% African (YRI) by ancestry; thus, ps and δ at the disease locus are determined by the actual SNPs in the HapMap data. In turn, we assume each SNP is a disease variant but not typed, and compute the sample size required to reject the SNP to the right after accounting for different genome-wide critical values. Figure (4) shows the log (base 2) ratio of sample size required by the ancestry-TDT to that required by the allele-TDT. For 67% of the genome, the allelic test requires smaller sample size than the ancestry test. However, for 22% of the variants, the ancestry test requires less than half the sample size needed by the allelic test.
Figure 4.
Figure (5) compares the sample size required for the combined test to that for the allelic test alone. Overall, power improves for 66% of the SNPs by using the combined test, and for 26% of the SNPs the combined test requires less than half of the sample size of the allelic test. For the remaining 34% of SNPs, the combined test is less efficient because the variants show little frequency variation between populations. However, the loss of efficiency never exceeds 6%.
Figure 5.
In an effort to describe settings where the combined test outperforms the allelic test, we compared various characteristics of both “disease” and marker loci. We find that the most predictive measures are the LD between the marker and disease locus and the ancestral allele frequency difference of the disease variant (but not the marker allele frequency). Compared to the entire set of SNPs considered, variants that do not benefit from the combined test tend to be in higher LD with the candidate marker and to have similar frequencies in the ancestral populations. As we noted earlier, (a) admixture mapping only has power to detect variants that are ancestry-specific; and (b) when the marker genotype is highly correlated with the genotype at the disease locus, ancestry information does not add much information even if the disease variant is ancestry-specific.
We also compared sample sizes required for the combined test to those for the ancestry test. For 83% of the SNPs the combined test is more powerful; in fact, for 40% of the SNPs the combined test requires less than 20% of the sample size needed for the ancestry test. However, as a consequence of the substantially lower genome-wide critical value for the ancestry test, the combined test can require up to 37% greater sample size. This is rare, though, since only for 14% of the SNPs does the combined test require as much as a 10% increase in sample size over the ancestry test.
In the previous analysis, we compute the power as the probability that a test can reject the SNP next to the true disease locus. In practice, however, it is helpful to reject any SNPs in a small neighborhood of the disease locus. Therefore, we also compared the power of the three tests, when we allow the tests to reject any SNPs in a six SNPs neighborhood (three to each side) of the disease locus. This does not alter the power and sample size of the ancestry test, because the ancestry seldom changes within such a small neighborhood. In contrast, the power of the allelic test can increase considerably if a neighboring marker (but not the marker immediately next to the disease variant) is in high LD with the disease variant. Using such a criterion, the combined test performs favorably on 37% of the SNPs when compared to the allelic test. When the neighborhood is expanded to 10 SNPs (five to each side), the combined test still outperforms the allelic test on 32% of the SNPs.
Table 1 compares the sample sizes required by the three tests under both criteria.
Table 1.
Relative efficiency of the three tests. Numbers represents the percentage of markers that fall into each category.
| 1-SNP | 6-SNPs | 10-SNPs | |
|---|---|---|---|
| Ncomb < Nallele | 66 | 37 | 32 |
| Ncomb < Nanc | 83 | 98 | 99 |
| Nallele < Nanc | 67 | 92 | 99 |
Discussion
In genome-wide association studies in admixed populations, tests based on geno-type and on ancestry have been developed for identifying markers that influence phenotype (Ewens and Spielman, 1995; McKeigue, 1997). We have shown that genotype and ancestry provide complementary information (except when disease predisposing and marker alleles are in essentially complete disequilibrium). Intuitively, the allele-TDT only uses information from genotypically heterozygous parents, while the ancestry-TDT only uses information from ancestrally heterozygous parents. Since some parents are genotypically heterozygous but not ancestrally heterozygous (or vice versa), the two tests are not identical. Simply performing both tests at each marker across the genome would exacerbate an already severe multiple comparison problem and is likely to decrease the power. Ideally, one might try to use the ancestry test if the disease allele is known to be population-specific, and use the genotype test in a neighborhood of high linkage disequilibrium. In the context of GWAS, however, one does not know the precise location of the disease locus, much less the local LD structure and the population allele frequencies of the risk variants. Therefore, it is not possible to choose an ideal test a priori.
We propose a statistical framework under a family-based design that integrates the two sources of information into one combined test. When neither marker genotype nor ancestry perfectly captures the transmission of the disease variant, the combined test achieves greater power than either the genotype or the ancestry test alone. In situations where either genotype or ancestry provides no information, the combined test is only slightly less powerful than the more informative test. For this reason, the combined test is a practical procedure for GWAS in admixed populations and improves power on average. It is our hope that the method and results presented here serve as a proof of principle example, which demonstrates the potential benefits of considering complementary sources of information in genetic association studies. Specifically, the general framework of combining genotype and ancestry information can be extended to the more commonly adopted population-based case-control designs, and it is likely that a combined testing procedure can improve the overall statistical efficiency in this setting as well. Since a joint ancestry-genotype test under a population-based design requires careful adjustment of population structure, we leave it for a future endeavor.
The sample size calculations reveal key elements that affect the relative efficiency of the allelic and the ancestry tests. Our findings are qualitatively consistent with the conclusions of Clarke and Whittemore (2007): the power of the allelic test depends on the LD between the marker and the disease locus, while the power of the ancestry test depends critically on the difference in the frequency of the risk variant between ancestral populations. Clarke and Whittemore (2007) compared the noncentrality parameters of the allelic and ancestry tests, which are most relevant in testing one candidate SNP. Since the genome-wide significance levels also depend on the correlation between markers, which differs substantially between the tests, the significance thresholds also differ. These critical values can be found either through simulation or through theoretical approximation. Incorporating these varying critical values, our analysis bears more relevance for GWAS. In particular, because of its substantially lower rejection threshold, the ancestry-based test competes more successfully with the allelic test in the GWAS than it would be for testing a single SNP.
A key parameter in determining the power of the allelic-TDT is the LD between marker and disease locus. Although this parameter is unknown a priori, with the decreasing cost of high-density genotyping, it is reasonable to assume that some markers on the array are in the vicinity of the disease locus, and hence are in high LD with the disease variant. In the context of high-throughput sequencing, we expect the disease variant itself to be genotyped (i.e. |r| = 1). Therefore, we expect that in the future most common disease variants can be detected more readily using an allelic test.
The power of ancestry-TDT depends on the allele frequency difference of the disease variants, for which we have little empirical data to date. Data from the HapMap project and the HGDP project reveal that a majority of alleles show similar frequencies even among the most distantly related human populations. Of the autosomal SNPs on the Illumina 550K arrays, the median absolute frequency difference between CEU and YRI individuals is .16, with a mere 5% of alleles showing a CEU-YRI frequency difference greater than 0.5. However, it is possible that the SNPs on this genotype array do not represent an unbiased collection of SNPs that influence phenotypes of interest.
The ancestry test may offer information in the presence of rare variants and allelic heterogeneity. Suppose, for example, that multiple rare variants occur in one ancestral population and they influence the phenotype in the same direction. This is likely the case for phenotypes that are under directional selection in ancestral populations, such as pigmentation genes and genes involved in metabolic traits (Pickrell et al., 2009). Because each variant is rare, the allelic test requires a large sample size to detect each variant unless a single marker is in high LD with all of them (in which case the associated allele of the marker will appear to be the variant). In contrast, the ancestry test (like a family based test for linkage) has the effect of grouping haplotypes carrying different risk variants, and thus implicates a genomic region rather than a single variant.
It should be emphasized that the correlation of the ancestry statistics along a chromosome, and thus the genome-wide critical value for the ancestry test and the success of a scan statistic to help in fine mapping the phenotype, depends on the admixing history of the population. In a recently admixed population, individuals inherit large segments of chromosome from a given ancestral population; and the ancestry statistics are highly correlated along the genome. As time goes on, recombination breaks large ancestry blocks into finer segments; and as a result, the correlation in ancestry statistics decreases. Under a population history comparable to that of the African Americans or the Mexican Americans, we found the genome-wide critical value for the ancestry test to be substantially lower than that for the allelic test. In contrast, for a population that has undergone more ancient admixture, such as the Uyghurs, who were estimated to have experienced admixture more than 100 generations ago (Xu and Jin, 2008; Li et al., 2009), ancestry test statistics are expected to be much less correlated, and therefore the genome-wide critical value is much higher, about 24.2 according to our analytic approximation.
Taking the factors discussed above into consideration, our general conclusion is that ancestry information can augment a genotype-based association testing procedure. In the context of GWAS or sequencing-based association studies, however, admixture mapping alone is usually ineffective. Our results also highlight the importance of studying diverse populations. In particular, variants that are fixed in a population cannot be detected by an association study in that population. Admixed populations offer unmatched opportunities to identify trait loci that are polymorphic in at least one ancestral population, or loci at which different alleles are fixed in ancestral populations.
Acknowledgements
The authors are grateful for helpful comments from Drs Josée Dupuis, Warren Ewens, Min Shi and Dmitri Zaykin. Research supported by NIGMS grant GM073059. NJ is supported by a NSF Predoctoral Fellowship. Subject enrollment and genotyping of the Mexican asthma study were supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (ZIA ES049019 and ZIA ES025045). Subject enrollment was supported in part by the National Council of Science and Technology (grant 26206-M), Mexico. Dr. Romieu was supported in part by the National Center for Environmental Health at the Centers for Disease Control. The authors acknowledge Deborah Nickerson and Joshua Smith at the University of Washington for genotyping services. For data management, we acknowledge the assistance of Grace Chiu at Westat Inc. (Research Triangle Park, NC) along with Shuangshuang Dai and John Grovenstein at the National Institute of Environmental Health Sciences.
References
- Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Whittemore AS. Comparison of admixture and association mapping in admixed families. Genet. Epidemiol. 2007;31:763–775. doi: 10.1002/gepi.20239. [DOI] [PubMed] [Google Scholar]
- Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision, and admixture. Am. J. Hum. Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owens SA, Weiss ST, Raby BA, Gao H, Eng C, Chapela R, Burchard EG, Tang H, Sullivan PF, London SJ. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner EO, Weinberg CR. Method for using complete and incomplete trios to identify genes related to a quantitative trait. Genet. Epidemiol. 2004;27:33–42. doi: 10.1002/gepi.20001. [DOI] [PubMed] [Google Scholar]
- Krieger H, Morton NE, Mi MP, Azevdo E, Freire-Maia A, Yasuda N. Racial admixture in north-eastern Brazil. Ann. Hum. Genet. 1965;29:113–125. doi: 10.1111/j.1469-1809.1965.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based methods for linkage and association analysis. Adv Genet. 2008;60(NIL):219–52. doi: 10.1016/S0065-2660(07)00410-5. [DOI] [PubMed] [Google Scholar]
- Li H, Cho K, Kidd JR, Kidd KK. Genetic landscape of Eurasia and “admixture” in Uyghurs. Am. J. Hum. Genet. 2009;85:934–937. doi: 10.1016/j.ajhg.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM. Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am. J. Hum. Genet. 1997;60:188–196. [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am. J. Hum. Genet. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, Pritchard JK. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am. J. Hum. Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redden DT, Divers J, Vaughan LK, Tiwari HK, Beasley TM, Fernndez JR, Kimberly RP, Feng R, Padilla MA, Liu N, Miller MB, Allison DB. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genet. 2006;2:e137. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Choudhry S, Via M, Basu A, Sebro R, Eng C, Beckman K, Thyne S, Chapela R, Rodriguez-Santana JR, Rodriguez-Cintron W, Avila PC, Ziv E, Burchard E. Ancestry-related assortative mating in latino populations. Genome Biology. 2009;10:132. doi: 10.1186/gb-2009-10-11-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Kimmel G, Halperin E, Jordan MI. On the inference of ancestries in admixed populations. Genome Res. 2008;18:668–675. doi: 10.1101/gr.072751.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund D, Yakir B. The Statistics of Gene Mapping. Springer; 2007. [Google Scholar]
- Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, Balam-Ortiz E, del Bosque-Plata L, Velazquez-Fernandez D, Lara C, Goya R, Hernandez-Lemus E, Davila C, Barrientos E, March S, Jimenez-Sanchez G. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O'Brien SJ, Reich D. A high-density admixture map for disease gene discovery in african americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist A, Fratkin E, Do CB, Batzoglou S. Effect of genetic divergence in identifying ancestral origin using HAPAA. Genome Res. 2008;18:676–682. doi: 10.1101/gr.072850.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Coram M, Wang P, Zhu X, Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 2006;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am. J. Hum. Genet. 2006;79:640–649. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am. J. Hum. Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, Long JC, Hanson RL, Sievers ML, Knowler WC. Individual estimates of European genetic admixture associated with lower body-mass index, plasma glucose, and prevalence of type 2 diabetes in Pima Indians. Am. J. Hum. Genet. 2000;66:527–538. doi: 10.1086/302773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jin L. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am. J. Hum. Genet. 2008;83:322–336. doi: 10.1016/j.ajhg.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen K, Seldin MF, Li H. A hidden Markov modeling approach for admixture mapping based on case-control data. Genet Epidemiol. 2004;27(3):225–39. doi: 10.1002/gepi.20021. [DOI] [PubMed] [Google Scholar]