Abstract
Species and biotype distribution was determined in 44 bovine viral diarrhea virus- (BVDV-) positive samples submitted to the Animal Disease Diagnostic Laboratory (ADDL) in Indiana during 2006–2008. BVDV RNA was detected in the 5′-untranslated region and Npro region using reverse transcriptase PCR followed by sequencing analysis of the PCR product. Additionally, cases were classified into one of six categories according to history and/or lesions: acute symptomatic, hemorrhagic, respiratory distress, reproductive, persistent infection (PI), and mucosal disease (MD). Of 44 BVDV-positive samples, 33 were noncytopathic (ncp), 10 were cytopathic (cp), and one presented both ncp and cp biotypes. Sequencing analysis demonstrated that all samples belonged to BVDV-1a, BVDV-1b, or BVDV-2. The most common isolate was ncp BVDV-1b, (44%) followed by ncp BVDV-2a (24%). Among the six categories, respiratory clinical signs were the most common (36%) followed by PI (25%) and MD (16%).
1. Introduction
For over half a century, bovine viral diarrhea virus (BVDV) has been known to cause significant disease in cattle herds and other ruminant populations worldwide, creating a substantial economic impact on both the beef and dairy industries [1].
Two types of BVDV infection are recognized in the literature: acute or transient infection and persistent infection (PI) [2]. Naïve animals infected with BVDV develop acute infection, clearing the virus from the body within 7–21 days and develop lifelong antibodies [3]. On the other hand, PI results as a consequence of fetal infection between 18 and 125 days of gestation with the fetus becoming immunotolerant to the virus [4]. If born, PI animals are BVDV antibody negative and BVDV positive, shedding large amounts of virus, and animals are viremic during their lifetime. In addition, PI animals may develop mucosal disease (MD) if they are superinfected with a homologous cytopathic (cp) strain of BVDV, or through a mutation of the infecting ncp BVD virus to the cp form [5]. In both types of infection, clinical signs vary between asymptomatic through mild transient signs to severe acute disease with signs from enteric, hematopoietic, reproductive, or respiratory systems. A severe form of clinical disease, later named hemorrhagic syndrome, was described for the first time during the 1990s associated with BVDV-2 [6]. This syndrome was characterized by fever, pneumonia, diarrhea, and lesions similar to the mucosal disease lesions, death [7]. Not all BVDV-2 species are associated with this severe form of clinical disease [8].
BVDV strains are classified into two species within the pestiviruses and two biotypes [9]. BVDV species classification is done by analyzing the 5′-untranslated region (5′-UTR) and the Npro region of the viral genome [10, 11]. Each BVDV species is divided into subgroups and currently 11 BVDV-1 and 2 BVDV-2 subgroups have been identified. Most authors agree that BVDV-1 is the most common isolate in the United States [12–15], although one study reported that, in the northwest of the US, BVDV-2 was the most common isolate1 [16]. In addition, each species is classified as one of two biotypes according to its ability to induce changes in cell cultures [17]. Cytopathic (cp) viruses are capable of causing cell death in cell culture, also known as the cytopathic effect (CPE); noncytopathic (ncp) viruses do not induce any visible cell changes [17]. Of these two biotypes, ncp viruses are more commonly isolated from field cases when compared to cp viruses [12, 14].
The objective of this study was to determine the biotype and species distribution of BVDV-positive samples submitted to the Indiana Animal Disease Diagnostic Laboratory (ADDL) from 2006 to 2008. A second objective was to correlate the biotype and species to the reported history and lesions in each case.
2. Material and Methods
Forty-four BVDV-positive samples submitted to ADDL during 2006–2008 for BVDV diagnosis were included in this study. Samples submitted included buffy coat, serum, fetus, placenta, brain, lung, mouth, esophagus, oral mucosa, oral tissue pool, mesenteric lymph nodes, thymus, coronary band, and intestine. Samples were inoculated in Madin-Darby bovine kidney (MDBK) cells and cultured in 48-well plates using 5% horse serum, 20 mM L-glutamine, and gentamicin 100 ug/mL incubated at 37°C (5% CO2 incubator) for three days. During this time cells were observed daily and CPE was recorded. Cell cultures in 48-well plates were fixed in 80% aqueous acetone, and the presence of the virus in the cell culture was detected by indirect fluorescent antibody assay using fluorescein isothiocyanate- (FITC-) labeled polyclonal antibodies raised against BVDV as described in [12].
Supernatants were harvested from inoculated plates, and viral RNA was extracted using a commercial kit (MagAttract Virus Mini M48 Kit (QIAGEN, Calif, USA)) and instrument (KingFisher instrument (Thermo Fisher Scientific Inc., Mass, USA)), followed by reverse transcriptase polymerase chain reaction (RT-PCR). The first set of primers used in the RT-PCR reaction, 103/326, amplified viral RNA in the 5′-UTR as described with some modification in [18]. The second set of primers, BD1/BD3, were utilized in this study to amplify viral RNA in the Npro region as described in [19]. Samples that appeared negative for viral RNA in the Npro region were retested by RT-PCR using primers BD1/BD4 [19].
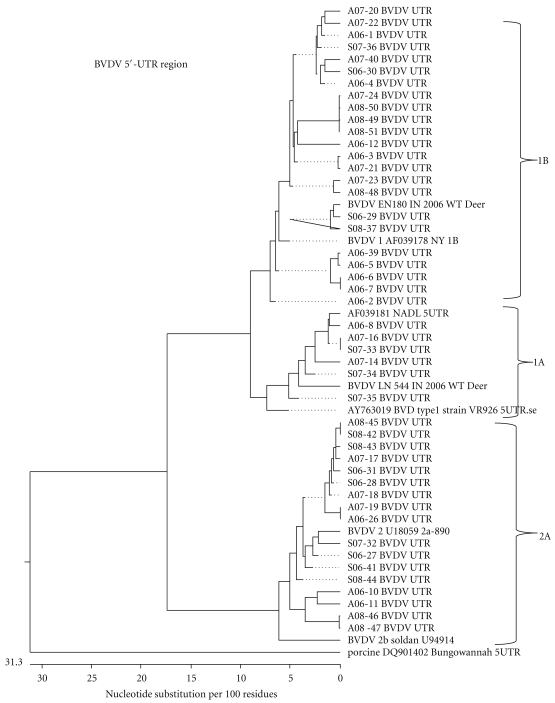
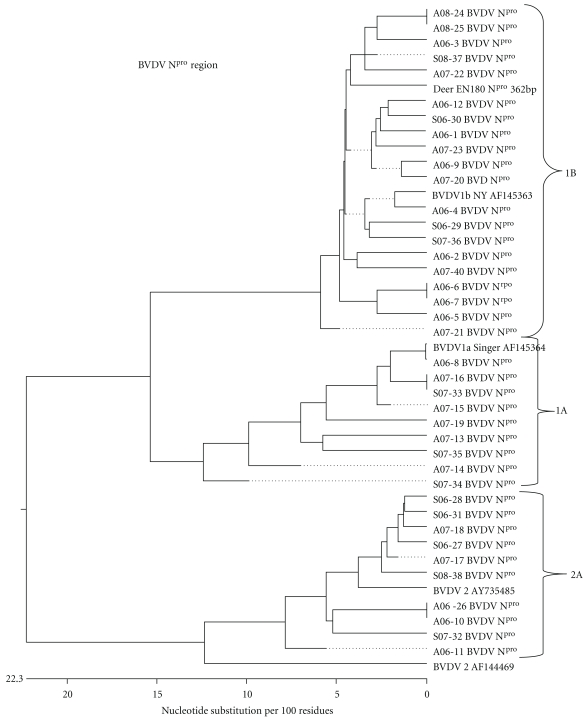
The amplified PCR products were purified using a commercial purification kit (QIAGEN (QIAGEN, Calif, USA)) according to the manufacturer's protocol. These products were sequenced, using an automated sequencer at the Purdue genomic core facilities, and analyzed, and their homology to other BVD viruses was determined based on published sequence information and reference control viruses. This analysis was performed by using computer software (DNASTAR (Madison, Wis USA)). The 5′-UTR and the Npro genome sequences of BVD viral isolates were evaluated for further subgenotype classification and compared to known BVDV reference sequence information to develop a phylogenetic tree (Figures 1 and 2).
Figure 1.
Phylogenetic tree of pestiviruses sequenced at 5′-UTR from cattle isolate samples submitted to the Indiana Animal Disease Diagnostic Laboratory between 2006 and 2008 compared to reference BVD viruses constructed by DNASTAR program, using Clustal W method.
Figure 2.
Phylogenetic tree of pestiviruses sequenced at Npro region from cattle isolated from samples submitted to the Indiana Animal Disease Diagnostic Laboratory between 2006 and 2008 compared to reference BVD viruses constructed by DNASTAR program, using Clustal W method.
Based on reported history and lesions, each case was assigned into one of the following six categories: acute symptomatic, hemorrhagic, respiratory distress, reproductive, persistent infection (PI), or mucosal disease (MD). A case was classified into the PI category if this was a second sample submission based on a previous positive result from our laboratory or if it was provided in the case history. A case was classified as mucosal disease if there were gross or histopathological lesions characteristic of mucosal disease as well as isolation of the cp biotype. Cases were assigned to the other 4 categories based on what has been previously described in [20].
3. Results
A total of 44 BVDV-positive samples from 27 of Indiana's 94 (29%) counties submitted to ADDL during 2006–2008 were included in this study. The ncp biotype was most commonly isolated comprising 33 (75%) of the 44 positive cases, followed by the cp biotype in 10 (22.7%) out of the 44 cases and finally one case (2.3%) in which both biotypes were isolated. Genetic analysis revealed that a total of three subgenotypes were present among these samples: BVDV-1a (16%), BVDV-1b (48%), and BVDV-2a (36%). Most of the samples were classified as ncp BVDV-1b (44%) and ncp BVDV-2a (24%) (Table 1). The most common BVDV infection was associated with respiratory signs which comprised 36% of the cases, followed by PI in 26% of the cases. In most of the cases categorized with respiratory clinical signs there were also other bacterial pathogens isolated that are commonly associated with bovine respiratory disease (BRD): Arcanobacterium pyogenes, Haemophilus somnus, Mycoplasma spp., Mannheimia haemolytica, and Pasteurella multocida. Vaccine history was not provided for all cases; 6 cases (13.7%) received a BVDV killed vaccine and 6 cases received a BVDV modified live (MLV) vaccine. In the case of MLV vaccines, if available, the vaccine strain sequence was compared in the Npro and 5′-UTR sequence region to the case isolate. Only one virus from a case had 99.6% homology to the vaccine strain, NADL.
Table 1.
Biotype and subgenotype distribution of BVDV samples submitted to Indiana Animal Disease Diagnostic Laboratory during 2006–2008.
| Biotype | Subgenotype | ||
|---|---|---|---|
| BVDV 1a | BVDV 1b | BVDV 2a | |
| ncp* | 3 | 19 | 11 |
| cp† | 4 | 1 | 5 |
| ncp and cp | 0 | 1 | 0 |
*Noncytopathic
†Cytopathic.
4. Discussion
This study determined that there exist several species and biotypes of the BVD virus from isolates across Indiana from 2006 to 2008 and supported the fact that there is wide diversity among BVDV strains. This paper is in accordance with previous reports in which the three most common subgenotypes isolated in the USA are BVDV-1a, BVDV-1b, and BVDV-2a, with BVDV-1b being the most common isolate [12–15].
Cases were most commonly categorized with respiratory clinical signs in this study. BVDV contributes to BRD as both a primary pathogen and immunosuppressor predisposing the animal to secondary bacterial infections [21–23]. In particular, BVDV-1 has been associated with BRD, specifically, BVDV-1b which was predominantly isolated from calves with respiratory disease [24]. However, in this study we cannot determine that there was an association of subgenotypes with particular clinical signs. From one respiratory case that was given an MLV vaccine, the isolated BVDV strain was homologous to the vaccine strain.
Animals categorized with persistent infections were based on previous submissions to our laboratory from the same case or from the history provided by the clinician. However, this number of PI cases could be an underestimation for two reasons: first, there is a possibility that PI animals were categorized into a different category because the PI diagnosis could not be made based on a single virus isolation finding, and, second, animals categorized as mucosal disease may have been PI animals at the beginning of disease.
As stated in previous studies, the majority of the vaccines available include either BVDV-1a only or BVDV-1a and BVDV-2a and, to the best of our knowledge, there is only one BVDV vaccine that includes BVDV-1b [11, 25]. The fact that BVDV-1b has been shown to be the most common isolate in the USA and evidence showing that BVDV-1a vaccines induce lower antibody titers against BVDV-1b when compared to BVDV-1a antibody titers raise the question of adequate protection from the vaccines available [25]. However, it is not known to what degree there is cross-reactivity between subgenotypes. Future successful BVDV eradication and control programs will rely on the development of new vaccines and current ongoing research and diagnostic work at Purdue Indiana ADDL and other laboratories.
Acknowledgment
The authors would like to thank the staff of the virology section in Indiana ADDL for help and support throughout this study.
References
- 1.Deregt D. BVD introduction and history. In: Goyal SMRJ, editor. Bovine Viral Diarrhea Virus Diagnosis, Management and Control. Oxford, UK: Blackwell; 2005. pp. 3–33. [Google Scholar]
- 2.Baker JC. The clinical manifestations of bovine viral diarrhea infection. Veterinary Clinics of North America. Food Animal Practice. 1995;11(3):425–445. doi: 10.1016/s0749-0720(15)30460-6. [DOI] [PubMed] [Google Scholar]
- 3.Nettleton PF, Entrican G. Ruminant pestiviruses. British Veterinary Journal. 1995;151(6):615–642. doi: 10.1016/S0007-1935(95)80145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelling CL. Evolution of bovine viral diarrhea virus vaccines. Veterinary Clinics of North America. Food Animal Practice. 2004;20(1):115–129. doi: 10.1016/j.cvfa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Bolin SR. The pathogenesis of mucosal disease. Veterinary Clinics of North America. Food Animal Practice. 1995;11(3):489–500. doi: 10.1016/s0749-0720(15)30463-1. [DOI] [PubMed] [Google Scholar]
- 6.Ridpath JF, Neill JD, Vilcek S, Dubovi EJ, Carman S. Multiple outbreaks of severe acute BVDV in North America occurring between 1993 and 1995 linked to the same BVDV2 strain. Veterinary Microbiology. 2006;114(3-4):196–204. doi: 10.1016/j.vetmic.2005.11.059. [DOI] [PubMed] [Google Scholar]
- 7.Carman S, Van Dreumel T, Ridpath J, et al. Severe acute bovine viral diarrhea in Ontario, 1993–1995. Journal of Veterinary Diagnostic Investigation. 1998;10(1):27–35. doi: 10.1177/104063879801000106. [DOI] [PubMed] [Google Scholar]
- 8.Liebler-Tenorio EM, Ridpath JF, Neill JD. Distribution of viral antigen and development of lesions after experimental infection of calves with a BVDV 2 strain of low virulence. Journal of Veterinary Diagnostic Investigation. 2003;15(3):221–232. doi: 10.1177/104063870301500303. [DOI] [PubMed] [Google Scholar]
- 9.Fauquet CM, Mayo MA, Maniloff J, et al. Virus Taxonomy: Eight Report of the International Committee on Taxonomy of Viruses. Amsterdam, The Netherlands: Elsevier Academic Press; 2005. Flaviviridae. [Google Scholar]
- 10.Avalos-Ramirez R, Orlich M, Thiel HJ, Becher P. Evidence for the presence of two novel Pestivirus species. Virology. 2003;286(2):456–465. doi: 10.1006/viro.2001.1001. [DOI] [PubMed] [Google Scholar]
- 11.Ridpath JF. Practical significance of heterogeneity among BVDV strains: impact of biotype and genotype on U.S. control programs. Preventive Veterinary Medicine. 2005;72(1-2):17–30. doi: 10.1016/j.prevetmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Chul Ahn B, Walz PH, Kennedy GA. Biotype, genotype and clinical presentation associated with bovine viral diarrhea virus (BVDV) isolates from cattle. International Journal of Applied Research in Veterinary Medicine. 2005;3(4):319–325. [Google Scholar]
- 13.Fulton RW, Hessman B, Johnson BJ, et al. Evaluation of diagnostic tests used for detection of bovine viral diarrhea virus and prevalence of subtypes 1a, 1b, and 2a in persistently infected cattle entering a feedlot. Journal of the American Veterinary Medical Association. 2006;228(4):578–584. doi: 10.2460/javma.228.4.578. [DOI] [PubMed] [Google Scholar]
- 14.Fulton RW, Ridpath JF, Ore S, et al. Bovine viral diarrhoea virus (BVDV) subgenotypes in diagnostic laboratory accessions: distribution of BVDV1a, 1b, and 2a subgenotypes. Veterinary Microbiology. 2005;111(1-2):35–40. doi: 10.1016/j.vetmic.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Tajima M, Dubovi EJ. Genetic and clinical analyses of bovine viral diarrhea virus isolates from dairy operations in the United States of America. Journal of Veterinary Diagnostic Investigation. 2005;17(1):10–15. doi: 10.1177/104063870501700104. [DOI] [PubMed] [Google Scholar]
- 16.Evermann JF, Ridpath JF. Clinical and epidemiologic observations of bovine viral diarrhea virus in the northwestern United States. Veterinary Microbiology. 2002;89(2-3):129–139. doi: 10.1016/s0378-1135(02)00178-5. [DOI] [PubMed] [Google Scholar]
- 17.Bolin SR, Grooms DL. Origination and consequences of bovine viral diarrhea virus diversity. Veterinary Clinics of North America. Food Animal Practice. 2004;20(1):51–68. doi: 10.1016/j.cvfa.2003.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Archives of Virology. 1994;136(3-4):309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- 19.Vilcek S, Paton DJ, Durkovic B, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Archives of Virology. 2001;146(1):99–115. doi: 10.1007/s007050170194. [DOI] [PubMed] [Google Scholar]
- 20.Everman JF, Barrington G. Clinical features. In: Goyal SM, Ridpath JF, editors. Bovine Viral Diarrhea Virus: Diagnosis, Management, and Control. Oxford, UK: Blackwell; 2005. pp. 105–119. [Google Scholar]
- 21.Baule C, Kulcsar G, Belak K, et al. Pathogenesis of primary respiratory disease induced by isolates from a new genetic cluster of bovine viral diarrhea virus type I. Journal of Clinical Microbiology. 2001;39(1):146–153. doi: 10.1128/JCM.39.1.146-153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines DM, Martin KM, Clark EG, Kee Jim G, Janzen ED. The immunohistochemical detection of Mycoplasma bovis and bovine viral diarrhea virus in tissues of feedlot cattle with chronic, unresponsive respiratory disease and/or arthritis. Canadian Veterinary Journal. 2001;42(11):857–860. [PMC free article] [PubMed] [Google Scholar]
- 23.Waldner CL, Kennedy RI. Associations between health and productivity in cow-calf beef herds and persistent infection with bovine viral diarrhea virus, antibodies against bovine viral diarrhea virus, or antibodies against infectious bovine rhinotracheitis virus in calves. American Journal of Veterinary Research. 2008;69(7):916–927. doi: 10.2460/ajvr.69.7.916. [DOI] [PubMed] [Google Scholar]
- 24.Fulton RW, Ridpath JF, Saliki JT, et al. Bovine viral diarrhea virus (BVDV) 1b: predominant BVDV subtype in calves with respiratory disease. Canadian Journal of Veterinary Research. 2002;66(3):181–190. [PMC free article] [PubMed] [Google Scholar]
- 25.Fulton RW, Ridpath JF, Confer AW, Saliki JT, Burge LJ, Payton ME. Bovine viral diarrhoea virus antigenic diversity: impact on disease and vaccination programmes. Biologicals. 2003;31(2):89–95. doi: 10.1016/s1045-1056(03)00021-6. [DOI] [PubMed] [Google Scholar]