Abstract
Purpose
The limitless invasive and proliferative capacities of tumor cells are associated with telomerase and expression of its catalytic component, human telomerase reverse transcriptase (hTERT). Interferon-γ (IFN-γ) modulates several cellular activities including signaling pathways and cell cycle through transcriptional regulation.
Experimental Design
Using a recombinant plasmid with hTERT siRNA cDNA, we down regulated hTERT during IFN-γ treatment in human glioblastoma SNB-19 and LN-18 cell lines and examined whether such a combination could inhibit angiogenesis and tumor growth in nude mice. In vitro angiogenesis assay was performed using co-culture of tumor cells with human microvascular endothelial cells. In vivo angiogenesis assay was performed using diffusion chambers under the dorsal skin of nude mice. In vivo imaging of intracerebral tumorigenesis and longitudinal solid tumor development studies were conducted in nude mice.
Results
In vitro and in vivo angiogenesis assays demonstrated inhibition of capillary-like network formation of microvascular endothelial cells and neovascularization under dorsal skin of nude mice, respectively. We observed inhibition of intracerebral tumorigenesis and subcutaneous solid tumor formation in nude mice after treatment with combination of hTERT siRNA and IFN-γ. Western blotting of solid tumor samples demonstrated significant down regulation of the molecules that regulate cell invasion, angiogenesis, and tumor progression.
Conclusions
Our study demonstrated that combination of hTERT siRNA and IFN-γ effectively inhibited angiogenesis and tumor progression through down regulation of molecules involved in these processes. Therefore, combination of hTERT siRNA and IFN-γ is a promising therapeutic strategy for controlling growth of human glioblastoma.
Keywords: angiogenesis, glioblastoma hTERT, IFN-γ, telomerase
Introduction
Glioblastomas are highly invasive and aggressive brain tumors with a dismal prognosis (1). In the United States, more than 20,000 new cases of primary malignant brain tumors are diagnosed every year accounting for 1.4% of all cancers and 2.3% of all cancer deaths (2). Since malignant brain tumor cells often infiltrate deep into the normal tissue, complete surgical removal of the brain tumor is almost impossible, contributing to the high incidence of recurrence (3). Although understanding of the glioblastoma pathophysiology has increased significantly over the past few years, an effective treatment has not yet been developed for this devastating cancer. Advance gene therapy in combination with traditional treatment techniques to prolong the lifespan of cancer patients and control or cure the disease is very promising (4, 5).
Tumor invasion, angiogenesis, and metastasis are complex mechanisms that involve a variety of biochemical and cellular processes, including proteolytic degradation of the extracellular matrix (ECM) (6). Studies focusing on matrix metalloproteinases (MMPs), especially MMP-9, have demonstrated that the overexpression of these proteolytic enzymes actively involves the degradation of ECM proteins, thereby promoting tumor invasion, angiogenesis, and metastasis of most solid tumors including brain tumors (7, 8). ECM degradation simultaneously stimulates expression of vascular endothelial growth factor (VEGF) and angiogenesis (9). Inhibition of these processes may not only suppress tumor growth and invasion but also improve the prognosis for recurrent malignant brain tumors. Methods to inhibit cell invasion and angiogenesis would likely prevent the growth of glioblastomas.
Telomerase adds repeats of specific DNA sequence (TTAGGG) to the 3′ end of DNA strands in the telomere regions. Human telomerase is upregulated in over 85% of primary cancers including glioblastomas and its activity is tightly controlled by expression of human telomerase reverse transcriptase (hTERT) (10-12). So knockdown of hTERT would be an appropriate strategy to control the growth of glioblastomas because telomerase plays the key role in conferring cellular immortality.
Interferon-γ (IFN-γ) is a multifunctional cytokine produced by T cells and natural killer cells. IFN-γ modulates several cellular activities, including signaling pathways, through transcriptional regulation (13, 14). It regulates more than 200 genes, producing a variety of physiological and cellular responses (13). One of the key elements of all tumor cells is evasion from immunosurveillance. Many investigators have indicated that either neutralization of IFN-γ or inhibition of IFN-γ–mediated pathways promotes spontaneous tumor formation in vivo (15, 16), strongly supporting the involvement of IFN-γ in the process of immunosurveillance. Therefore exposure of cancer cells to IFN-γ would be an ideal strategy to regulate tumor cell growth.
Using the technique of RNA interference, successful gene silencing can be achieved either through introduction of synthetic, small interfering RNA (siRNA) oligo nucleotides (17) or their expression through a plasmid vector carrying a specific siRNA cDNA (18). The purpose of this investigation was to down regulate telomerase activity through knockdown of hTERT using a plasmid vector carrying the cognate siRNA cDNA in combination with IFN-γ treatment in two highly invasive human glioblastoma SNB-19 and LN-18 cell lines and to examine whether such a combination could inhibit angiogenesis and tumor growth in nude mice. Furthermore, we wanted to elucidate the molecular mechanisms of inhibition of angiogenesis and tumor growth in vivo after treatment with hTERT siRNA and IFN-γ.
Materials and Methods
Cell culture conditions
Human glioblastoma SNB-19 cells were procured from the National Cancer Institute (Frederick, MD). Human glioblastoma LN-18 cells were purchased from American Type Culture Collection (Manassas, VA). We selected these cell lines because PTEN is mutated and not expressed in SNB-19 cells and PTEN is wild-type in LN-18 cells. We propagated SNB-19 cells were in 50:50 mixture of Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 whereas LN-18 cells in DMEM (Mediatech, Herndon, VA), supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and antibiotics in a humidified incubator containing 5% CO2 at 37°C. Human IFN-γ (Sigma, St. Louis, MO) was diluted in serum-free medium to a final concentration of 10 ng/ml.
Construction of hTERT siRNA expression vector
The hTERT siRNA cDNA with sense and antisense strands was constructed into a mammalian expression vector, pRNAT-CMV3.2/Neo (GenScript, Piscataway, NJ), between BamH I and Xho I sites. We prepared three siRNA sequences to select the most effective one on the basis of percent knockdown of hTERT both at the mRNA and protein levels. The selected siRNA sequence targeting human hTERT mRNA began at nucleotide 2035 (NM_198253), 5′-GGC ACT GTT CAG CGT GCT Ctt-3′ (sense) and 3′-GAG CAC GCT GAA CAG TGCC-5′ (antisense). The scrambled siRNA sequence used was, 5′-CCG TCG ACG CGT ACT TGG Ttt-3′ (sense) and 3′-CGG TCC AGA GCA TCA ACGG-5′ (antisense). The loop selected was 5′-TTC AAG AGA-3′. The linear siRNA construct (with the sense and antisense strand, loop, termination signal, BamH I and Xho I restriction sites) was annealed to the complimentary strand and ligated into the siRNA expression vector (pRNAT-CMV3.2/Neo) between the BamH I and Xho I sites. In this vector, the cytomegalovirus (CMV) promoter drives the expression of siRNA, and the SV40 promoter drives the expression of the resistance gene, neomycin. This vector also carries coral Green Fluorescence Protein (cGFP) for tracking of transfection efficiency in cell cultures. The siRNA sequence was confirmed by DNA sequencing. The plasmid vector carrying the hTERT siRNA cDNA was transformed into JM109 competent cells (Promega, Madison WI), and the positive colonies were screened using the Qiagen miniprep plasmid DNA purification kit (Qiagen, Valencia, CA). The highly expressing colony was selected and propagated in LB broth containing neomycin. The plasmid vector expressing hTERT siRNA was purified using maxiprep plasmid DNA purification kit (Qiagen, Valencia, CA) and used for both cell culture and animal experiments.
Treatment of glioblastoma cells with hTERT siRNA plasmid vector and IFN-γ
About 80% confluent cultures of SNB-19 and LN-18 cells were transfected with the plasmid vector carrying hTERT siRNA cDNA or treated with a final concentration of 10 ng IFN-γ/ml (10 ng IFN-γ corresponded to100 units) or both agents together in serum-free medium. The plasmid vector was transfected with Fugene HD (Roche Diagnostics, IN, USA) in a mixture of 3:1 (3 μl Fugene and 1 μg DNA). Transfection efficiency was monitored with the expression of cGFP using a phase contrast fluorescent microscope (Olympus IX71, Tokyo, Japan). A set of cultures was also transfected with the plasmid vector carrying the scrambled siRNA cDNA sequences for hTERT. A dose of 10 ng/ml IFN-γ was selected based on a dose-response study for cell viability as determined by the 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) assay (19). Higher concentrations of IFN-γ did not significantly decrease cell viability, while lower concentrations found to be less effective. After 24 h, the medium was replaced with regular serum-medium and the cultures were incubated for another 24 h.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) for hTERT mRNA
We carried out real-time RT-PCR experiments to determine the down regulation of hTERT mRNA after transfection with the plasmid vector carrying hTERT siRNA cDNA or treatment with IFN-γ or both agents together. Total cellular RNA was isolated using the Aurum kit (Bio-Rad, Hercules, CA). We used the following primer sequences for PCR amplification of hTERT gene (NM_198253): forward 5′-CAC CAA GAA GTT CAT CTC C-3′, reverse 5′-CAA GTG CTG TCT GAT TCC-3′). Real-time RT-PCR was carried out using a one step RT-PCR kit with SYBR green (Bio-Rad, Hercules, CA) on a real-time PCR machine (iCycler iQ5, Bio-Rad, Hercules, CA) with the following reaction conditions: cDNA synthesis, 10 min at 50°C; reverse transcriptase inactivation at 95°C for 5 min; PCR cycling and detection at 95°C for 10 sec; and data collection at 56°C for 30 sec. We used 100 ng of total isolated RNA for transcription.
Western blotting for hTERT
Western blotting was carried out for hTERT protein levels using hTERT antibody (Santa Cruz Biotechnology, Santa Cruz, CA) as described below. The membranes were re-probed with Western re-probe buffer (Gbiosciences, St. Louis, MO) and analyzed for glyceraldehydes-3-phosphate dehydrogenase (GAPDH) content using a GAPDH monoclonal antibody (Novus Biologicals, Littleton, CO). The Western blotting images were quantified using Gel-Pro analyzer software (Media Cybernetics, Silver Spring, MD).
In vitro angiogenesis assay
In vitro angiogenesis assay (20) was carried out to evaluate the effect of hTERT siRNA and/or IFN-γ on network formation of human microvascular endothelial (HME) cells (Cascade Biologics, Portland, OR) in co-culture. SNB-19 and LN-18 cells (1×104) were seeded into 4-well chamber slides. After 24 h, the cells were transfected with the plasmid vector carrying hTERT siRNA cDNA or treated with IFN-γ or both agents together in serum-free medium. The cells were incubated for 24 h and then co-cultured with 2×104 HME cells in a 50:50 mixture of serum-free medium and HME medium (Medium 131, Cascade Biologics, Portland, OR). The co-cultures were terminated after 72 h, cells were fixed in 95% cold ethanol and treated with Von Willebrand Factor (Factor VIII) antibody, followed by biotinylated second antibody. After washings, the slides were further treated with horse radish peroxidase-labeled streptavidin. The final stain was developed using 3% 3-amino-9-ethylcarbzole in N,N-dimethylformamide. The cells were viewed under a microscope (Olympus BH2, Tokyo, Japan) and photographed. The images were quantified for network formation using Image-Pro Discovery software (Media Cybernetics, Silver Spring, MD).
In vivo angiogenesis assay (dorsal skinfold chamber model)
Dorsal skinfold chamber model (21) was used to examine the anti-angiogenic effect of hTERT siRNA and/or IFN-γ on SNB-19 and LN-18 cells. The diffusion chamber rings (Millipore, Bedford, MA) were prepared with Millipore membrane filters (0.45 μm), sterilized by UV irradiation and then injected with 200 μl of cell suspension (2×105 cells) after treatment with either the hTERT siRNA plasmid vector or IFN-γ or both agents together for 48 h. The opening of the chamber was sealed with sterile bone wax and the chambers were surgically implanted under the dorsal skin of nude mice (Charles River Laboratories, MA). After 10 days, the chambers were removed surgically and the superficial fascia exposed to the chamber was harvested. The formation of new vasculature (neovascularization) was distinguished from pre-existing vessels as curved thin structures in zigzag pattern using a stereomicroscope (Olympus SZX12, Tokyo, Japan) equipped with a Spot RT Slider digital camera (Meyer Instruments, Houston, TX) and photographed. The tumor-induced neovasculature was measured and quantified with the help of an ocular micrometer.
Orthotopic tumorigenesis in nude mice
SNB-19 and LN-18 cells were stably transfected with a mammalian expression vector carrying the luciferase gene phCMV-FSR (Genlantis, San Diego, CA) and propagated in medium containing G-418 (500 μg/ml). The cells were highly homogeneous and propagated from a single cell. The cells were harvested, counted, and 1×106 cells suspended in 10 μl of serum-free medium were injected into the intracerebrum of nude mice using a 25 µl Hamilton syringe with the help of a digital stereotaxic apparatus (Stoelting, Wood Dale, IL) after drilling a small hole on the skull. The animals were left for three days without any treatment. Afterwards, the mice received intrathecal injection of either the hTERT siRNA plasmid vector (5 μg DNA/injection/mouse) or IFN-γ (103 units/injection/mouse) or both agents on alternate days for 20 days. The plasmid vector carrying hTERT siRNA cDNA was suspended in RNAse-free sterile water (25 μg DNA/10 μl) and mixed (1:4, v/v) with i-Fect transfection reagent (Neuromics, Edina, MN) to obtain 5 μg DNA/10 μl of injection volume. The injections were given using a Hamilton syringe with the help of a stereotaxic apparatus at the site of tumor cell plantation. In the case of combination treatment, IFN-γ was injected first (10 μl in serum-free medium) followed by siRNA plasmid vector with an interval of 10 min. One set of mice received similar injections of scrambled siRNA vector while another set was left untreated. On day 21, the mice were injected intraperitoneally with 100 μl (50 mg/ml) luciferin (Genlantis, San Diego, CA). After 10 min, the mice were visualized for luciferase activity using the Xenogen IVIS-200 (Hopkinton, MA) imaging system. A total of 6 mice were used in each group. All animal experiments were performed in compliance with our Institutional Animal Care and Use Committee (IACUC).
Solid tumor development in nude mice
The effect of hTERT siRNA and/or IFN-γ on solid tumor development under the dorsal skin of nude mice was studied. Since SNB-19 cells grow very slowly to form a subcutaneous solid tumor, we have used only LN-18 cells to study subcutaneous solid tumor development. About 80% confluent cultures of LN-18 cells were harvested, counted, and 6×107 cells suspended in 300 μl of serum-free medium were mixed with 300 μl of high concentration Matrigel (22.1 mg/ml, BD Biosciences, San Jose, CA). We used 100 μl of this cell suspension (1×107 cells) in Matrigel to inject under the dorsal skin of nude mice. The animals were left for 4 weeks without any treatment for development of visible tumors with approximate volume of 150 to 300 mm3. Animals were then divided into 5 groups of 6 mice each. The mice were injected at the tumor site either with the hTERT siRNA plasmid vector (50 μg DNA/injection/mouse) or IFN-γ (104 units/injection/mouse) or both agents on alternate days for 6 weeks. The siRNA plasmid vector was mixed with in vivo-jetPEI (Polyplus-transfection, New York, NY) in 5% glucose solution (100 μl/injection). In the case of combination treatment, the animals received IFN-γ injections first followed by siRNA plasmid vectors with an interval of 10 min. One group of animals received scrambled siRNA vector and was considered as the treated controls. Tumor volume was measured beginning from week 4 using a digital vernier caliper. Tumor volume was calculated using the formula [(smallest diameter2 × widest diameter)/2] and the growth curves were plotted for each treatment group (22). At the end of the 10th week, the animals were anesthetized with a mixture of ketamine and xylazine and then photographed. The solid tumors were surgically removed, tumor weight was recorded, and the tumors were photographed. Tumor samples were stored at -80°C for further analysis.
Western blotting for molecules involved in cell proliferation, angiogenesis, and cell cycle regulation in solid tumors
In order to elucidate the molecular mechanisms of the inhibition of angiogenesis and tumor growth by knockdown of hTERT and/or treatment with IFN-γ, we assayed protein levels of the molecules involved in these processes in the solid tumors. The tumor samples were weighed, cut into small pieces, and homogenized using a Omni Ruptor 400 Ultrasonic homogenizer (Omni International, Marietta, GA). The homogenized tumor samples were centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatants were collected. Protein concentration in the supernatant was determined using the Coomassie-Plus protein assay (Pierce Biotechnology, Rockford, IL) and the samples were stored at -20°C until used. The proteins were resolved on 4-20% polyacrylamide gradient gels (Bio-Rad, Hercules, CA) and electroblotted to an activated PVDF membrane (Millipore, Bedford, MA). Non-specific binding sites were blocked with 5% non-fat milk, and the membranes were incubated overnight on a rocker at 4°C with specific antibodies for various protein molecules. The antibodies for proliferating cell nuclear antigen (PCNA) and CD31 were obtained from BD Biosciences (San Jose, CA), VEGF, basic fibroblast growth factor (bFGF), antibodies for CDK2, CDK4, and cyclin D1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the antibodies for matrix metalloproteinase-9 (MMP-9), p27Kip1 and p21Waf1 were procured from Cell Signaling Technology (Danvers, MA). After incubation, the membranes were washed and treated with horseradish peroxidase (HRP)-conjugated respective secondary antibodies (Biomeda, Foster City, CA) at room temperature for 2 h. The membranes were washed again, treated with chemiluminescence reagent (Amersham, Buckinghamshire, UK), and exposed to Kodak film (BioMax XAR, New Haven, CT) for autoradiography. The membranes were re-probed using Western re-probe buffer (Gbiosciences, St. Louis, MO) for GAPDH content with a GAPDH monoclonal antibody (Novus Biologicals, Littleton, CO) to demonstrate that equal amount of protein was loaded in each lane. The Western blotting images were quantified using Gel-Pro analyzer software (Media Cybernetics, Silver Spring, MD).
Statistical analysis
Arithmetic mean and standard deviation (SD) were calculated for all quantitative data. The results were statistically evaluated using one-way analysis of variance (ANOVA). The least significant difference method was used to compare the mean values of control or scrambled siRNA treated groups with those of hTERT siRNA or IFN-γ treated groups. The individual hTERT siRNA or IFN-γ mean values were also compared with the combination treatment mean values. A value of P < 0.05 was considered as statistically significant.
Results
Knockdown of hTERT mRNA and protein in glioblastoma SNB-19 and LN-18 cells
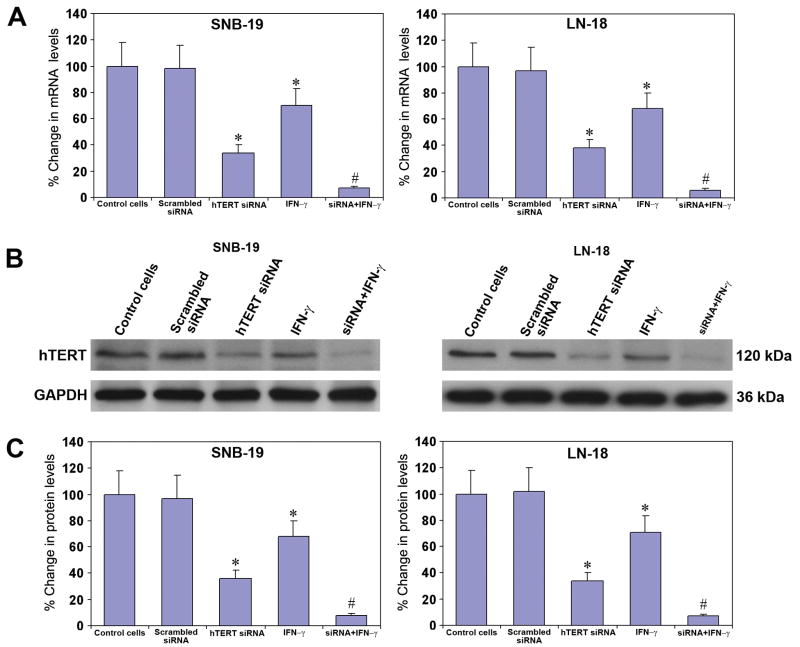
We examined the knockdown of hTERT mRNA and protein levels in glioblastoma SNB-19 and LN-18 cells (Fig. 1). Transfection of SNB-19 and LN-18 cells with a plasmid vector carrying hTERT siRNA cDNA resulted in a marked down regulation of cognate mRNA (Fig. 1A) and protein (Fig. 1B) levels in both cell lines. Transfection of cells with a plasmid vector carrying the scrambled hTERT siRNA cDNA did not alter hTERT mRNA and protein levels. Treatment with IFN-γ also resulted in significant down regulation of hTERT mRNA and protein levels. Treatment with combination of both agents depicted remarkable knockdown of hTERT mRNA and protein levels in both cell lines. GAPDH was used as a loading control for protein levels. Quantification of Western images demonstrated more than 90% knockdown of hTERT protein levels after the combination treatment (Fig. 1C).
Fig. 1.
Alteration in hTERT mRNA and protein levels in SNB-19 and LN-18 cells after transfection with a plasmid vector carrying hTERT siRNA cDNA or treatment with IFN-γ (10 ng/ml) or both agents together for 48 h. A, Real-time RT-PCR analysis for hTERT mRNA levels. Values indicate mean ± SD of 6 assays in each group (*, P < 0.001 compared to the control mean values and #, P < 0.001 compared to hTERT siRNA or IFN-γ mean values). B, Western blotting for hTERT protein levels. The data are representative of 6 independent experiments. C, Quantitative evaluation of Western blotting. Values are mean ± SD of 6 assays in each group (*, P < 0.001 compared with the control mean values and #, P < 0.001 compared with hTERT siRNA or IFN-γ mean values).
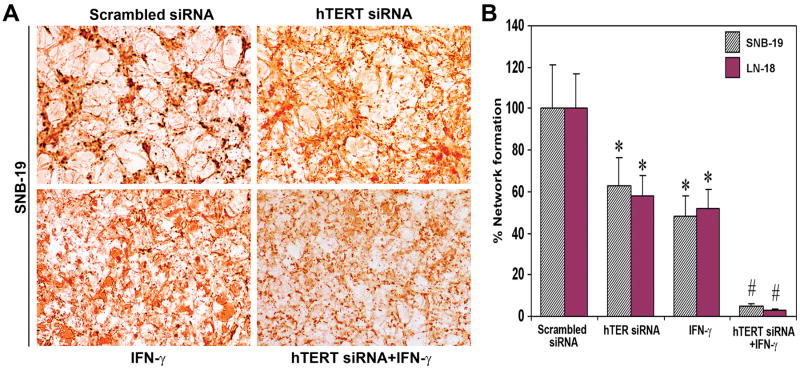
Combination of hTERT siRNA and IFN-γ inhibited in vitro angiogenic network formation
We examined the in vitro angiogenic network formation in co-culture of HME and glioblastoma cells after the treatments (Fig. 2). Von Willebrand factor is a characteristic marker for endothelial cells. The co-culture of HME cells with SNB-19 or LN-18 cells or cells transfected with scrambled siRNA produced capillary-like network formation of HME cells (Fig. 2A). Such a network formation of endothelial cells was significantly reduced when co-cultured with SNB-19 and LN-18 cells transfected with a plasmid vector expressing hTERT siRNA or treated with IFN-γ (Fig. 2A). Similar network formation was almost completely inhibited after treatment with combination of both agents. Quantitative evaluation of in vitro network formation of HME cells demonstrated that treatment with hTERT siRNA, IFN-γ, and combination of both agents caused, respectively, 37, 52, and 95% inhibition in case of SNB-19 cells and 42, 48, and 95% in case of LN-18 cells (Fig. 2B). Since there was no difference between untreated controls and the scrambled siRNA treated samples, the scrambled siRNA results were considered as the controls.
Fig. 2.
Effect of hTERT siRNA or/and IFN-γ on in vitro network formation of HME cells. A, SNB-19 cells on chamber slides were transfected with hTERT siRNA vector or treated with IFN-γ or both agents together. After 24 h, HME cells were co-cultured with glioblastoma cells. The co-cultures were terminated at 72 h and immunohistochemically stained for Von Willebrand factor. B, Quantitation of in vitro network formation by HME cells. Values indicate mean ± SD of 6 experiments in each group of both SNB-19 and LN-18 cells (*, P <0.001 compared with the control mean values and #, P <0.001 compared with hTERT siRNA or IFN-γ mean values).
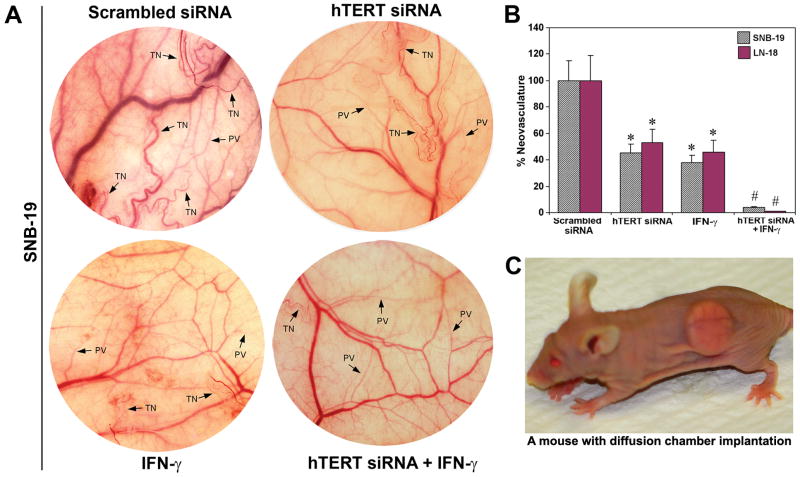
hTERT siRNA and IFN-γ inhibited in vivo angiogenesis
The effect of hTERT siRNA and/or IFN-γ on neovascularization was studied under the dorsal skin of nude mice (Fig. 3). The implantation of diffusion chambers containing SNB-19 and LN-18 untreated cells and cells transfected with a plasmid vector carrying hTERT scrambled siRNA cDNA resulted in the development of microvessels as indicated by the thin and curved structures arising from the pre-existing vessels in a zigzag manner (Fig. 3A). The formation of such microvessels was significantly reduced in the cells treated with hTERT siRNA or IFN-γ and almost completely inhibited in the cells treated with both agents together (Fig. 3A). This inhibition of neovascularization indicated that the cells treated with combination of hTERT siRNA and IFN-γ failed to secrete the potent angiogenic factors such as VEGF and bFGF. Quantitative measurement revealed that treatment with hTERT siRNA, IFN-γ, and both agents together caused, respectively, 55, 62, and 96% inhibition of neovascularization in SNB-19 cells (Fig. 3B) and 47, 54, and 99% inhibition in LN-18 cells (Fig. 3B). The scrambled siRNA results were considered as the treated controls. We showed a mouse bearing surgically implanted diffusion chamber for in vivo angiogenesis assay (Fig. 3C). The diffusion of angiogenic factors through the chamber membrane induces neovascularization in the superficial fascia of nude mice. This process was effectively inhibited due to treatment of cells with combination of hTERT siRNA and IFN-γ.
Fig. 3.
In vivo angiogenesis assay. A, SNB-19 cells transfected with a plasmid vector expressing hTERT siRNA or treated with IFN-γ or both agents together were suspended in 200 μl of serum-free medium, injected into a diffusion chamber, implanted under the dorsal skin of nude mice and left for 10 days. Strong development of tumor-induced neovasculature (TN) with curved thin structures in zigzag pattern arising from pre-existing vasculature (PV) was observed in SNB-19 control and scrambled siRNA transfected cells. B, Quantitative presentation of neovasculature to indicate the extent of in vivo angiogenesis. Values show mean ± SD of 6 experiments in each group of SNB-19 and LN-18 cells (*, P <0.001 compared with the control mean values and #, P <0.001 compared with hTERT siRNA or IFN-γ mean values). C, A nude mouse bearing a diffusion chamber implanted under the dorsal skin for the in vivo angiogenesis assay.
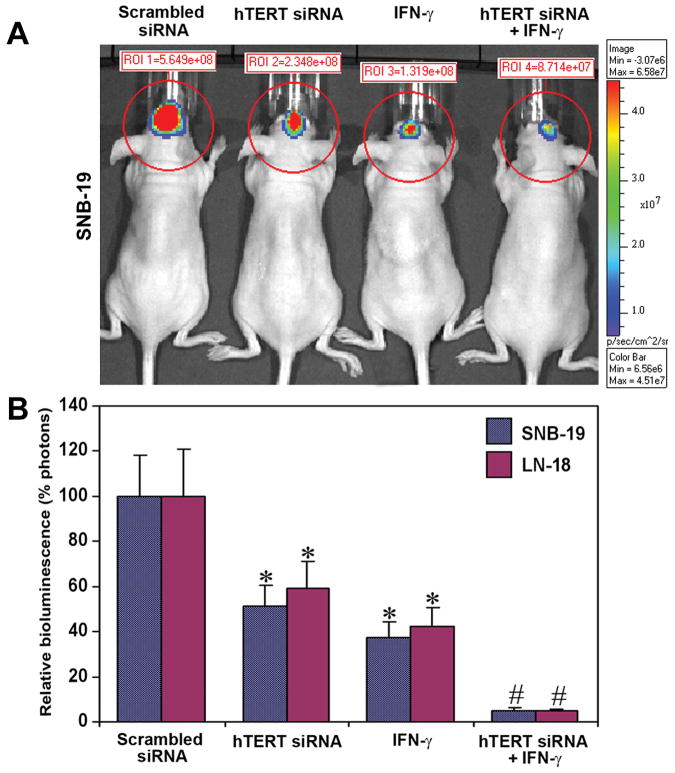
Inhibition of orthotopic tumorigenesis in nude mice
The effect of hTERT siRNA and/or IFN-γ on inhibition of orthotopic tumorigenesis in nude mice was evaluated (Fig. 4). We stably transfected SNB-19 and LN-18 cells with luciferase gene, injected into the intracerebrum of nude mice, and allowed the cells to multiply for 3 weeks during treatment. The untreated group of mice and the mice injected with scrambled siRNA vector showed multiplication and tumorigenesis as indicated by the large bioluminescence image produced by the tumor cells carrying the luciferase gene (Fig. 4A). Production of this bioluminescent image was partially inhibited in animals injected with hTERT siRNA plasmid vector or treated with IFN-γ and was almost completely inhibited in animals treated with both agents together (Fig. 4A). Quantitative evaluation of the inhibition of intracerebral tumorigenesis demonstrated 47.8, 62.5, and 94.8% inhibition in SNB-19 cells and 41, 58, and 96.2% inhibition in LN-18 cells, respectively, after the treatment with hTERT siRNA, IFN-γ, and combination of both agents (Fig. 4B). The scrambled siRNA treated mice were considered as the treated controls.
Fig. 4.
Inhibition of intracerebral tumorigenesis in nude mice after treatment with hTERT siRNA or/and IFN-γ. A, SNB-19 cells were stably transfected with luciferase gene and implanted into the intracerebrum of nude mice. Beginning from day 3, the mice were injected at the site of tumor cell implantation either with hTERT siRNA vector or IFN-γ or both agents together on alternate days for 20 days. Then mice were injected with luciferin and visualized for the effect of treatments using Xenogen IVIS-200 imaging system. The data are representative of 6 animals in each group. The background signal for bioluminescence from an untreated mouse is about 1.5e+05 photons on Xenogen IVIS-200 imaging machine. B, Quantitation of relative bioluminescence as percent photons in both SNB-19 and LN-18 cells after treatment with hTERT siRNA or/and IFN-γ. Data show mean ± SD of 6 animals in each group (*, P < 0.001 compared with the scrambled siRNA mean values and #, P < 0.001 compared with hTERT siRNA or IFN-γ mean values).
Inhibition of solid tumor growth in nude mice
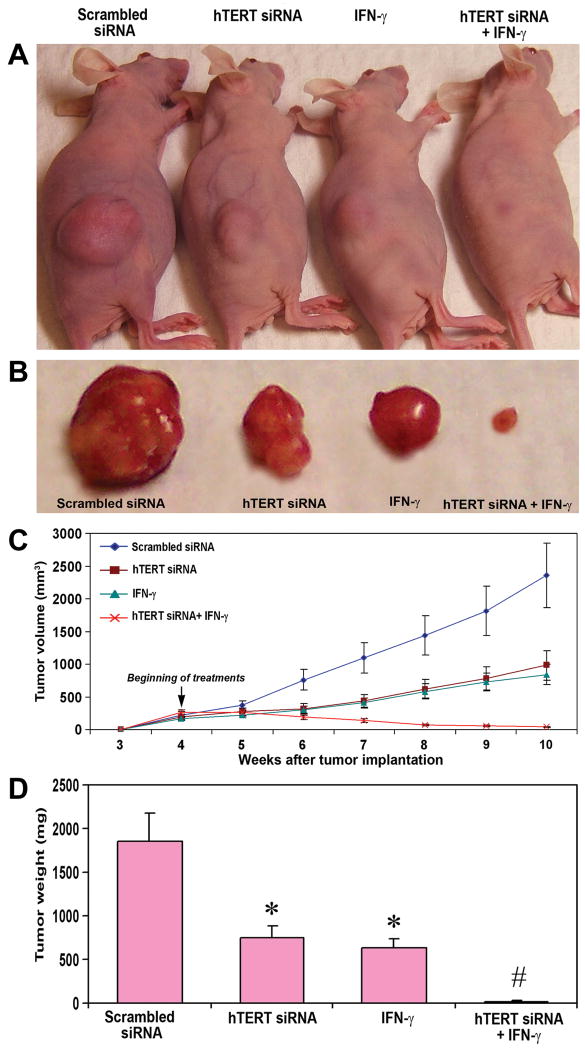
We studied the effect of hTERT siRNA and/or IFN-γ on subcutaneous solid tumor development in nude mice (Fig. 5). The control and scrambled siRNA groups of mice developed large tumors within a period of 10 weeks (Fig. 5A and B). The tumor size was considerably reduced in the animals treated with hTERT siRNA or IFN-γ. The treatment with combination of both agents resulted in a remarkable reduction in the solid tumor size (Fig. 5A and B). At the end of the 10th week there was no visible tumor, except a reddish patch, after treatment with combination of both agents. Longitudinal measurement of tumor volume revealed steady state growth of solid tumors in control and the scrambled siRNA vector transfected animals (Fig. 5C). The growth curve was significantly switched downwards after treatment with hTERT siRNA or IFN-γ and was almost straight after treatment with combination of both agents (Fig. 5C). Measurement of tumor weight demonstrated 60 and 66% reduction after treatment with hTERT siRNA and IFN-γ, respectively (Fig. 5D). The treatment with combination of both agents resulted in 99% reduction of tumor weight. The scrambled siRNA treated mice were considered as the treated controls.
Fig. 5.
Inhibition of subcutaneous solid tumor development in nude mice after treatment with hTERT siRNA or/and IFN-γ. A, SNB-19 cells were harvested, counted, suspended in an equal volume of high concentration Matrigel and 100 μl of the suspension (5×106 cells) was injected under the dorsal skin of nude mice. The animals were left for 4 weeks without any treatment for uniform development of visible tumors. Afterwards, the mice were injected at the tumor site with either hTERT siRNA vector, IFN-γ, or both agents together on alternate days for 6 weeks. At the end of 10th week, the animals were anesthetized with ketamine and xylazine and then photographed. B, The tumors were surgically removed, weighed, and photographed. The data are representative of 6 animals in each group. C, Longitudinal measurement of tumor volume using a digital vernier caliper in nude mice after treatment with hTERT siRNA or/and IFN-γ. Data show mean ± SD of 6 animals in each group. D, Quantitation of tumor weight. Data show mean ± SD of 6 animals in each group (*, P < 0.001 compared with the scrambled siRNA mean values and #, P < 0.001 compared with hTERT siRNA or IFN-γ mean values).
Combination of hTERT siRNA and IFN-γ down regulated the molecules involved in cell proliferation, angiogenesis, and cell cycle in solid tumors
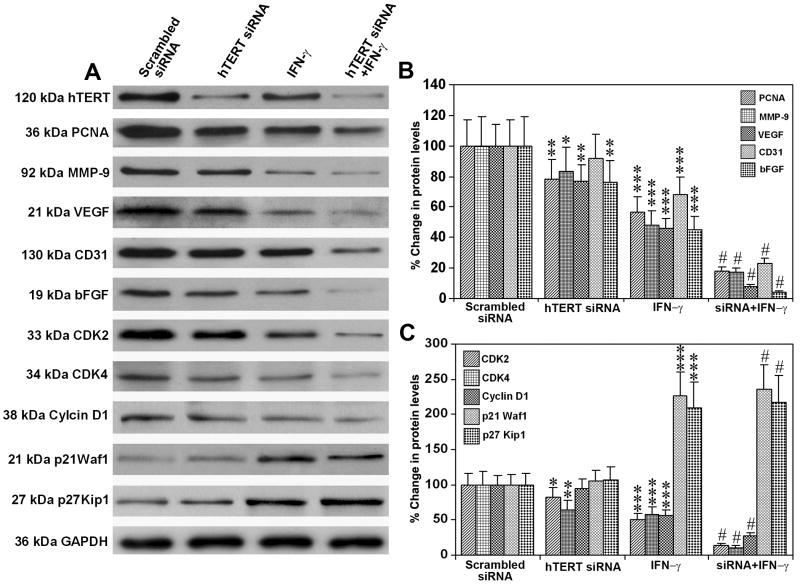
In order to elucidate the molecular mechanisms of the inhibition of glioblastoma cell invasion, angiogenesis, and tumor progression by knockdown of hTERT siRNA and/or treatment with IFN-γ, we performed Western blotting for the protein levels of the prominent molecules involved in these processes in solid tumors (Fig. 6). The hTERT protein levels in solid tumor samples showed more than 60% knockdown after injection with hTERT siRNA alone and more than 80% knockdown after treatment with combination of hTERT siRNA and IFN-γ (Fig. 6A). There was significant (P < 0.001) down regulation of PCNA and MMP-9, the molecules involved in tumor cell proliferation and invasion, after treatment with combination of hTERT siRNA and IFN-γ (Fig. 6A and B). The key angiogenic factors, VEGF and bFGF, were also markedly decreased after treatment with hTERT siRNA, IFN-γ, or both agents (Fig. 6A and B). There was a significant reduction in CD31 molecule after treatment with IFN-γ or combination of hTERT siRNA and IFN-γ (Fig. 6A and B), indicating decrease in proliferation of endothelial cells and angiogenesis. The molecules CDK2, CDK4, and cyclin D1, known to be involved in progression of cell cycle, were also down regulated after treatment with IFN-γ or combination of both agents (Fig. 6A and C). In contrast, the major CDK inhibitors p21Waf1 and p27Kip1 were remarkably upregulated after treatment with IFN-γ or combination of both agents (Fig. 6A and C). However, no alteration was noticed in the protein level of any of these molecules after treatment with hTERT siRNA plasmid vector. There was no significant alteration in the protein levels of all the molecules in the animals injected with plasmid vector carrying the scrambled hTERT siRNA, compared with the parental cells, and thus scrambled hTERT siRNA treated animals were considered as the treated controls. Re-probing for GAPDH demonstrated equal loading of protein sample in each lane.
Fig. 6.
Western blotting for molecules involved in cell proliferation, angiogenesis, and cell cycle in solid tumors. A, Representative Western blots for hTERT, PCNA, MMP-9, VEGF, CD31, bFGF, CDK2, CDK4, Cyclin D1, p21Waf1 and p27Kip1 in the xenograft solid tumors of LN-18 cells. B and C, Quantitation of Western blots. Values show mean ± SD of 6 experiments in each group (*, P < 0.05; **, P < 0.01; and ***, P <0.001 compared with the scrambled siRNA mean values; and #, P <0.001 compared with hTERT siRNA or IFN-γ mean values).
Discussion
The present study demonstrated that treatment with combination of hTERT siRNA and IFN-γ inhibited cell proliferation, angiogenesis, and tumor progression in glioblastomas due to down regulation of the molecules involved in these processes and upregulation of p27Kip1 and p21Waf1. Treatment with IFN-γ alone suppressed to some extent the expression of hTERT and other proteins involved in cell proliferation and angiogenesis. The combined effects of hTERT siRNA and IFN-γ most effectively down regulated the expression of the molecules involved in cell proliferation and angiogenesis and thus caused inhibition of tumor progression. However, further studies are necessary to delineate the other molecular mechanisms of inhibition of glioblastomas following treatment with combination of hTERT siRNA and IFN-γ.
The ability of most tumor cells to grow indefinitely relies on the presence of functional telomerase to maintain telomere length, thus circumventing normal cellular senescence or apoptosis to promote tumor growth (23, 24). Telomerase activity in majority of human cancers is tightly controlled by its catalytic component hTERT. In the present study, treatment with hTERT siRNA resulted in more than 60% suppression and treatment with combination of hTERT siRNA and IFN-γ resulted in more than 90% suppression of hTERT mRNA and protein levels. The suppression of hTERT was correlated with decreases in angiogenesis and tumor growth. In addition, analysis of xenograft tumor samples demonstrated decreases in expression of PCNA, MMP-9, VEGF, and bFGF, the major molecules involved in angiogenesis and tumor progression. PCNA is directly involved in DNA replication and cell multiplication, while MMP-9 paves the way for tumor cell proliferation and angiogenesis through degradation of the connective tissue matrix.
In glioblastoma, hTERT expression is a survival predictor and correlated with a poor survival rate (25). Thus, suppression of telomerase expression has enormous therapeutic potential in various cancers including glioblastomas, since this enzyme plays the key role in conferring cellular immortality (26). Inhibition of telomerase activity has been shown to impair both subcutaneous and intracranial tumor growth in glioblastoma xenografts (27). In the present study, we have observed about 50% decrease in angiogenesis as well as in tumor progression after knockdown of hTERT using cognate siRNA.
Treatment with IFN-γ has been found to down regulate hTERT expression and telomerase activity in human cervical cancer due to upregulation of p27Kip1 (28). In the present study, we confirmed similar down regulation of hTERT mRNA and protein levels after IFN-γ treatment. This indicated that IFN-γ could down regulate hTERT expression through upregulation of p27Kip1. Our data depicted potent effect of IFN-γ on inhibition of angiogenesis and tumor progression in glioblastoma, coinciding with the significant decreases in the protein levels of VEGF, bFGF and CD31. These findings prompted us to examine the dual role of hTERT knockdown and IFN-γ treatment as a combination therapy for effectively controlling growth of glioblastoma. Our studies demonstrated that combination of hTERT knockdown with IFN-γ treatment resulted in a marked reduction in tumor progression of glioblastoma xenografts.
Angiogenesis, the formation of new blood vessels (neovascularization) from pre-existing ones, is a significant event in cell invasion and tumor progression. Neovascularization is one of the major rate-limiting events in the invasion and progression of glioblastomas, since the presence of blood vessels not only sustains tumor growth but also facilitates penetration of tumor cells deep inside normal brain tissue. Development of appropriate molecular strategies to prevent neovascularization is an important milestone in the therapeutic intervention against glioblastomas. VEGF is the key angiogenic stimulant and the major driving force behind not only tumor angiogenesis but also normal blood vessel formation (29, 30). VEGF is highly upregulated in glioblastomas and is responsible for endothelial cell proliferation and vascular permeability in primary human brain tumors (31). Vascular endothelial cells present in astrocytic tumors express telomerase and hTERT and are involved in tumor angiogenesis (32). Inhibition of telomerase in the endothelial cells disrupts tumor angiogenesis in glioblastoma xenografts (33). In vitro capillary-like networking of endothelial cells requires special angiogenic factors such as VEGF released by the tumor cells in culture. In this study, we observed a marked inhibition of capillary-like network formation in vitro and a remarkable decrease of neovascularization under the dorsal of skin of nude mice after treatment with combination of hTERT siRNA and IFN-γ. This suggested that the cells treated with combination of hTERT siRNA and IFN-γ failed to secrete potent angiogenic factors such as VEGF and bFGF.
There has been no previous report on the suppression of intracranial or subcutaneous tumorigenesis of glioblastoma after treatment with combination of hTERT siRNA and IFN-γ. However, it has been observed that suppression of hTERT mRNA inhibits tumorigenicity and motility of HCT116 human colon cancer cells (34). Additionally, replication-defective adenovirus mediated transfer of IFN-γ gene has been shown to repress brain tumor growth by inhibiting angiogenesis (35). In the present study, we observed near complete growth inhibition of both intracranial and subcutaneous tumors in nude mice after treatment with combination of hTERT siRNA and IFN-γ. Our studies also demonstrated down regulation of the molecules involved in cell cycle and angiogenesis after treatment with combination of hTERT siRNA and IFN-γ.
In conclusion, the present study demonstrated that combination of hTERT knockdown and IFN-γ treatment effectively inhibited angiogenesis and tumor growth in glioblastomas through down regulation of molecules involved in these processes and could offer a potential therapeutic strategy for treatment of human glioblastomas.
Acknowledgments
Grant support: This work was supported in part by the R01 grants (CA-91460 and NS-57811) from the National Institutes of Health (Bethesda, MD).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Translational Relevance: The invasive and proliferative abilities of tumor cells are dependent on telomerase, which is tightly regulated by expression of human telomerase reverse transcriptase (hTERT). Telomerase is active in 90% human tumors including glioblastoma but inactive in most normal cells, making it an ideal target for cancer therapy. Since interferon-γ (IFN-γ) modulates more than 200 molecules both at transcriptional and translational level, down regulation of hTERT in glioblastoma in the presence of IFN-γ may reduce angiogenesis and tumor growth. In our study, we down regulated hTERT in two human glioblastoma cell lines using plasmid vector based siRNA during IFN-γ treatment and found significant inhibition of angiogenesis both in vitro and in vivo. There was marked inhibition of orthotopic tumorigenesis and solid tumor formation in the subcutaneous region of immunocompromised mice after the treatment with combination of hTERT siRNA and IFN-γ. Therefore, down regulation of hTERT during IFN-γ treatment is a promising therapeutic strategy to control the malignant growth of human glioblastoma.
References
- 1.Pulkkanen KJ, Yla-Herttuala S. Gene therapy for malignant glioma: current clinical status. Mol Ther. 2005;12:585–98. doi: 10.1016/j.ymthe.2005.07.357. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson SS, Laningham F, Fisher PG. Advances toward an understanding of brainstem gliomas. J Clin Oncol. 2006;24:1266–72. doi: 10.1200/JCO.2005.04.6599. [DOI] [PubMed] [Google Scholar]
- 3.Combs SE, Widmer V, Thilmann C, Hof H, Debus J, Schulz-Ertner D. Stereotactic radiosurgery (SRS): treatment option for recurrent glioblastoma multiforme (GBM) Cancer. 2005;104:2168–73. doi: 10.1002/cncr.21429. [DOI] [PubMed] [Google Scholar]
- 4.George J, Gondi CS, Dinh DH, Gujrati M, Rao JS. Restoration of tissue factor pathway inhibitor-2 in a human glioblastoma cell line triggers caspase-mediated pathway and apoptosis. Clin Cancer Res. 2007;13:3507–17. doi: 10.1158/1078-0432.CCR-06-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George J, Banik NL, Ray SK. Combination of taxol and Bcl-2 siRNA induces apoptosis in human glioblastoma cells and inhibits invasion, angiogenesis, and tumour growth. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2008.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 7.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 8.Cairns RA, Khokha R, Hill RP. Molecular mechanisms of tumor invasion and metastasis: an integrated view. Curr Mol Med. 2003;3:659–71. doi: 10.2174/1566524033479447. [DOI] [PubMed] [Google Scholar]
- 9.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–27. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–3. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 11.Carpentier C, Lejeune J, Gros F, Everhard S, Marie Y, Kaloshi G, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Sanson M. Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol. 2007;84:249–53. doi: 10.1007/s11060-007-9378-3. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara T. Telomerase-specific virotherapy for human squamous cell carcinoma. Expert Opin Biol Ther. 2009;9:321–9. doi: 10.1517/14712590802715731. [DOI] [PubMed] [Google Scholar]
- 13.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 14.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 15.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 16.Blanck G. Components of the IFN-γ signaling pathway in tumorigenesis. Arch Immunol Ther Exp (Warsz) 2002;50:151–58. [PubMed] [Google Scholar]
- 17.George J, Banik NL, Ray SK. Bcl-2 siRNA augments taxol mediated apoptotic death in human glioblastoma U138MG and U251MG cells. Neurochem Res. 2009;34:66–78. doi: 10.1007/s11064-008-9659-z. [DOI] [PubMed] [Google Scholar]
- 18.George J, Tsutsumi M. siRNA-mediated knockdown of connective tissue growth factor prevents N-nitrosodimethylamine-induced hepatic fibrosis in rats. Gene Ther. 2007;14:790–803. doi: 10.1038/sj.gt.3302929. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 20.Amyot F, Camphausen K, Siavosh A, Sackett D, Gandjbakhche A. Quantitative method to study the network formation of endothelial cells in response to tumor angiogenic factors. IEE Proc Syst Biol. 2005;152:61–6. doi: 10.1049/ip-syb:20045036. [DOI] [PubMed] [Google Scholar]
- 21.Sckell A, Leunig M. Dorsal skinfold chamber preparation in mice. In: Murray JC, editor. Angiogenesis Protocols: Methods in Molecular Medicine. Vol. 46. Humana Press; Totowa, New Jersey: 2001. pp. 95–105. [DOI] [PubMed] [Google Scholar]
- 22.Wachsberger PR, Burd R, Marero N, Daskalakis C, Ryan A, McCue P, Dicker AP. Effect of the tumor vascular-damaging agent, ZD6126, on the radioresponse of U87 glioblastoma. Clin Cancer Res. 2005;11:835–42. [PubMed] [Google Scholar]
- 23.Stern JL, Bryan TM. Telomerase recruitment to telomeres. Cytogenet Genome Res. 2008;122:243–54. doi: 10.1159/000167810. [DOI] [PubMed] [Google Scholar]
- 24.Osterhage JL, Friedman KL. Chromosome end maintenance by telomerase. J Biol Chem. 2009 Mar 12; doi: 10.1074/jbc.R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Wei Q, Wang LE, Aldape KD, Cao Y, Okcu MF, Hess KR, El-Zein R, Gilbert MR, Woo SY, Prabhu SS, Fuller GN, Bondy ML. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24:1627–32. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- 26.Belgiovine C, Chiodi I, Mondello C. Telomerase: cellular immortalization and neoplastic transformation. Multiple functions of a multifaceted complex. Cytogenet Genome Res. 2008;122:255–62. doi: 10.1159/000167811. [DOI] [PubMed] [Google Scholar]
- 27.Falchetti ML, Fiorenzo P, Mongiardi MP, Petrucci G, Montano N, Maira G, Pierconti F, Larocca LM, Levi A, Pallini R. Telomerase inhibition impairs tumor growth in glioblastoma xenografts. Neurol Res. 2006;28:532–37. doi: 10.1179/016164106X116818. [DOI] [PubMed] [Google Scholar]
- 28.Lee SH, Kim JW, Oh SH, Kim YJ, Rho SB, Park K, Park KL, Lee JH. IFN-γ/IRF-1-induced p27Kip1 down regulates telomerase activity and human telomerase reverse transcriptase expression in human cervical cancer. FEBS Lett. 2005;579:1027–33. doi: 10.1016/j.febslet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Plank MJ, Sleeman BD. Tumour-induced angiogenesis: a review. J Theor Med. 2003;5:137–53. [Google Scholar]
- 30.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–8. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 31.Machein MR, Kullmer J, Fiebich BL, Plate KH, Warnke PC. Vascular endothelial growth factor expression, vascular volume, and, capillary permeability in human brain tumors. Neurosurgery. 1999;44:732–40. doi: 10.1097/00006123-199904000-00022. [DOI] [PubMed] [Google Scholar]
- 32.Pallini R, Pierconti F, Falchetti ML, D'Arcangelo D, Fernandez E, Maira G, D'Ambrosio E, Larocca LM. Evidence for telomerase involvement in the angiogenesis of astrocytic tumors: expression of human telomerase reverse transcriptase messenger RNA by vascular endothelial cells. J Neurosurg. 2001;94:961–71. doi: 10.3171/jns.2001.94.6.0961. [DOI] [PubMed] [Google Scholar]
- 33.Falchetti ML, Mongiardi MP, Fiorenzo P, Petrucci G, Pierconti F, D'Agnano I, D'Alessandris G, Alessandri G, Gelati M, Ricci-Vitiani L, Maira G, Larocca LM, Levi A, Pallini R. Inhibition of telomerase in the endothelial cells disrupts tumor angiogenesis in glioblastoma xenografts. Int J Cancer. 2008;122:1236–42. doi: 10.1002/ijc.23193. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Zhang YW, Zhang ZX, Miao ZH, Ding J. hTERT-targeted RNA interference inhibits tumorigenicity and motility of HCT116 cells. Cancer Biol Ther. 2008;7:228–236. doi: 10.4161/cbt.7.2.5259. [DOI] [PubMed] [Google Scholar]
- 35.Fathallah-Shaykh HM, Zhao LJ, Kafrouni AI, Smith GM, Forman J. Gene transfer of IFN-gamma into established brain tumors represses growth by antiangiogenesis. J Immunol. 2000;164:217–22. doi: 10.4049/jimmunol.164.1.217. [DOI] [PubMed] [Google Scholar]