Abstract
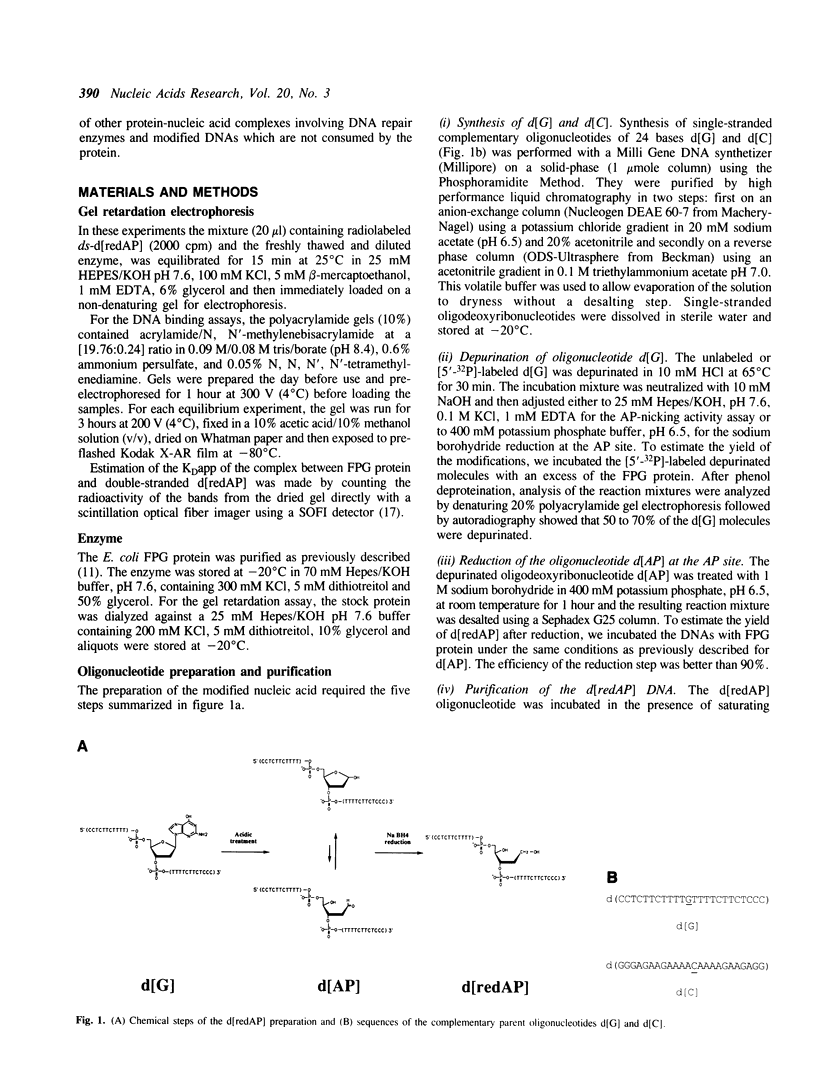
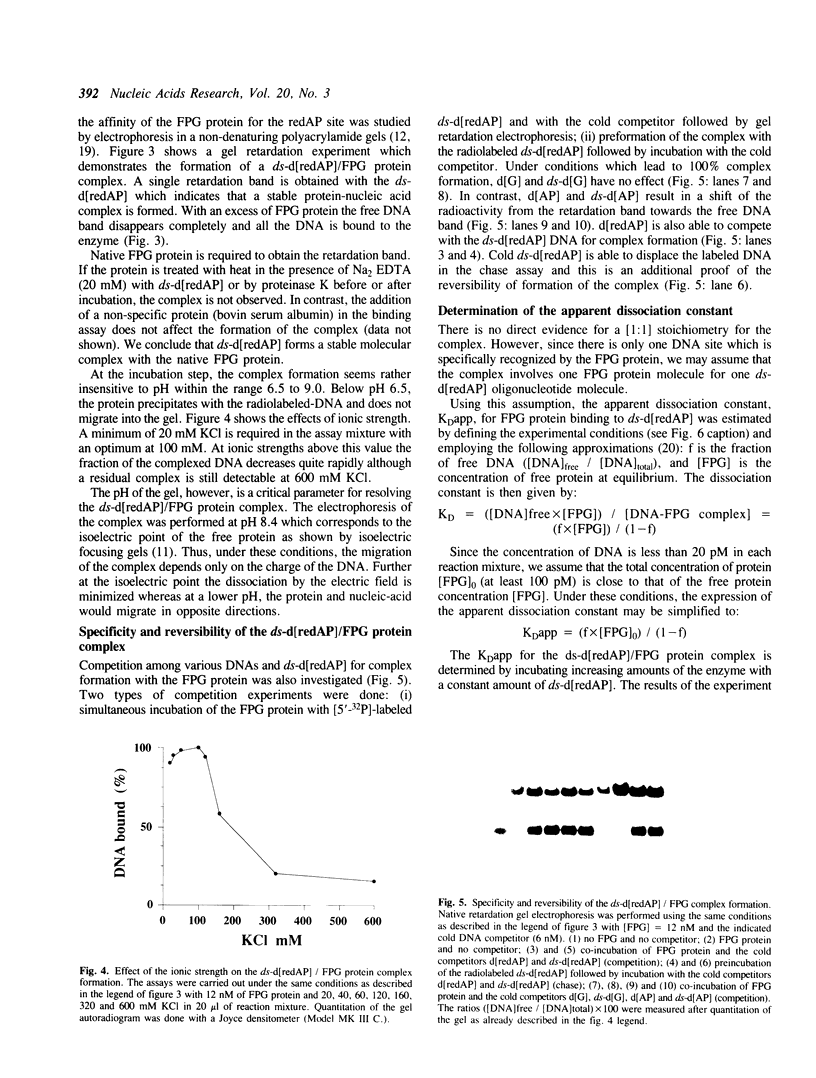
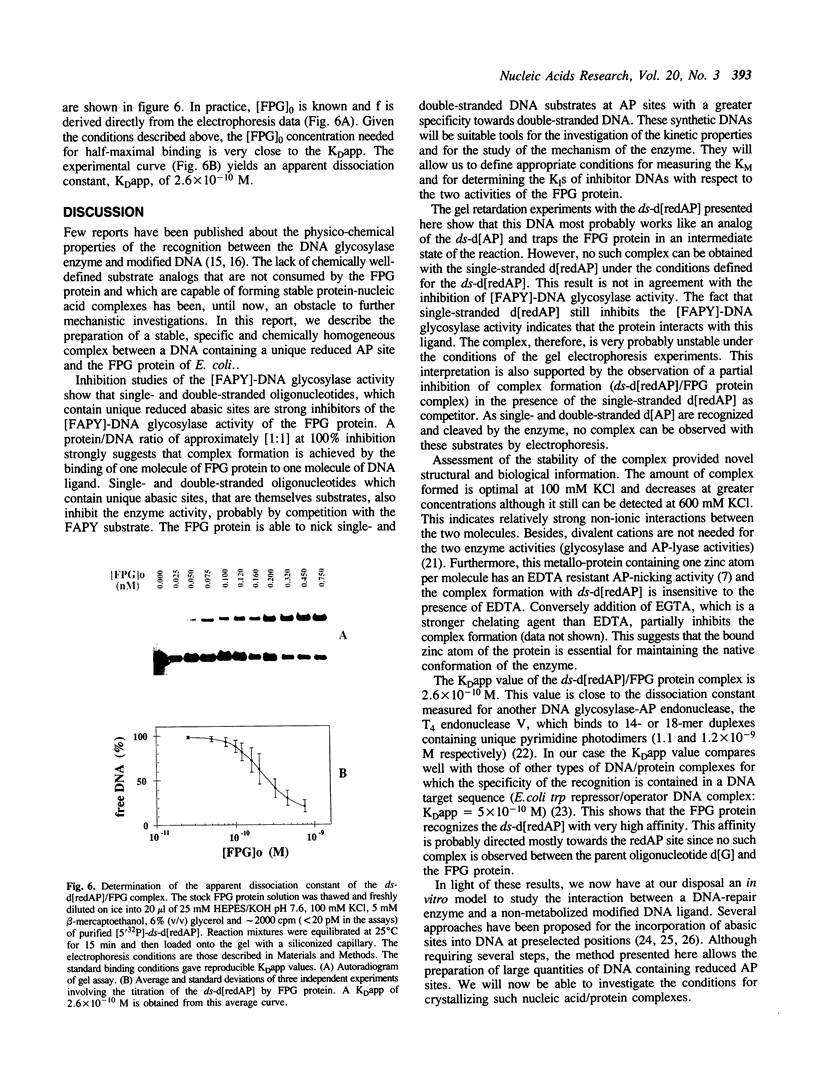
The E. coli Formamidopyrimidine-DNA Glycosylase (FPG protein), a monomeric DNA repair enzyme of 30.2 kDa, was purified to homogeneity in large quantities. The FPG protein excises imidazole ring-opened purines and 8-hydroxyguanine residues from DNA. Besides DNA glycosylase activity, the FPG protein is endowed with an EDTA-resistant activity which nicks DNA at apurinic/apyrimidic sites (AP sites). In contrast, DNAs containing chemically reduced AP sites are not incised by the FPG protein. However, the DNA glycosylase activity of the FPG protein is strongly inhibited in the presence of a purified synthetic 24 base-pair double-stranded oligonucleotide which contains a single apurinic site transformed chemically through borohydride reduction into a ring-opened deoxyribose derivative. The ability of the FPG protein to form a complex with this synthetically modified DNA was studied by electrophoresis in non-denaturing polyacrylamide gels. The FPG protein specifically binds the double-stranded oligonucleotide containing an apurinic site previously reduced in the presence of sodium borohydride. The complex was identified as a single retardation band on non-denaturing polyacrylamide gel electrophoresis. Complex formation is reversible and an apparent dissociation constant, KDapp, of 2.6 x 10(-10) M was determined. In contrast, no such retardation band was obtained between the FPG protein and double-stranded DNA containing an intact apurinic site or single-stranded DNA containing either an intact or a reduced apurinic site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Verly W. G., O'Connor T., Laval J. Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine]DNA glycosylase. Biochem J. 1989 Sep 1;262(2):581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Belleney J., Roques B. P., Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984 Jul 11;12(13):5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Bichara M., Fuchs R. P., Laval J. Excision of the imidazole ring-opened form of N-2-aminofluorene-C(8)-guanine adduct in poly(dG-dC) by Escherichia coli formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1989 Oct;10(10):1905–1909. doi: 10.1093/carcin/10.10.1905. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987 Oct;6(10):3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Lederer F., Gouyette A., Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990 Mar 5;265(7):3916–3922. [PubMed] [Google Scholar]
- Breimer L. H. Enzymatic excision from gamma-irradiated polydeoxyribonucleotides of adenine residues whose imidazole rings have been ruptured. Nucleic Acids Res. 1984 Aug 24;12(16):6359–6367. doi: 10.1093/nar/12.16.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci U S A. 1988 Feb;85(4):975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeczot H., Tudek B., Lambert B., Laval J., Boiteux S. Escherichia coli Fpg protein and UvrABC endonuclease repair DNA damage induced by methylene blue plus visible light in vivo and in vitro. J Bacteriol. 1991 Jun;173(11):3419–3424. doi: 10.1128/jb.173.11.3419-3424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M. L., Lloyd R. S. Structure-function studies of the T4 endonuclease V repair enzyme. Mutat Res. 1989 Sep;218(2):49–65. doi: 10.1016/0921-8777(89)90011-6. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 1983 Jan 11;11(1):141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoka T., Ishida M., Ohtsuka E. Affinity of single- or double-stranded oligodeoxyribonucleotides containing a thymine photodimer for T4 endonuclease V. J Biol Chem. 1989 Feb 15;264(5):2609–2614. [PubMed] [Google Scholar]
- Joachimiak A., Kelley R. L., Gunsalus R. P., Yanofsky C., Sigler P. B. Purification and characterization of trp aporepressor. Proc Natl Acad Sci U S A. 1983 Feb;80(3):668–672. doi: 10.1073/pnas.80.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J., Boiteux S., O'Connor T. R. Physiological properties and repair of apurinic/apyrimidinic sites and imidazole ring-opened guanines in DNA. Mutat Res. 1990 Nov-Dec;233(1-2):73–79. doi: 10.1016/0027-5107(90)90152-t. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revzin A. Gel electrophoresis assays for DNA-protein interactions. Biotechniques. 1989 Apr;7(4):346–355. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Urata H., Yamamoto K., Akagi M., Hiroaki H., Uesugi S. A 2-deoxyribonolactone-containing nucleotide: isolation and characterization of the alkali-sensitive photoproduct of the trideoxyribonucleotide d(ApCpA). Biochemistry. 1989 Dec 12;28(25):9566–9569. doi: 10.1021/bi00451a002. [DOI] [PubMed] [Google Scholar]






