Abstract
Nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) are large enzymes responsible for the biosynthesis of medically and ecologically important secondary metabolites. In a previous report we described a proteomics approach to screen for expressed NRPSs or PKSs from bacteria with or without sequenced genomes. Here we use this proteome mining approach to discover a novel natural product arising from rare adenylation (A) and reductase (Red) domains in its biosynthetic machinery. We also clone the entire gene cluster and elucidate the biosynthesis of the new compound produced by an unsequenced Bacillus sp. isolated from soil collected in Koran, Louisiana.
Keywords: Nonribosomal peptide synthetase, natural products, proteomics, Fourier-transform mass spectrometry
The potent bioactivities of peptides produced by non-ribosomal biosynthesis range from antibiotics such as penicillins and vancomycin to anticancer drugs such as bleomycin1. Members of this chemical family include immunosuppressants like cyclosporin2, pathogenicity determinants such as yersiniabactin3 and aureusimine4, and powerful mycotoxins like HC-toxin5. With the deluge of microbial genome sequences becoming available, it is apparent that secondary metabolite gene clusters are widely dispersed, largely uncharacterized, and often encode NRPSs or PKSs6. Despite some successes7, 8 harnessing the genetic potential seen in microbial genomes, forced expression of the 30–80 kilobases of an entire gene cluster has not evolved into a generally robust method for exploiting their rich biosynthetic capacity. The proteomic approach described below instead seeks to screen native hosts in a large number of culture conditions. Recently, proteomic approaches to study secondary metabolism have proven useful for the detection and characterization of thiotemplate biosynthetic pathways9–10. In this study, 23 unsequenced environmental isolates were cultured in nine different conditions and were sampled at three time points before screening by simple SDS-PAGE, as described previously10. Twenty of the 23 strains showed some evidence of protein bands >150 kDa which tend to be large NRPS or PKS enzymes, often >1300 amino acids in length.
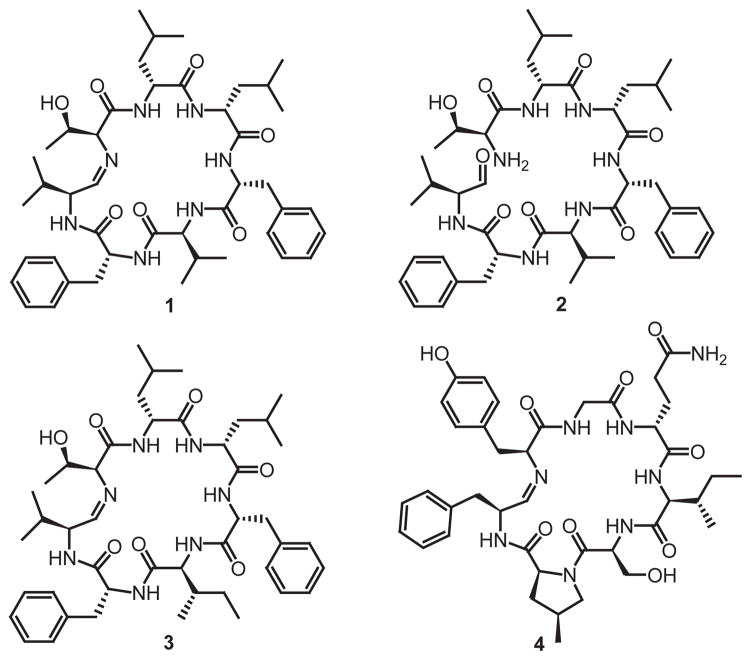
Mass spectrometric analysis of four high molecular weight protein bands from one environmental Bacillus isolate is shown in Figure S1a. Direct peptide sequencing by tandem mass spectrometry (e.g., the spectrum of Figure S1b) gave the amino acid subsequences that guided design of PCR primers for amplification of DNA corresponding to the region between identified peptides and conserved core regions of NRPS adenylation domains (Figure S1c). A single amplicon was our hook into the unknown gene cluster. Based upon the low sequence identity of this amplicon’s translated amino acid sequence (42%) to the closest sequence in GenBank, it was possible that we had uncovered a yet-unsequenced biosynthetic pathway. Eventual DNA sequencing of this >30 kilobase gene cluster and prediction of its functional elements led us quickly to predict and then detect a new peptide natural product named koranimine (1, m/z 804.502; Figure 1).
Figure 1.
Koranimine (1), is the major product. Compound 2, is the direct result of reductive release from the assembly line, which forms the imine structure (1). Compound 3 is a minor product that has isoleucine substituted for valine at the fifth amino acid in the peptide. Compound 4 is nostocyclopeptide A1.
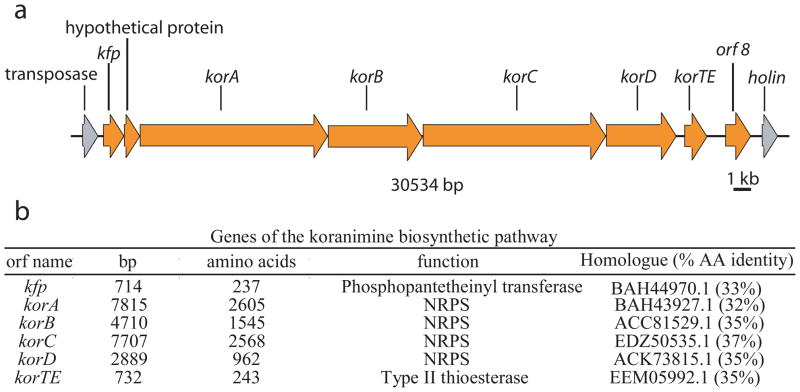
When the entire 30 kb cluster (Figure 2) was cloned from our environmental strain (NK2003) and heterologously expressed in Bacillus subtilis 168, a time-dependent increase of 1 was observed at 25 minutes in the total ion chromatogram of a LC-FTMS run (Figure S2; m/z 804.502). Additionally, the species at m/z 804.502 was not present in the culture supernatant of the control strain (B. subtilis 168, Figure S2). These data proved the association between the koranimine biosynthetic gene cluster (30 kb) and its small molecule product. The gene cluster of 1 is not highly related to any other NRPS clusters present in GenBank at this time, a noteworthy result after more than a decade of shotgun sequencing of bacterial genomes.
Figure 2.
(a) ORF map for the koranimine gene cluster. (b) Annotation of the koranimine genes.
Initial tandem mass spectrometric analysis of 1 identified the six amino acid sequence tag Thr-Leu/Ile-Leu/Ile-Phe-Val-Phe (Figure S3). Stable isotope incorporation studies revealed the presence of one threonine residue, two leucine residues, two phenylalanine residues and two valine residues in the koranimine molecule (Figure S4). Only minor levels of mis-incorporation of Ile instead of Leu were observed (Figure S4c). However, the feeding results using 13C6-labeled isoleucine verified the production of a related compound that incorporates Ile instead of Val at the fifth amino acid position (Figure 1, 3). Analysis of D10-leucine and D8-15N1-phenylalanine labeled koranimine revealed complete abstraction of the α-deuterium of both amino acids, indicating that the domains predicted to have epimerization function were in fact converting all instances of L-Leu and L-Phe to their D-Leu and D-Phe stereoisomers, respectively (Figure S5).
Figure 3.
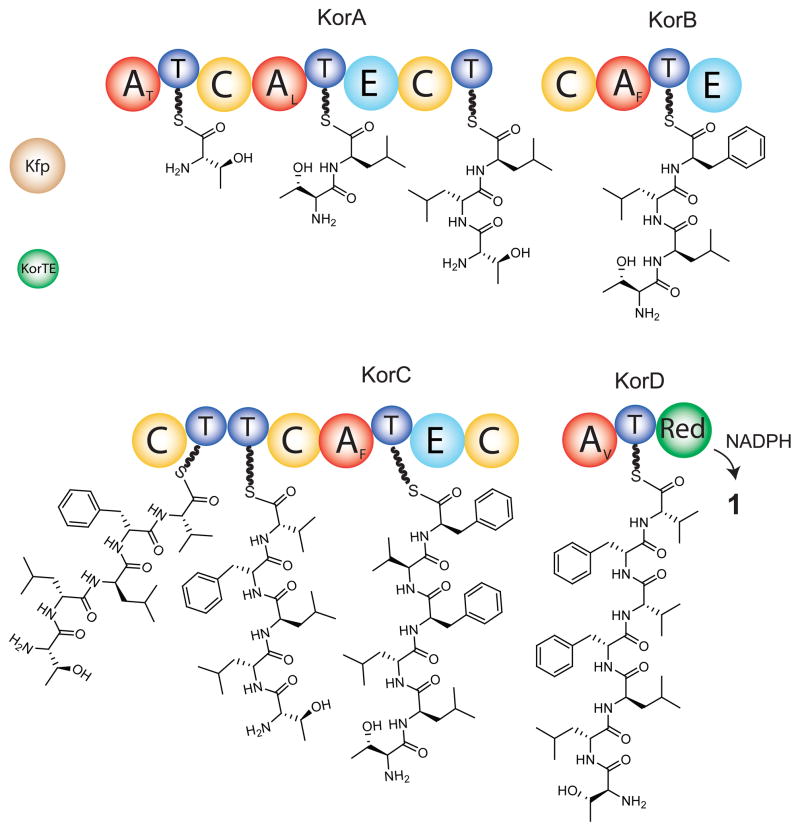
Domain structure of synthetases and biosynthetic scheme for production of koranimine; A-adenylation, T-thiolation, C-condensation, E-epimerization, Red-reductase.
Isolation of 1.1 mg of 1 from 20 L of culture followed by structural interpretation using 2D NMR techniques confirmed the presence of Val, Phe, Leu, and some minor incorporation of Ile vs. Val (Figure S6), all consistent with the mass spectrometric data in Figures S3, S4, and S5. Based on limited material, indirect detect gHMBC experiments were used to detect signatures for Phe, Leu, and Val (Figure S6). A main structural aspect of koranimine exhibiting apparent chemical variation by both MS and NMR is the terminal imine functionality, which arises from decidedly non-standard NRPS termination chemistry (vide infra). By MS, signals consistent with both the cyclic imine (1) and open aldehyde form (2, +18.01 Da) were observed (Figure 1, 1 and 2). The NMR data did not show the expected imine 1H at ~6.8 ppm or the imine carbon at ~167 ppm (Figure S6). Subjecting the purified product to ICP-MS showed no trace of any bound mono-, di-, or tri-valent metal. However, reduction by NaCNBH3 showed a clear increase of 2.012 Da (Figure S7), leading us to assign confidently the imine structure for compound 1.
The biosynthetic mechanism for koranimine production is shown in Figure. 3, with several unusual aspects of the domains and their overall organization. Only after the structure of 1 was determined were we able to make a confident proposal for the entire biosynthetic pathway. The first peculiarity is that the natural product was determined to be a heptapeptide, but the gene cluster contains only five adenylation domains. Prediction of substrates for adenylation domains is routine in the NRPS field11. This exercise accurately predicted only the first, second and seventh amino acids to be threonine, leucine and valine installed by adenylation domains KorA-A1, KorA-A2 and KorD, respectively. The adenylation domains of KorB and KorC are both only 42% identical at the amino acid level to their nearest homologues (gb: ACC81021.1, and gb: AAO23333.1, respectively), and standard algorithms produced no substrate predictions, indicating that these enzymes contain highly unusual adenylation domain signatures. The adenylation domains in KorB and KorC were ultimately implicated in the incorporation of Phe by metabolic labeling (Figure S4d).
Another peculiarity in the koranimine system is KorA, where a condensation-thiolation (C-T) di-domain lacks a directly linked adenylation or epimerization (or E) domain. Deductions from the koranimine structure indicate that KorA incorporates and epimerizes the second and third amino acids (both leucine). Therefore, a single adenylation domain likely services two carrier sites. While an adenylation domain acting in this way is unconventional, it is not without precedent12. This particular example opens up two mechanistic possibilities, both unprecedented. In path a of Figure S9, the adenylation domain services both carrier sites (either in cis or in trans), forcing the single epimerization domain to act at both sites. Alternatively, in path b of Figure S8 the D-Leu-acyl-S-KorA-T2-enzyme intermediate would form followed by an unprecedented shuttling of this intermediate by the condensation domain to move the D-Leu-intermediate to KorA-T3, opening up KorA-T2 for a second round of adenylation and epimerization. Then, KorA-C2 would switch roles to catalyze the standard type of condensation and form the amide bond between two D-Leu residues.
In KorC, there is yet another unusual domain structure containing a T-T-C tri-domain, also with no directly linked adenylation domain. Biochemical experiments showed that the tandem T-domains of KorC are both charged by the adenylation domain of KorD (Figure S9). This long-range transactivation, similar to long-range transactivation of HMWP1 by HMWP2 in the yersiniabactin pathway12, may be a rate limiting step and could explain the presence of the tandem T-domains in the fourth module. Tandem T-domains have previously been implicated in increasing flux through thiotemplate systems13, 14.
We propose KorTE has an editing function like other type II thioesterases, namely removing mis-acylated carrier sites from the pathway and increasing the efficiency of the pathway15, 16. KorTE did decrease the occupancy of acyl-S-KorC (Figure S9g). We envisioned that the long range transaminoacylation of KorC by KorD might be facilitated by the action of a shuttling function of the orf 8 enzyme, predicted to be an α/β hydrolase family enzyme (in a similar fashion as SyrC of the syringomycin pathway17). In vitro experiments showed that the orf 8 enzyme does have hydrolytic activity toward acyl-S-enzyme intermediates (Figure S10f). However, it did not stimulate substrate transfer from KorD to KorC in vitro (Figure S9f).
The sixth and final NRPS module, in addition to charging the fourth module and incorporating the seventh residue, valine, performs a two-electron, NADPH-dependent reduction of the C-terminal acyl-S-thioester acid to the aldehyde. This terminal aldehyde functionality has been previously shown to be involved in non-enzymatic macrocyclization18 yielding the final imine product (1). The only precedent for this is the nostocyclopeptide system19 (nostocyclopeptide A1, 4), where the first reductive chain release was shown. However, the nostocyclopeptides have completely different amino acid sequences (Figure 1). The imine functionality for nostocyclopeptide has been shown to be reversible in a NMR tube18 and evidence points to a spontaneous formation upon reductive release of an aldehyde from the NRPS assembly-line. The functional impact of having a dissimilar sequence to nostocyclopeptide 18, 20 is unknown at this time, though a functional study in 2010 demonstrated that a nostocyclopeptide antagonizes the toxic effect of microcystin on hepatocytes21 and blocks hepatocyte drug trransporters22. Koranimine was not found to inhibit the growth of Escherichia coli, Staphylococcus aureus, Enterococcus faecalis, Klebsiella pneumonia or Saccharomyces cerevisiae. Further bioassessment of 1 is underway to probe the limited structural discrepancies between 1 and 4.
In sum, the structure of a new natural product was determined using multistage tandem mass spectrometry (MSn), feeding studies with stable isotope tracers, nuclear magnetic resonance (NMR), and in vitro enzyme reconstitution. The original discovery of the compound began with a new “proteome-first” strategy that scans microbial proteomes for expressed gene clusters, even those not yet uncovered by genome sequencing efforts such as koranimine. Further, the approach can be used to screen conditions where typically cryptic gene clusters can be teased into expression. Future efforts for expression screening at the protein level will become more efficient and sensitive to serve as a high value sieve for large numbers of environmental isolates in the discovery of novel natural products and their biosynthetic pathways. This discovery platform is structure-based, does not rely on bioassays, and can even be applied to exploration of cross- and mixed-systems of microbial ecology (i.e., “metaproteomics”).
Supplementary Material
Acknowledgments
We thank William Metcalf, Huimin Zhao, Wilfred van der Donk, Satish Nair, Doug Mitchell and Karl Scheidt for comments. We also thank the Roy J. Carver Charitable Trust, the University of Illinois and the National Institutes of Health (GM R01 067725 and GM P01 077596) which have supported the development of PrISM.
Footnotes
Supporting Information Available: Supplemental figures, methods for molecular biological techniques, compound isolation and mass spectrometry as well as supplemental references are available; this material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Du LC, Sanchez C, Chen M, Edwards DJ, Shen B. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 2.Weber G, Schorgendorfer K, Schneiderscherzer E, Leitner E. Curr Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- 3.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyatt MA, Wang W, Roux CM, Beasley FC, Heinrichs DE, Dunman PM, Magarvey NA. Science. 2010;329:294–296. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- 5.Walton JD. Phytochemistry. 2006;67:1406–1413. doi: 10.1016/j.phytochem.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 6.Donadio S, Monciardini P, Sosio M. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 7.Knappe TA, Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 8.Lautru S, Deeth RJ, Bailey LM, Challis GL. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 9.Meier JL, Niessen S, Hoover HS, Foley TL, Cravatt BF, Burkart MD. ACS Chem Biol. 2009;11:948–957. doi: 10.1021/cb9002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bumpus SB, Evans BS, Thomas PM, Ntai I, Kelleher NL. Nat Biotechnol. 2009;27:951–956. doi: 10.1038/nbt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suo Z, Tseng CC, Walsh CT. Proc Natl Acad Sci U S A. 2001;98:99–104. doi: 10.1073/pnas.021537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang H, Zirkle R, Metz JG, Braun L, Richter L, Van Lanen SG, Shen B. J Am Chem Soc. 2008;130:6336–6337. doi: 10.1021/ja801911t. [DOI] [PubMed] [Google Scholar]
- 14.Rahman AS, Hothersall J, Crosby J, Simpson TJ, Thomas CM. J Biol Chem. 2005;280:6399–6408. doi: 10.1074/jbc.M409814200. [DOI] [PubMed] [Google Scholar]
- 15.Kim BS, Cropp TA, Beck BJ, Sherman DH, Reynolds KA. J Biol Chem. 2002;277:48028–48034. doi: 10.1074/jbc.M207770200. [DOI] [PubMed] [Google Scholar]
- 16.Scwarzer D, Mootz HD, Linne U, Marahiel MA. Proc Natl Acad Sci U S A. 2002;99:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh GM, Vaillancourt FH, Yin J, Walsh CT. Chem Biol. 2007;14:31–40. doi: 10.1016/j.chembiol.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Enck S, Kopp F, Marahiel MA, Geyer A. Chembiochem. 2008;9:2597–2601. doi: 10.1002/cbic.200800314. [DOI] [PubMed] [Google Scholar]
- 19.Becker JE, Moore RE, Moore BS. Gene. 2004;325:35–42. doi: 10.1016/j.gene.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 20.Kopp F, Mahlert C, Grunewald J, Marahiel MA. J Am Chem Soc. 2006;128:16478–16479. doi: 10.1021/ja0667458. [DOI] [PubMed] [Google Scholar]
- 21.Jokela J, Herfindal L, Wahlsten M, Permi P, Selheim F, Vasconcelos V, Doskeland SO, Sivonen K. Chembiochem. 2010;11:1594–1599. doi: 10.1002/cbic.201000179. [DOI] [PubMed] [Google Scholar]
- 22.Herfindal L, Myhren L, Kleppe R, Krakstad C, Selheim F, Jokela J, Sivonen K, Doskeland SO. Mol Pharm. doi: 10.1021/mp1002224. dx.doi.org/10.1021/mp1002224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.