Abstract
Tnfrsf9 gene expression has previously been characterized in the late pregnant mouse uterus and placenta. However, little is known about its expression in the uterus during the implantation phase of early pregnancy. Since TNFRSF9 is known to play a potentially important role in immune function in general, the purpose of this study was to assess the levels and localization of Tnfrsf9 expression in the mouse uterus and conceptus during implantation. Relative Tnfrsf9 mRNA levels were significantly increased in implantation compared to non-implantation site tissue on Days 6.5- 8.5 of pregnancy. This increase did not depend on the presence of the conceptus, as mRNA levels were not significantly different between pregnant implantation sites and artificially-induced deciduomas. Localization using in situ hybridization revealed that a subpopulation of endothelial as well as uterine natural killer cells expressed Tnfrsf9 in the endometrium during implantation. In the developing conceptus, primary trophoblast giant and ectoplacental cells expressed Tnfrsf9 on Days 6.5-8.5. This was followed by expression in the trophoblast giant cell layers surrounding the conceptus on Day 9.5 of pregnancy. Since 2 main splice forms of Tnfrsf9 mRNA exist and encode proteins which have distinct biological functions, we also determined their presence in these tissues. Both mRNA splice forms were present in uterine and conceptus tissues as determined by RT-PCR. In conclusion, both membrane and soluble forms of Tnfrsf9 are expressed in specific cell types of the uterus and conceptus during the progression of implantation in mice and thus may play an important role in this process.
Introduction
Mammalian implantation is the process by which the conceptus comes into intimate contact with the maternal blood supply. This process is necessary for the mother to adequately provide nutrients for fetal development and successful completion of pregnancy. Implantation in mammals is initiated with the attachment of the conceptus to the uterine wall and is complete with the formation of the functional placenta. One of the first maternal changes during implantation in rodents and humans is a dramatic transformation of the endometrial tissue into decidual tissue (Abrahamsohn and Zorn, 1993, Gellersen, et al., 2007, Mori, et al., 1991). The hallmark of this change is the differentiation of endometrial fibroblasts into decidual cells, commonly referred to as decidualization or decidual cell differentiation. However, other changes during the development of the decidual tissue include alterations of immune cell populations, extracellular matrix composition and vascular structure. On the side of the conceptus, it is the hatched blastocyst composed of trophectoderm cells surrounding the inner cell mass and blastocoel that comes into initial contact with the endometrium at the initiation of implantation (Cross, et al., 1994). After the onset of implantation the trophectoderm cells give rise to trophoblast giant cells, ectoplacental cone cells and spongiotrophoblast cells all of which contribute to the developing placenta (Cross, 2000).
The tumor necrosis factor receptor superfamily member 9 (Tnfrsf9) gene (4-1BB, Cd137, ILA, Ly63) encodes a 30 kDa membrane glycoprotein (TNFRSF9) which belongs to the tumor necrosis factor receptor (TNFR) family. Although originally thought to be expressed only in activated T cells (Pollok, et al., 1993), it is now known that many immune cell types can express Tnfrsf9 including: activated NK (Marvel and Walzer, 2010), NKT (Kim, et al., 2008), B (Zhang, et al., 2010), eosinophil (Heinisch, et al., 2001), mast (Nishimoto, et al., 2005), neutrophil (Heinisch, et al., 2000), mature Treg (Williams and Rudensky, 2007), activated monocyte (Sollner, et al., 2007) and dendritic cells (Wilcox, et al., 2002). TNFRSF9 plays roles in many aspects of innate and adaptive immunity, including cancer immunology and autoimmune disease about which the reader is referred to several recent reviews for further information (Chen, 2006, Cheuk, et al., 2004, Croft, 2003, Myers and Vella, 2005, Nam, et al., 2005, Vinay, et al., 2006). It is notable that Tnfrsf9 expression may also occur in non-immune cell types such as endothelial (Drenkard, et al., 2007), neuron, astrocyte and microglia (Reali, et al., 2003) cells. However, to the best of our knowledge, the function of this expression is presently not known.
Immunological aspects of embryo implantation have been studied in detail, especially since Sir Peter Medawar originated several exciting hypotheses almost 60 years ago of why the maternal immune system does not reject the semi-allogenic conceptus during implantation and pregnancy (Medawar, 1953). However, since that time all the work has revealed that these hypotheses and others generated in the interim are not sufficient and thus more work is necessary (Robertson, 2010, Yoshinaga, 2008). In that regard, although TNFRSF9 seems to play many roles in the immune system, very little is known about Tnfrsf9 expression at implantation sites in the uteri of pregnant mammals. Recently, Tnfrsf9 expression in the late mouse uterus and placenta was partially characterized (Zhao, et al., 2007). However, to the best of our knowledge, little is known about the expression of this gene in the uterus or conceptus during the implantation phase of early pregnancy. Since TNFRSF9 is known to play a potentially important role in immunity in general, the purpose of this study was to determine if and where Tnfrsf9 is expressed in the mouse uterus and conceptus during implantation in mice.
Materials and Methods
Animals
CD1 mice purchased from Charles River (Charles River, Wilmington, MA) and Il15 knockout mice (Germantown, NY) were used in this study. The Il15 knockout mice in a C57/bl6 background were backcrossed to mice of the CD1 strain for over 15 generations and the generations thereafter were used in the present study.
CD1 female mice (8-12 weeks old) were mated to CD1 male mice to obtain pregnancies. In some cases, Il15-knockout and wild-type female mice were mated to wild-type and Il15-knockout males, respectively. This generated pregnant mice with conceptuses of all the same (heterozygous) genotype regardless of the maternal genotype. For all mice, the morning a vaginal plug was discovered was taken as Day 0.5 of pregnancy. Mice were killed on day 3.5 (just prior to the onset of implantation) or days 4.5-9.5 after the onset of implantation. Uterine tissues were collected from nonimplantation (NIS) and implantation segments (IS) of the uterus as previously described (Bany and Schultz, 2001). For some experiments we used a bead-induced decidualization model as described elsewhere (Herington and Bany, 2007, Herington, et al., 2009). Briefly, female mice were mated with vasectomized male mice and the morning a vaginal plug was found was considered Day 0.5 of pseudopregnancy. Small beads were then transferred into the uteri of Day 2.5 pseudopregnant mice which results in the formation of deciduoma containing segments of the uterus. In a similar fashion to uteri of pregnant mice, non-stimulated (NS) and bead-induced deciduoma (BID) segments of the uteri were collected from Days 3.5 to 9.5 of pseudopregnancy.
Tissue Collection and Processing
For total RNA isolation, tissues were collected and homogenized in Trizol Reagent (InVitrogen, Carlsbad, CA ). For IS tissues, conceptuses were separated using fine forceps prior to homogenization. For in situ hybridization, the tissues were fixed overnight in 4% paraformaldehyde then dehydrated, cleared in xylene and blocked in paraffin using standard techniques. Paraffin tissue sections (5 μm) were prepared and mounted onto Superfrost Plus slides (VWR, Radnar, PA).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RNA isolation followed by quantitative RT-PCR (qRT-PCR) was used to measure the relative steady-state levels of Tnfrsf9 mRNA normalized to the housekeeping target 18 S rRNA. The approach and precise method used for qRT-PCR has been described in detail previously (Herington, Underwood, McConaha and Bany, 2009). The 18S rRNA oligonucleotide primers (5’- CGGCTACCACATCCAAGGAA-3’ and 5’-GCTGGAATTACCGCGGCT-3’) were designed based on Genbank accession number NR_003278 and the annealing temperature used was 61.8 °C to obtain the 187 base pair amplicon. The Tnfrsf9 oligonucleotide primers (5’-TTGGGAACATTTAATGACCAGA-3’ and 5’-TCCCGGTCTTAAGCACAGAC -3’) were designed based on Genebank accession number NM_011612 and the annealing temperature used was 62 °C. The 91 base pair amplicon for Tnfrsf9 qRT-PCR was common for both splice variants.
Non-quantitative RT-PCR was used to assess the presence of each of the two splice variants encoding the transmembrane and soluble forms of Tnfrsf9 mRNA. The primers used were previously described (Shao, et al., 2005) and produce 771 and 636 bp amplicons from the alternatively spliced transmembrane and secreted forms of Tnfrsf9, respectively. RT-PCR was carried out using Superscript II reverse transcriptase (Promega, Madison, WI) and TopTaq PCR master mix (Qiagen, Valencia, CA) following the procedures recommended by the manufacturers. PCR was carried out using an Eppendorf Mastercycler thermocycler (Fisher Scientific, Pittsburgh, PA ) programmed for 36 cycles of 94 C melt, 58 C annealing and 72 C extension steps for 15, 45 and 20 seconds, respectively. After a final incubation for 7 minutes at 72 C the samples were subjected to agarose gel electrophoresis in the presence of ethidium bromide. The resultant gels were imaged using a UV transilluminator and a Kodak EDAS290 Gel Documentation system (Fisher Scientific).
In situ Hybridization
Tnfrsf9 mRNA was localized in the tissue sections using colorimetric in situ hybridization. An amplicon was generated using Tnfrsf9-specific primers 5’-AGGTGGACAGCCGAACTGTAACAT-3’ and 5’-TTCTTCTTCCTGTGGACATCGGCA-3’ by RT-PCR as described above. The resulting amplicon was then cloned into the PGEM-T Easy vector as recommended by the manufacturer (Promega). The sequence and orientation of the clone were verified by sequencing with T7 and SP6 promoter primers (UIUC DNA Sequencing Center). Linear templates were prepared using the appropriate restriction enzymes and sense and antisense digoxigenin (DIG) labeled riboprobes were generated as previously described (Bany and Cross, 2006). In situ hybridization was carried out as described previously (Simmons, et al., 2008) except that the final color development with BCIP/NBT substrate was in the presence of 5% polyvinyl alcohol (De Block and Debrouwer, 1993). Slides were counterstained using nuclear fast red. The resulting stained slides stained purple for Tnfrsf9 mRNA with light red nuclei. Use of sense probes revealed no hybridization signals in all tissue types examined (data not shown).
Dolichos biflorus Agglutinin (DBA) Lectin histochemistry
In some experiments, after completing the in situ hybridization and color development for Tnfrsf9 mRNA localization, the sections were subjected to three 10 minute washes in PBS and stained for DBA lectin binding. DBA lectin staining was carried out exactly as described previously (Herington and Bany, 2007). Finally, the slides were counterstained with nuclear fast red prior to mounting cover slips. The resulting slides stained purple for Tnfrsf9 mRNA, brown for DBA lectin and red for nuclei.
Statistical Analysis
The effects of day and tissue type on relative Tnfrsf9 mRNA levels between NIS and IS tissues on Days 4.5-8.5 of pregnancy were determined by repeated-measures analysis of variance. Differences between means were determined by t-Test and Duncan's multiple range test. The effects of day and tissue-type on relative Tnfrsf9 mRNA between pregnant and pseudopregnant uteri on Days 3.5-8.5 were determined by a 2-way analysis of variance. Differences between days for BID and IS tissue were determined using Duncan's multiple range test. Statistical analysis was carried out using Excel (Microsoft, Redmond, WA) and Sigmastat (Systat Inc., San Jose, CA) software.
Results
Tnfrsf9 mRNA Levels increase in the Uterus During Decidualization
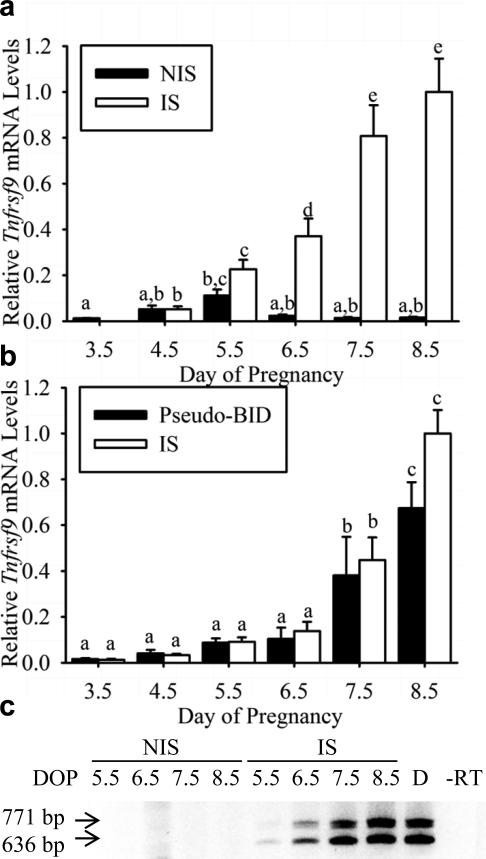
Relative Tnfrsf9 mRNA levels were determined in uteri of pregnant mice on Days 3.5-8.5 of pregnancy using qRT-PCR (Figure 1a). Tnfrsf9 mRNA levels were low on day 3.5 of pregnancy, just prior to the onset of implantation. After the onset of implantation, mRNA levels in NIS tissues were significantly (P< 0.01) increased only on Day 5.5, but not Days 4.5 plus 6.5-8.5, compared to that of Day 3.5 pre-implantation tissue. On the other hand, Tnfrsf9 mRNA levels in IS tissues were significantly (P< 0.02) increased on Days 4.5-8.5 compared to that of Day 3.5 pre-implantation tissue. Finally, mRNA levels between NIS and IS tissues were significantly greater only on Days 6.5 (P<0.02), 7.5 (P<0.0005) and 8.5 (P<0.005). The differences between post-implantation samples are depicted by letters over the bars in Figure 1a. To determine if expression of Tnfrsf9 in uterine tissue during decidualization requires the presence of a conceptus, we utilized a bead-induced deciduoma model described previously (Herington and Bany, 2007, Herington, Underwood, McConaha and Bany, 2009). A comparison of mRNA levels between the tissue-types on each given day did not reveal a significant (P<0.05) difference in Tnfrsf9 mRNA levels (Figure 1b). Tnfrsf9 mRNA levels were detected at low levels in Day 3.5 pseudopregnant and pregnant tissues. After the onset of decidualization, mRNA levels significantly (P< 0.001) increased but in a similar fashion in both BID and IS tissues as revealed by a lack of interaction (P>0.05) between the effects of day and tissue type. The differences between days within BID and IS tissues are depicted by letters over the bars in Figure 1b.
Figure 1.
qRT-PCR and RT-PCR analysis of steady-state Tnfrsf9 mRNA levels and presence of alternatively spliced forms of Tnfrsf9 in the mouse uterus during implantation and decidualization . (a) qRT-PCR analysis of relative mRNA levels in nonimplantation site (NIS) compared to implantation site (IS) tissue segments on Day 3.5-8.5 of pregnancy. Bars in the graphs bearing similar letters are not significantly different and represent mean ± SEM (N=4). (b) qRT-PCR analysis of relative mRNA levels (mean ± SEM, N=4) in bead-induced deciduoma (BID) compared to IS tissue segments on Days 3.5 to 8.5 of pseudopregnancy and pregnancy, respectively. (c) RT-PCR analysis of Tnfrsf9 splice variant expression in non-implantation site (NIS) compared to implantation site (IS) tissue segments on Day 5.5-8.5 of pregnancy and Day 7.5 BID deciduoma tissue. RT-PCR results are representative of several (N=3) independent samples. Abbreviations: DOP, day of pregnancy; bp, base pair; D, deciduoma; and – RT, no-reverse transcriptase control.
It should be noted that the primers used for the qRT-PCR above were designed to produce the same 91 base pair amplicon, regardless of the splice form from which it is amplified. Therefore, the quantitative signals measured did not discern between splice variants. To determine which splice variants are expressed in the uteri of pregnant mice, we utilized previously developed RT-PCR strategy (Shao, et al., 2008). As shown in Figure 1c, both the 771 and 636 bp splice variants which encode the membrane and soluble forms of TNFRSF9 were readily detected in the IS tissues on days 5.5-8.5. Similar findings were found for that of Day 7.5 BID tissue (Figure 1c, Lane D). Finally, control samples where RNA subjected to RT conditions in the absence of reverse transcriptase followed by PCR showed no amplicons (Figure 1c, Lane -RT).
Localization of Tnfrsf9 mRNA in the Uterus During Decidualization
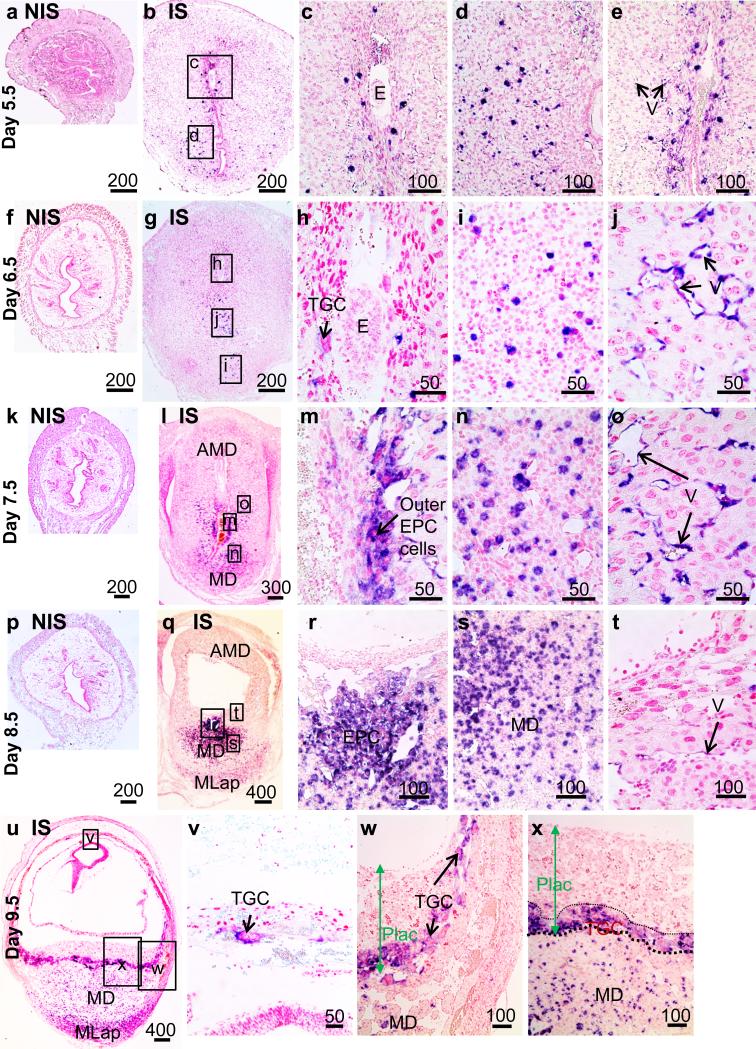
In situ hybridization was used to localize mRNA in the uterus during implantation on Days 5.5-9.5 of pregnancy (Figure 2). Tnfrsf9 mRNA hybridization signals were not detected in Day 3.5 and 4.5 tissue (data not shown) nor in NIS tissue on Days 5.5-8.5 (Figure 2a,f,k,p). However, in IS tissue on Day 5.5 (Figure 2b), strong hybridization signals in a small number of large cells in the endometrium were found in both the antimesometrial region just adjacent to the conceptus (Figure 2c) and in the mesometrial region (Figure 2d). Strong hybridization signals were also detected in vascular endothelial cells near the luminal epithelium at the mesometrial-antimesometrial region boundary (Figure 2e). In IS tissues from the uteri of Days 6.5 (Figure 2g-j) and 7.5 (Figure 2l-o) pregnant mice, hybridization signals were seen in similar scattered large cells of the endometrium but the numbers of these cells dramatically increased. Hybridization signals were also seen in endothelial cells in a similar region found in Day 5.5 IS tissue Figure 2 j,o). Unlike all other Days examined the number of endothelial cells staining positive on Day 7.5 appeared to be greater. One notable difference in localization of signals in Day 6.5 and 7.5 IS tissues compared to that of Day 5.5 was consistent staining of primary trophoblast giant cells on Day 6.5 (Figure 2h) and in outer ectoplacental cone (EPC) cells on Day 7.5 (Figure 2m). Localization of Tnfrsf9 mRNA throughout the EPC was then seen in IS tissue from Day 8.5 uteri (Figure 2q,r). Further, hybridization signals were detected in a larger number of large cells scattered in the mesometrial decidua and to a lesser extent to cells in the mesometrial lymphoid aggregate of pregnancy (MLap) region which is starting to form (Figure 2q,s). Notably, although hybridization signal was seen in some endothelial cells in the antimesometrial decidua close to the conceptus (Figure 2t), the signals were weak compared to previous days. Finally, hybridization signals at mid-pregnancy in IS tissue on Day 9.5 (Figure 2u) were seen in virtually all TGC's adjacent to the decidua capsularis (Figure 2v) and throughout the TGC cell layer of the late developing placenta adjacent to the mesometrial decidua (Figure 2w,x). In a similar fashion to that of Day 8.5, scattered cells in the mesometrial decidua of Day 9.5 uteri showed strong hybridization signals. However, the numbers of these cells appeared to be more numerous in the MLap region (Figure 2u).
Figure 2.
Localization of Tnfrsf9 mRNA in non-implantation (NIS) and implantation (IS) tissue segments of mouse uterine tissue from Day 5-5-9.5 pregnant mice. Photomicrographs show tissues from Day 5.5 NIS (a) or IS (b-e), Day 6.5 NIS (f) or IS (g-j), Day 7.5 NIS (k) or IS (l-o), Day 8.5 NIS (p) or IS (q-t), and Day 9.5 IS (u-x). These are representative of at least 3 independent samples. Global linear adjustments of the brightness and color level were made on the photomicrographs to more accurately represent what was seen on the slides under the microscope. Numbers above scale bars are in microns. Abbrev: AMD, antimesometrial decidua; E, embryo; EPC, ectoplacental cone; MD, mesometrial decidua; MLap, mesometrial lymphoid aggregate of pregnancy; Plac, developing placenta; TGC, trophoblast giant cells; V, vascular endothelium.
Uterine Tnfrsf9 Expression in Il15-Knockout Mice
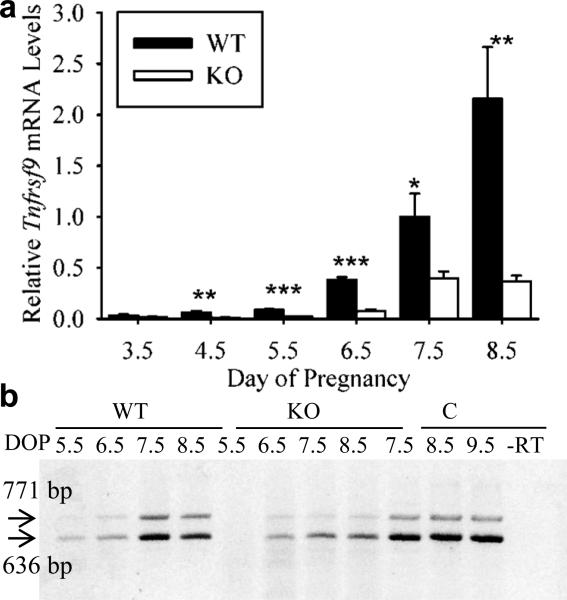
Previous work has shown that the majority of immune cells within the endometrium during implantation are uterine natural killer (uNK) cells. Previously, we evaluated the numbers and localization of DBA lectin-positive uNK cells in IS tissues in the mouse uterus during implantation (Herington and Bany, 2007). Since the pattern of localization for Tnfrsf9 mRNA seen in scattered cells of the endometrium in this study looked similar to the localization of uNK cells, we assessed Tnfrsf9 mRNA levels and localization in the uteri of Il15-deficient (knockout) mice. These mice lack uNK cells (Barber and Pollard, 2003). As shown in Figure 3a, steady-state mRNA levels were dramatically and significantly greater in IS tissues from the uteri of wild-type compared to Il15-knockout mice on Days 4.5, 5.5, 6.5, 7.5 and 8.5 of pregnancy.
Figure 3.
qRT-PCR and RT-PCR analysis of steady-state mRNA levels and presence of alternatively spliced mRNA forms of Tnfrsf9 in implantation site tissue segments of pregnant wild-type (WT) and Il15-knockout (KO) mice. (a) qRT-PCR analysis of relative mRNA levels of Tnfrsf9 mRNA in WT compared to KO IS tissue segments on Day 3.5-8.5 of pregnancy. Stars represent a significant difference in mRNA levels on a given Day of pregnancy between samples from WT and KO mice (*, P<0.05; **, P<0.005; ***, P<0.0005). (b) RT-PCR analysis of Tnfrsf9 splice variant expression implantation site (IS) tissue segments from the uteri of WT and KO mice on Day 5.5-8.5 of pregnancy and in conceptus tissues of WT mice from Days 7.5-9.5. RT-PCR results are representative of several (N=3) independent samples. Abbreviations: bp, base pair; DOP, day of pregnancy; –RT, no-reverse transcriptase control.
Next we assessed the presence of Tnfrsf9 mRNA splice variants in the wild-type compared to Il15-knockout mice (Figure 3b). Both the 771 and 636 bp splice variants were readily detected in the IS tissues of Day 5.5-8.5 pregnant wild-type mice. Similar findings were observed for the IS tissues from Il15 knockout mice with the noted exception that we did not consistently see the two amplicons in Day 5.5 samples. We also assessed the presence of splice variants in samples of the developing placenta/conceptus of wild-type Day 7.5-9.5 pregnant mice (Figure 3b). Both the 771 and 636 bp splice variants of Tnfrsf9 were readily detected in these tissues.
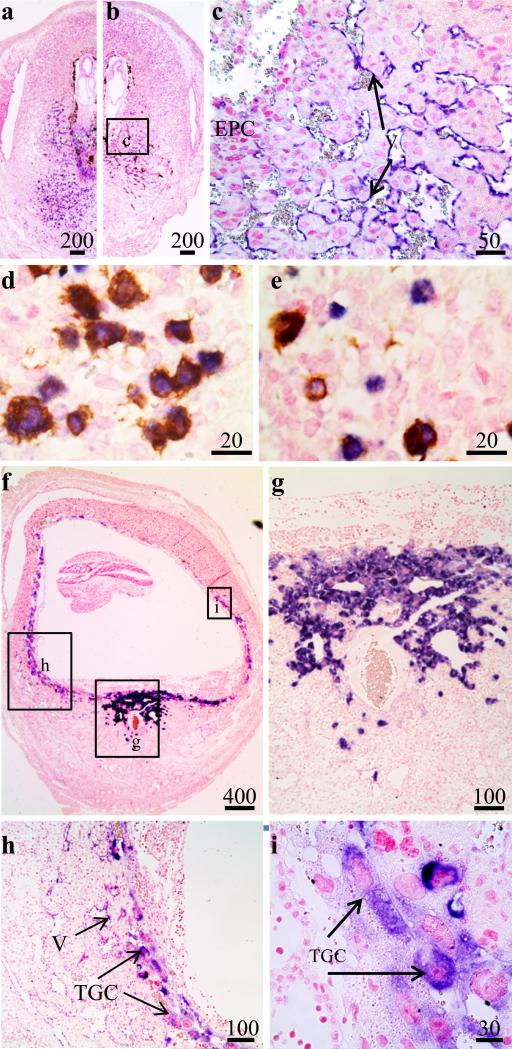
To determine if the scattered cells found in the mesometrial decidua were immune cells, we compared Tnfrsf9 mRNA localization in wild-type verses Il15-knockout mice in IS tissue of Day 7.5 pregnant mice. Hybridization signals for Tnfrsf9 mRNA were seen in the scattered cells of the mesometrial decidua in IS tissues from the uteri of Day 7.5 pregnant wild-type mice (Figure 4a). These hybridization signals were not seen in IS tissue from Il15-knockout mice (Figure 4b). On the other hand, hybridization signals remained positive for what is mainly in endothelial cells in the decidua of Il15-knockout mice (Figure 4c). The qRT-PCR and in situ localization results strongly suggested that a significant amount of Tnfrsf9 expression in the mouse uterus during implantation is from uNK cells. Therefore, we also carried out double staining using a combination of in situ hybridization for Tnfrsf9 mRNA and DBA lectin histochemistry for uNK cells. Most, but not all, Tnfrsf9-positive cells also stained positive for DBA lectin (Figure 4d,e) on Day 7.5 of pregnancy. The potential identity of the Tnfrsf9-positive but DBA lectin-negative cells was not pursued further. However, they may be the subpopulation of DBA-negative but periodic acid Schiff-positive uNK cells previously shown to be in the endometrium at this time (Zhang, et al., 2009). On the side of the conceptus, it was noted that ectoplacental cells of heterozygous Il15 conceptuses in the uteri of Day 7.5 pregnant Il15-knockout mice did not consistently stain positive for Tnfrsf9 mRNA. This is in contrast to conceptuses of the same genotype in the uteri of wild-type mice (Figure 4a-c). Finally, Tnfrsf9 mRNA was localized in IS tissue of Day 9.5 pregnant Il15-knockout mice (Figure 4f-i). Little hybridization signal was detected in the Day 9.5 decidua basalis and MLap while strong localization signals were seen in the trophoblast giant cells surrounding the conceptus.
Figure 4.
Localization of Tnfrsf9 mRNA and DBA lectin binding positive cells in implantation (IS) tissue segments of pregnant uteri. Photomicrographs show Day 7.5 IS tissue in wild-type (WT) (a,d,e) and Il15-knockout (KO) (b,c) mice. Sections shown in panels D and E were doubly stained for Tnfrsf9 mRNA and DBA lectin. Photomicrographs show Day 9.5 IS tissue in Il15-knockout (f-i) mice. These are representative of at least 3 independent samples. Global linear adjustments of the brightness and color level were made on the photomicrographs to more accurately represent what was seen on the slides under the microscope. Numbers above scale bars are in microns. Abbrev: E, embryo; EPC, ectoplacental cone; TGC, trophoblast giant cells; V, vascular endothelium.
Discussion
The Tnfrsf9 gene is believed to play key roles in several aspects of immunity (Croft, 2003, Gellersen, Brosens and Brosens, 2007, Heinisch, Bizer, Volgger and Simon, 2001, Heinisch, Daigle, Knopfli and Simon, 2000, Kim, Chang, Lee, Lee, Kim, Kwon and Kang, 2008, Marvel and Walzer, 2010, Michel and Schwarz, 2000, Myers and Vella, 2005, Nishimoto, Lee, Hong, Potter, Maeda-Yamamoto, Kinoshita, Kawakami, Mittler, Kwon, Ware, Croft and Kawakami, 2005, Pollok, Kim, Zhou, Hurtado, Kim, Pickard and Kwon, 1993, Salih, et al., 2002, Vinay, Cha and Kwon, 2006, Zhang, Voskens, Sallin, Maniar, Montes, Zhang, Lin, Li, Burch, Tan, Hertzano, Chapoval, Tamada, Gastman, Schulze and Strome, 2010) but little is known about its role, if any, in the immunology of early pregnancy. Recently Zhao, et al. showed that Tnfrsf9 mRNA can be detected in the implantation sites of pregnant mice and levels decrease between Days 13.5 and 18 of pregnancy (Zhao, Koon, Curtis, Soper and Bethin, 2007). The present study extends this work and shows that Tnfrsf9 expression dramatically increases in implantation sites of the uterus during the implantation phase of pregnancy. Localization of this expression revealed that the major cell types of the uterus that express Tnfrsf9 are a subset of endothelial and uNK cells in the mesometrial decidua. Alternatively, expression in the conceptus was localized to trophoblast giant and ectoplacental cone cells. These results suggest that Tnfrsf9 expression may play a key immune-related role in mouse implantation, but the precise function needs to be further evaluated.
Previous experiments showed that mouse trophoblast cells express Tnfrsf9 in vitro and expression levels increase in response to interferon gamma (IFNG) treatment (Hoshida, et al., 2007). It has been estimated that, in vivo, uNK cells provide about 90% of the IFNG on the mesometrial side of the implantation site in mice (Croy, et al., 2002). Therefore, it is tempting to speculate that IFNG from uNK cells may drive the expression of the trophoblast cell expression of Tnfrsf9 shown in the present study. Indeed, the reduced/lack of Tnfrsf9 expression in trophoblast cells of Day 7.5 conceptuses in the uteri of Il15-knockout mice (lacking uNK cells) support this conjecture. However, the possibility that trophoblast cell- and thus placental-development is delayed in the Il15-knockout mice also cannot be ruled out at present.
The specific ligand of TNFRSF9 is normally expressed on antigen-presenting cells and is called tumor necrosis factor subfamily 9 ligand (TNFSF9) (Reviewed in (Salih, Kiener and Nussler, 2002, Schwarz, 2005, Schwarz, 2006, Shao and Schwarz, 2010)). TNFSF9, encoded by the Tnfsf9 gene, is a transmembrane glycoprotein of the tumor necrosis factor family of proteins and is found mainly on the surface of antigen presenting cells. Upon binding, TNFSF9 activates membrane TNFRSF9 and initiates a complex set of intracellular signaling pathways (Lee and Kwon, 2006), TNFRSF9 also provides a reverse signal upon binding and initiates intracellular signaling pathways in the TNFSF9 expressing cells (Schwarz, 2006, Shao and Schwarz, 2010). Therefore, like other members of the TNF superfamily of receptors and ligands (Eissner, et al., 2004), membrane TNFRSF9-TNFSF9 signaling is bidirectional. With the major finding of this study that Tnfrsf9 is expressed in the conceptus and endometrium during implantation, further work is warranted to determine if Tnfsf9 is expressed in the uterus or conceptus during implantation.
Two forms of TNFRSF9 exist due to alternative splicing of the Tnfrsf9 transcript (Michel and Schwarz, 2000), but little is known regarding their function. One form is the fully functional transmembrane protein while the other is soluble that lacks a transmembrane domain and is secreted by the cells. Since soluble TNFRSF9 is secreted, it could possibly act in an autocrine, paracrine or endocrine fashion. Although very little is known about the functions of soluble TNFRSF9, it is commonly believed to be immunosuppressive by acting as a competitive antagonist of membrane TNFRSF9 and by binding to but not activating TNFSF9 signaling (Schwarz, 2006). In the present study, we found both splice forms are expressed in the maternal and conceptus tissues during implantation. We speculate that production of soluble TNFRSF9 might be involved, at least in part, in immunosuppressive activities that suppress rejection of the conceptus by the maternal immune system. This could be at the level of the uterus or perhaps the lymphoid organs which are also believed to play an important role in immune tolerance to the conceptus (Taglauer, et al., 2010). Indeed, blockade of TNFRSF9-TNFSF9 interactions increases allograft survival in cardiac transplants (Cho, et al., 2004, Saiki, et al., 2008). However, such an immunosuppressive role for secreted TNFRSF9 by the uterus or trophoblast to obtain maternal immune tolerance to the conceptus is currently pure conjecture. Clearly, more work is required to understand the potential roles of membrane verses soluble Tnfrsf9 expression in the mouse uterus and conceptus during pregnancy. Perhaps enthusiasm to conduct such work might be reduced since Tnfrsf9 KO mice have been noted to be completely fertile (Kwon, et al., 2002). However, to the best of our knowledge, data of detailed assessments of the fertility and pregnancies of these animals cannot be found in the literature. Further, removing the expression of a Tnfrsf9 in the entire mouse may be immunosuppressive itself, so alternate strategies may be needed to study its function in maternal immune tolerance to the conceptus.
TNFRSF9 is believed to be important in cancer biology and in some autoimmune diseases, which has provided interest in developing TNFRSF9 immunotherapies (Cheuk, Mufti and Guinn, 2004, Kim and Broxmeyer, 2001, Nam, Kang, Kwon, Kim and Lee, 2005, Shao, Sun, Koh and Schwarz, 2008, Sun, et al., 2003). It is important to determine the role of Tnfrsf9 expression in implantation biology and pregnancy to determine potential harmful or helpful effects of such therapy may have on pregnant patients or those trying to become pregnant. Further, knowledge about TNFRSF9 signaling in pregnancy might help devise new approaches to treat some forms of immune-related infertility in females.
Acknowledgements
This work was supported in part by Southern Illinois University School of Medicine and NIH - Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD049010) grants (to BB). Personnel support from Southern Illinois University was received in the form of an Undergraduate Research Assistantship and Undergraduate REACH Awards provided to Ms. Kirsten Eckstrum. The authors gratefully acknowledge Ms. Sheila Scillufo and Jen Herington for technical assistance in maintaining the mouse colonies and PCR step of the qRT-PCR work, respectively.
References
- Abrahamsohn PA, Zorn TM. Implantation and decidualization in rodents. J Exp Zool. 1993;266:603–628. doi: 10.1002/jez.1402660610. [DOI] [PubMed] [Google Scholar]
- Bany BM, Cross JC. Post-implantation mouse conceptuses produce paracrine signals that regulate the uterine endometrium undergoing decidualization. Dev Biol. 2006;294:445–456. doi: 10.1016/j.ydbio.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Bany BM, Schultz GA. Increased expression of a novel heat shock protein transcript in the mouse uterus during decidualization and in response to progesterone. Biol Reprod. 2001;64:284–292. doi: 10.1095/biolreprod64.1.284. [DOI] [PubMed] [Google Scholar]
- Barber EM, Pollard JW. The uterine NK cell population requires IL-15 but these cells are not required for pregnancy nor the resolution of a Listeria monocytogenes infection. Journal of immunology. 2003;171:37–46. doi: 10.4049/jimmunol.171.1.37. [DOI] [PubMed] [Google Scholar]
- Chen L, editor. CD137 Pathway: Immunology and Disease. Springer Science + Business Media, LLC.; New York: 2006. [Google Scholar]
- Cheuk AT, Mufti GJ, Guinn BA. Role of 4-1BB:4-1BB ligand in cancer immunotherapy. Cancer Gene Ther. 2004;11:215–226. doi: 10.1038/sj.cgt.7700670. [DOI] [PubMed] [Google Scholar]
- Cho HR, Kwon B, Yagita H, La S, Lee EA, Kim JE, Akiba H, Kim J, Suh JH, Vinay DS, Ju SA, Kim BS, Mittler RS, Okumura K, Kwon BS. Blockade of 4-1BB (CD137)/4-1BB ligand interactions increases allograft survival. Transpl Int. 2004;17:351–361. doi: 10.1007/s00147-004-0726-3. [DOI] [PubMed] [Google Scholar]
- Croft M. Costimulation of T cells by OX40, 4-1BB, and CD27. Cytokine Growth Factor Rev. 2003;14:265–273. doi: 10.1016/s1359-6101(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Cross JC. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin Cell Dev Biol. 2000;11:105–113. doi: 10.1006/scdb.2000.0156. [DOI] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Croy BA, Chantakru S, Esadeg S, Ashkar AA, Wei Q. Decidual natural killer cells: key regulators of placental development (a review). J Reprod Immunol. 2002;57:151–168. doi: 10.1016/s0165-0378(02)00005-0. [DOI] [PubMed] [Google Scholar]
- De Block M, Debrouwer D. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal Biochem. 1993;215:86–89. doi: 10.1006/abio.1993.1558. [DOI] [PubMed] [Google Scholar]
- Drenkard D, Becke FM, Langstein J, Spruss T, Kunz-Schughart LA, Tan TE, Lim YC, Schwarz H. CD137 is expressed on blood vessel walls at sites of inflammation and enhances monocyte migratory activity. FASEB J. 2007;21:456–463. doi: 10.1096/fj.05-4739com. [DOI] [PubMed] [Google Scholar]
- Eissner G, Kolch W, Scheurich P. Ligands working as receptors: reverse signaling by members of the TNF superfamily enhance the plasticity of the immune system. Cytokine Growth Factor Rev. 2004;15:353–366. doi: 10.1016/j.cytogfr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Gellersen B, Brosens IA, Brosens JJ. Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med. 2007;25:445–453. doi: 10.1055/s-2007-991042. [DOI] [PubMed] [Google Scholar]
- Heinisch IV, Bizer C, Volgger W, Simon HU. Functional CD137 receptors are expressed by eosinophils from patients with IgE-mediated allergic responses but not by eosinophils from patients with non-IgE-mediated eosinophilic disorders. J Allergy Clin Immunol. 2001;108:21–28. doi: 10.1067/mai.2001.116864. [DOI] [PubMed] [Google Scholar]
- Heinisch IV, Daigle I, Knopfli B, Simon HU. CD137 activation abrogates granulocyte-macrophage colony-stimulating factor-mediated anti-apoptosis in neutrophils. Eur J Immunol. 2000;30:3441–3446. doi: 10.1002/1521-4141(2000012)30:12<3441::AID-IMMU3441>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Herington JL, Bany BM. Effect of the conceptus on uterine natural killer cell numbers and function in the mouse uterus during decidualization. Biol Reprod. 2007;76:579–588. doi: 10.1095/biolreprod.106.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herington JL, Underwood T, McConaha M, Bany BM. Paracrine signals from the mouse conceptus are not required for the normal progression of decidualization. Endocrinology. 2009;150:4404–4413. doi: 10.1210/en.2009-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida MS, Gorjao R, Lima C, Daher S, Curi R, Bevilacqua E. Regulation of gene expression in mouse trophoblast cells by interferon-gamma. Placenta. 2007;28:1059–1072. doi: 10.1016/j.placenta.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Kim DH, Chang WS, Lee YS, Lee KA, Kim YK, Kwon BS, Kang CY. 4-1BB engagement costimulates NKT cell activation and exacerbates NKT cell ligand-induced airway hyperresponsiveness and inflammation. J Immunol. 2008;180:2062–2068. doi: 10.4049/jimmunol.180.4.2062. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Broxmeyer HE. Therapeutic potential of 4-1BB (CD137) as a regulator for effector CD8(+) T cells. J Hematother Stem Cell Res. 2001;10:441–449. doi: 10.1089/15258160152509064. [DOI] [PubMed] [Google Scholar]
- Kwon BS, Hurtado JC, Lee ZH, Kwack KB, Seo SK, Choi BK, Koller BH, Wolisi G, Broxmeyer HE, Vinay DS. Immune responses in 4-1BB (CD137)-deficient mice. J Immunol. 2002;168:5483–5490. doi: 10.4049/jimmunol.168.11.5483. [DOI] [PubMed] [Google Scholar]
- Lee H- W, Kwon BS. Cd137 signal transduction. In: Chen L, editor. Cd137 Pathway. Springer; New York: 2006. pp. 15–27. [Google Scholar]
- Marvel J, Walzer T. CD137 in NK cells. Blood. 2010;115:2987–2988. doi: 10.1182/blood-2010-01-261404. [DOI] [PubMed] [Google Scholar]
- Medawar PB. Some immunological and endocrinological problems raised by the evolution of vivaparity in vertebrates. Symp Soc Exp Biol. 1953;7:320–338. [Google Scholar]
- Michel J, Schwarz H. Expression of soluble CD137 correlates with activation-induced cell death of lymphocytes. Cytokine. 2000;12:742–746. doi: 10.1006/cyto.1999.0623. [DOI] [PubMed] [Google Scholar]
- Mori T, Takakura K, Narimoto K, Kariya M, Imai K, Fujiwara H, Okamoto N, Kariya Y, Shiotani M, Umaoka Y, et al. Endocrine and immune implications of human endometrial decidualization in implantation. Ann N Y Acad Sci. 1991;626:321–330. doi: 10.1111/j.1749-6632.1991.tb37927.x. [DOI] [PubMed] [Google Scholar]
- Myers LM, Vella AT. Interfacing T-cell effector and regulatory function through CD137 (4-1BB) co-stimulation. Trends Immunol. 2005;26:440–446. doi: 10.1016/j.it.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Nam KO, Kang WJ, Kwon BS, Kim SJ, Lee HW. The therapeutic potential of 4-1BB (CD137) in cancer. Curr Cancer Drug Targets. 2005;5:357–363. doi: 10.2174/1568009054629681. [DOI] [PubMed] [Google Scholar]
- Nishimoto H, Lee SW, Hong H, Potter KG, Maeda-Yamamoto M, Kinoshita T, Kawakami Y, Mittler RS, Kwon BS, Ware CF, Croft M, Kawakami T. Costimulation of mast cells by 4-1BB, a member of the tumor necrosis factor receptor superfamily, with the high-affinity IgE receptor. Blood. 2005;106:4241–4248. doi: 10.1182/blood-2005-04-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollok KE, Kim YJ, Zhou Z, Hurtado J, Kim KK, Pickard RT, Kwon BS. Inducible T cell antigen 4-1BB. Analysis of expression and function. J Immunol. 1993;150:771–781. [PubMed] [Google Scholar]
- Reali C, Curto M, Sogos V, Scintu F, Pauly S, Schwarz H, Gremo F. Expression of CD137 and its ligand in human neurons, astrocytes, and microglia: modulation by FGF-2. J Neurosci Res. 2003;74:67–73. doi: 10.1002/jnr.10727. [DOI] [PubMed] [Google Scholar]
- Robertson SA. Immune regulation of conception and embryo implantation-all about quality control? J Reprod Immunol. 2010;85:51–57. doi: 10.1016/j.jri.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Saiki H, Suzuki J, Kosuge H, Haraguchi G, Ishihara T, Haga T, Maejima Y, Isobe M, Uede T. Blockade of the 4-1BB pathway attenuates graft arterial disease in cardiac allografts. Int Heart J. 2008;49:105–118. doi: 10.1536/ihj.49.105. [DOI] [PubMed] [Google Scholar]
- Salih HR, Kiener PA, Nussler V. 4-1 BB ligand--just another costimulating molecule? Int J Clin Pharmacol Ther. 2002;40:348–353. doi: 10.5414/cpp40348. [DOI] [PubMed] [Google Scholar]
- Schwarz H. Biological activities of reverse signal transduction through CD137 ligand. J Leukoc Biol. 2005;77:281–286. doi: 10.1189/jlb.0904558. [DOI] [PubMed] [Google Scholar]
- Schwarz H. Significance of reverse signal transduction for the biology of the CD137 receptor/lignad system. In: Chen L, editor. CD137 Pathway. Springer; New York: 2006. pp. 29–45. [Google Scholar]
- Shao H, Fu Y, Liao T, Peng Y, Chen L, Kaplan HJ, Sun D. Anti-CD137 mAb treatment inhibits experimental autoimmune uveitis by limiting expansion and increasing apoptotic death of uveitogenic T cells. Invest Ophthalmol Vis Sci. 2005;46:596–603. doi: 10.1167/iovs.04-0835. [DOI] [PubMed] [Google Scholar]
- Shao Z, Schwarz H. CD137 ligand, a member of the tumor necrosis factor family, regulates immune responses via reverse signal transduction. J Leukoc Biol. 2010 doi: 10.1189/jlb.0510315. [DOI] [PubMed] [Google Scholar]
- Shao Z, Sun F, Koh DR, Schwarz H. Characterisation of soluble murine CD137 and its association with systemic lupus. Mol Immunol. 2008;45:3990–3999. doi: 10.1016/j.molimm.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Simmons DG, Rawn S, Davies A, Hughes M, Cross JC. Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC Genomics. 2008;9:352. doi: 10.1186/1471-2164-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner L, Shaqireen DOKMM, Wu JT, Schwarz H. Signal transduction mechanisms of CD137 ligand in human monocytes. Cell Signal. 2007;19:1899–1908. doi: 10.1016/j.cellsig.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Sun Y, Subudhi SK, Fu YX. Co-stimulation agonists as a new immunotherapy for autoimmune diseases. Trends Mol Med. 2003;9:483–489. doi: 10.1016/j.molmed.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Taglauer ES, Adams Waldorf KM, Petroff MG. The hidden maternal-fetal interface: events involving the lymphoid organs in maternal-fetal tolerance. Int J Dev Biol. 2010;54:421–430. doi: 10.1387/ijdb.082800et. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinay DS, Cha K, Kwon BS. Dual immunoregulatory pathways of 4-1BB signaling. J Mol Med. 2006;84:726–736. doi: 10.1007/s00109-006-0072-2. [DOI] [PubMed] [Google Scholar]
- Wilcox RA, Chapoval AI, Gorski KS, Otsuji M, Shin T, Flies DB, Tamada K, Mittler RS, Tsuchiya H, Pardoll DM, Chen L. Cutting edge: Expression of functional CD137 receptor by dendritic cells. J Immunol. 2002;168:4262–4267. doi: 10.4049/jimmunol.168.9.4262. [DOI] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K. Review of factors essential for blastocyst implantation for their modulating effects on the maternal immune system. Semin Cell Dev Biol. 2008;19:161–169. doi: 10.1016/j.semcdb.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Yamada AT, Croy BA. DBA-lectin reactivity defines natural killer cells that have homed to mouse decidua. Placenta. 2009;30:968–973. doi: 10.1016/j.placenta.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Zhang X, Voskens CJ, Sallin M, Maniar A, Montes CL, Zhang Y, Lin W, Li G, Burch E, Tan M, Hertzano R, Chapoval AI, Tamada K, Gastman BR, Schulze DH, Strome SE. CD137 promotes proliferation and survival of human B cells. J Immunol. 2010;184:787–795. doi: 10.4049/jimmunol.0901619. [DOI] [PubMed] [Google Scholar]
- Zhao B, Koon D, Curtis AL, Soper J, Bethin KE. Identification of 9 uterine genes that are regulated during mouse pregnancy and exhibit abnormal levels in the cyclooxygenase-1 knockout mouse. Reprod Biol Endocrinol. 2007;5:28. doi: 10.1186/1477-7827-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]