Abstract
The DNA damage response (DDR) is a signal transduction pathway that decides the cell's fate either to repair DNA damage or to undergo apoptosis if there is too much damage. Post-translational modifications modulate the assembly and activity of protein complexes during the DDR pathways. MicroRNAs (miRNAs) are emerging as a class of endogenous gene modulators that control protein levels, thereby adding a new layer of regulation to the DDR. In this review, we describe a new role for miRNAs in regulating the cellular response to DNA damage with a focus on DNA double-strand break damage. We also discuss the implications of miRNA's role in the DDR to stem cells, including embryonic stem cells and cancer stem cells, stressing the potential applications for miRNAs to be used as sensitizers for cancer radiotherapy and chemotherapy.
Keywords: microRNA, DNA damage response, radiosensitivity, stem cells
Introduction
The DNA damage response (DDR) is a molecular mechanism that cells have evolved to sense DNA damage, transduce these signals and promote their repair (Harper and Elledge, 2007). The essential cellular role of DDR is to stop the cell cycle so as to allow cells to repair damaged DNA or to undergo apoptosis if too much damage has occurred. Therefore, DDR is considered as a defense mechanism against tumor development (Bartek et al., 2007). In addition to this essential role, DDR is also involved in many other physiological processes, such as meiosis, telomere homeostasis and virus infection (Jackson and Bartek, 2009).
Every cell in the human body experiences DNA lesions as a result of constant insults from environmental agents, such as ionizing radiation, or from intrinsic factors like reactive oxygen species. The resulting DNA damages, including base adducts, DNA mismatch, insertion/deletion, O6 alkyguanine, inter-strand DNA crosslinking, single-strand breaks (SSBs) and double-strand breaks (DSBs), can be repaired by cells using different repair mechanisms (Jackson and Bartek, 2009). DSBs, despite their infrequent occurrence, are the most difficult to repair and are extremely detrimental to the cells. Cells employ two major pathways to fix DSBs: non-homologous end-joining (NHEJ) which is a fast but error-prone process in G0/G1 cell-cycle phase, and homologous recombination repair (HRR), which is a slow and error-free process in late S/G2-phase (Lieber, 2008; San Filippo et al., 2008).
Tremendous progress has been made in the elucidation of the underlying mechanisms of cellular responses to DSB-type damage. Protein post-translational modifications, such as phosphorylation, acetylation, methylation or ubiquitinylation, have been shown to play crucial roles in the assembly and disassembly of repair complexes (Huen and Chen, 2010). MicroRNAs (miRNAs) have emerged as endogenous gene regulators that downregulate protein expression by mRNA cleavage or translation repression (Bartel, 2009), adding another dimension of protein regulation. In this review, we will summarize the role of miRNAs in DDR with special focus on the miRNAs that are either induced by DSB or that regulate DSB-type damage response.
DNA damage-regulated miRNAs
DNA DSB damage affects many cellular processes, including gene transcription. A genetic linkage and association study has characterized a cohort of regulators that modulate irradiation (IR)-responsive gene expression and are thought to mediate the cellular response to radiation (Smirnov et al., 2009). Recent studies show that miRNAs can themselves be regulated by DNA damage as described below with IR-induced DSB as an example.
Biogenesis of miRNAs
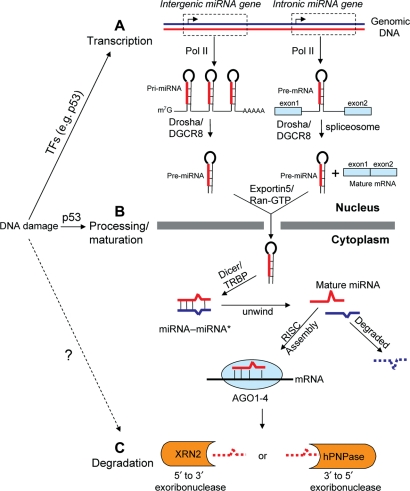
miRNAs are single-stranded small RNAs of 19–25 nt in length that downregulate gene expression by either cleaving target mRNA or repressing translation. The biogenesis of miRNAs comprises transcription, processing/maturation and degradation (Figure 1). Depending on the genomic location, miRNAs are transcribed differently: intergenic miRNAs are transcribed into pri-miRNAs by RNA polymerase II as they contain their own promoter and regulatory units (Lagos-Quintana et al., 2001; Lau et al., 2001), whereas intronic miRNAs are co-transcribed with their host genes from a common promoter (Rodriguez et al., 2004; Baskerville and Bartel, 2005). Pri-miRNAs from intergenic miRNA are 5′ capped (m7G) and 3′ polyadenylated and further cleaved into pre-miRNA by Drosha/DGCR8 microprocessor complex, while the intronic miRNA is directly cleaved by Drosha/DGCR8 complex into pre-miRNA without affecting the splicing step of host genes (Han et al., 2004; Kim and Kim, 2007; Morlando et al., 2008).
Figure 1.
DNA damage affects the biogenesis of miRNAs. (A) DNA damage regulates specific miRNA's expression through transcription, and p53 is an exemplary transcription factor that mediates miRNAs’ transcription. (B) DNA damage also regulates a subset of miRNAs’ by modulating the processing and maturation of miRNA biogenesis. p53 could interact with the Drosha/DGCR8 complex through p68 helicase to enhance the miRNAs’ expression. (C) Whether DNA damage influences miRNAs expression by modulating the degradation step of miRNAs needs further investigation. The steps of miRNA biogenesis and degradation are described in more detail in the text.
Pre-miRNA is then exported from nucleus to cytoplasm by exportin 5 and Ran-GTP (Yi et al., 2003; Lund et al., 2004) and cleaved by Dicer/TRBP to an imperfect miRNA/miRNA* duplex around 20–25 nt in size (Chendrimada et al., 2005). Only one strand of the duplex (red in Figure 1) is incorporated into an RNA-induced silencing complex (RISC/Argonaute 1–4) to bind to 3′ UTR of target genes and suppress expression, while the other strand is normally degraded. The RISC-loaded mature miRNA is protected from degradation by Argonaute proteins (Meister et al., 2004; Okamura et al., 2004; Rand et al., 2005). However, after finishing its task, the mature single-strand miRNA will also be degraded by the 5′-3′ exoribonuclease XRN2 (Chatterjee and Grosshans, 2009) or the 3′-5′ exoribonucleases, for example, human polynucleotide phosphorylase (Das et al., 2010) and nuclear exosome (Pawlicki and Steitz, 2010). Other protein factors or RNA modifications that regulate the stability and degradation of specific miRNAs are reviewed elsewhere (Kai and Pasquinelli, 2010; Siomi and Siomi, 2010).
DNA damage regulates miRNA transcription
DNA damage can regulate miRNA expression at the transcriptional level. p53 plays a critical role in this regulation (Figure 1A). miRNA expression profiles of wild-type and p53-deficient cells have been compared to identify the miRNA components of p53 transcriptional pathways. A family of miRNAs, miR-34a-c, is induced by DNA damage and oncogenic stress, in a p53-dependent manner (He et al., 2007). Ectopic expression of miR-34 induces cell-cycle arrest and downregulates a program of genes promoting cell-cycle progression (He et al., 2007). The induction of miR-34a by p53 is further confirmed by two other studies using genome-wide miRNA screening for p53-dependent regulation following DNA damage (Chang et al., 2007; Tarasov et al., 2007). In both studies, ectopic expression of miR-34a induces apoptosis or cell-cycle arrest in the G1-phase. Most of the protein targets for miR-34a are involved in cell-cycle progression, apoptosis and DNA repair. Loss of function mutations in Caenorhabditis elegan miR-34 result in the abnormal cellular survival response to radiation and these C. elegans are highly radiosensitive in soma but radioresistant in the germ cells (Kato et al., 2009), similar to the function of C. elegan p53 (Cep1). Cep1 loss of function mutants display protection from apoptosis in germ cells but are sensitized to radiation-induced apoptosis in somatic cells (Derry et al., 2001), suggesting that the p53-miR-34 pathway is required for a normal response to DNA damage in vivo.
miR-34c, another member of miR-34 family, is also induced by p53 following DNA damage. However, in the absence of p53, the induction of miR-34c still occurs although to a lesser extent; this is thought to be mediated by an alternative ATM-dependent pathway that involves p38 MAPK signaling to MK2 (Cannell et al., 2010). Overexpression of miR-34c suppresses c-Myc expression, whereas inhibition of miR-34c activity prevents DNA damage-induced S-phase arrest and leads to increased DNA synthesis which is reversed by subsequent c-Myc depletion (Cannell et al., 2010). These data suggest that DNA damage-induced miR-34c upregulation, in either p53-dependent or p38 MAPK-dependent manner, serves to suppress c-Myc and prevent inappropriate replication which may otherwise lead to genomic instability.
Genome-wide screens for DNA damage-responsive miRNAs have identified a cohort of miRNAs that exhibit p53-dependent regulation (Chang et al., 2007; Tarasov et al., 2007). Like the miR-34 family, two homologous miRNAs, miR-192 and miR-215, are upregulated by genotoxic stress and dependent on p53 activation (Braun et al., 2008; Georges et al., 2008). Ectopic expression of miR-192/215 induces cell-cycle arrest and the protein targets for miR-192/miR-215 include a number of transcripts that regulate G1/S and G2/M checkpoints (Georges et al., 2008). Interestingly, miR-192 and miR-215 are detected at high levels in normal colon tissue but severely reduced in many colon cancer samples (Braun et al., 2008), suggesting of their tumor suppressor roles, perhaps through p21 accumulation and cell-cycle arrest (Braun et al., 2008; Georges et al., 2008).
These findings suggest that p53-activated miRNAs may act in concert with other p53 transcriptional gene targets to modulate the cellular response to DNA damage. As DNA damage also modulates the activity of other transcription factors, such as NF-κB, CREB and E2F1, which are known to regulate miRNA expression (Taganov et al., 2006; Petrocca et al., 2008; Nudelman et al., 2010), it would not be surprising if these transcription factor-regulated miRNAs also contribute to the cellular response to DNA damage. It should be noted that because p53 is activated by both SSB and DSB, these p53-activated miRNAs may play a greater role in DNA repair that has been hitherto appreciated.
DNA damage modulates miRNA processing/maturation
DNA damage can also regulate miRNA expression through modulating the miRNA processing and maturation steps (Figure 1B). This has been shown in a recent study in which several miRNAs (including miR-16-1, miR-143 and miR-145) are upregulated by IR-induced DNA damage (Suzuki et al., 2009). The underlying mechanism is that p53 interacts with the Drosha/DGCR8 processing complex through an association with RNA helicase p68 (a.k.a. DDX5) and facilitates the processing of pri-miRNAs to pre-miRNAs. p53 mutants interfere with a functional assembly between Drosha complex and p68, leading to attenuation of miRNA processing activity (Suzuki et al., 2009). These findings suggest a new transcription-independent regulatory mode for miRNA expression in response to DNA damage (Figure 1B).
p63 and p73, together with p53, have been noted in computation predictions to function as regulators of miRNA processing and maturation. In addition to direct interaction with the miRNA processing/maturation complex through p68 helicase, two other venues have been suggested by which p53/p63/p73 can regulate miRNA processing: (i) p53-regulated miRNAs could target most of the components (mRNAs’ 3′ UTR) of the miRNA processing complex, such as Drosha/DGCR8, Dicer/TRBP2 and Argonaute proteins; (ii) a number of components of the miRNA processing machinery, including Dicer, could serve as direct transcriptional targets of p53/p63/p73. In particular, p53/p63/p73 appear to regulate the processing of miRNAs, such as let-7, miR-200c, miR-143, miR-107, miR-16, miR-145, miR-134, miR-449a, miR-503 and miR-21 (Boominathan, 2010). This study provides mechanistic insights into how p53, p63 and p73 may regulate the components of the miRNA processing.
Taken together, DNA damage can induce the transcription of specific miRNAs and, thereby, regulate specific cellular functions or influence the expression of a subset of miRNA by modulating miRNA processing. Whether DNA damage regulates miRNA expression through interfering with miRNA degradation and turnover also warrants further investigation (Figure 1C).
miRNA profiling in cells responding to IR
miRNA arrays have been used to profile miRNA expression in different cells responding to IR. When a human lymphoblastic cell line, IM9, is treated with IR and miRNA profiling is examined, expression level changes (>2 folds) are noted for 73 (1 Gy) and 33 (10 Gy) human miRNAs. Many predicted genes targeted by these IR-responsive miRNAs are involved in the regulation of apoptosis, cell cycle and DNA repair (Cha et al., 2009). Using A549, a human non-small cell lung cancer cell line, microarray analyses identify 12 (20 Gy) and 18 (40 Gy) miRNAs that exhibit more than 2-fold changes in their expression levels (Shin et al., 2009). Again, the predicted protein targets include many known genes for the DDR. By using multiplexed quantitative real-time PCR to screen global miRNA expression in prostate cancer cells after IR (6 Gy), 15 miRNAs are identified with significant alteration in their expression levels. Among them, miR-521 is the most downregulated and further confirmed to modulate radiosensitivity of prostate cancer cells by suppressing the expression of Cockayne syndrome protein A, a DNA repair protein (Josson et al., 2008). The miRNA profile in primary human dermal microvascular endothelial cells after 2 Gy radiation was measured using oligo-microarrays covering 361 miRNAs. Eleven miRNAs are significantly upregulated or downregulated and are shown to modulate radiosensitivity by clonogenic survival and proliferation assays (Wagner-Ecker et al., 2010).
Disturbingly, no obvious overlap of IR-responsive miRNA profiles has been noted among different cell lines, including primary cells, solid cancer cells and blood cells. This suggests either that: (i) IR-responsive miRNA profiles are cell type-specific, (ii) they might be dose-dependent, or (iii) the reproducibility of miRNA profiling needs further optimization.
miRNAs regulate DDR
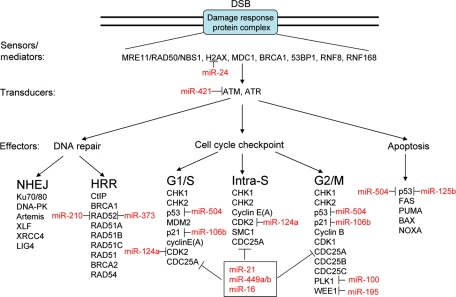
miRNAs have emerged as endogenous gene regulators that affect protein stability and, therefore, offer another degree of regulation for DDRs. DSBs activate a signal transduction process that leads to cell-cycle arrest, followed by either repair or apoptosis. Protein modifications, such as phosphorylation or ubiquitinylation, regulate the stability and speed of assembly of the machineries that control the cell cycle, DNA repair or apoptosis. Before discussing the role of miRNA in regulating DSB DDR, we briefly review the core protein components for these DDR pathways (Figure 2).
Figure 2.
miRNAs regulate DSB DDR through modulating the core protein components of various pathways. The key proteins during the DDR pathway are summarized in the order of sensors/mediators, transducers and effectors, while the effectors include DNA repair, cell-cycle checkpoint (G1/S, intra-S and G2/M) and apoptosis mechanisms. Interactions with miRNAs that regulate the expression of DDR protein components are indicated in red.
Sensors and mediators of DSB signaling
The MRE11-RAD50-NBS1 (MRN) complex acts as a sensor of DSB to recruit ATM. Binding to a DNA damage site is achieved through MRE11/RAD50 (MR) heterotetramers while NBS1 influences the DNA binding and nuclease activities of the MR complex. ATM-dependent phosphorylation of histone variant H2AX seems to be the initial signal for subsequent accumulation of DNA damage-response proteins. MDC1 binds to both γH2AX and the MRN complex, acting to further recruit ubiquitin ligase RNF8 so as to form a scaffold to facilitate the accumulation of RNF168, BRCA1 and 53BP1 around the break sites (van Attikum and Gasser, 2009).
Transducers: ATM/ATR
ATM and ATR are serine/theronine kinases that transduce the DNA damage signals to downstream proteins. ATM is primarily activated in response to DSBs whereas ATR responds primarily to SSBs and stalled replication forks. ATM/ATR coordinate downstream events, such as cell cycle, DNA repair and apoptosis, by phosphorylating a wide assortment of protein substrates (Lavin, 2008; Shiloh, 2003).
Effectors of cell-cycle checkpoints
G1/S checkpoint
The ATM/CHK2 (ATR/CHK1)-p53/MDM2-p21 pathway is the major one that controls the DNA damage-induced G1/S checkpoint. ATM/CHK2 and ATR/CHK1 are activated by DSBs and SSBs, respectively, to stabilize p53 and induce p21 expression. p21 inhibits cyclin-dependent kinase to silence the G1/S-promoting cyclinE(A)/CDK2 complex and thereby causes G1 arrest. Another p53-independent pathway is also responsible for G1/S checkpoint, in which activated CHK1/2 directly downregulates CDC25A phosphatase through ubiquitin-dependent proteasome-mediated turnover; this consequently inhibits the cyclinE(A)/CDK2 complex. The p53-independent CHK1/2-CDC25A pathway delays the G1/S transition only for a few hours while the p53-dependent checkpoint pathway prolongs G1 arrest (Kastan and Bartek, 2004).
Intra-S-phase checkpoint
Two parallel signaling pathways are reported to mediate the DNA damage-induced intra-S checkpoint: CHK1/2-CDC25A-cyclinE(A)/CDK2 and ATM-NBS1-SMC1. In the first pathway, CDC25A degradation leads to the inhibition of cyclinE(A)/CDK2 and blocks the loading of CDC45 onto chromatin which is required for the recruitment of DNA polymerase α for DNA synthesis (Bartek et al., 2004). In the second pathway, ATM phosphorylates NBS1 and recruits NBS1 to form a complex that regulates S-phase checkpoint; the precise mechanism is still not clear (Yazdi et al., 2002).
G2/M checkpoint
DNA damage-induced G2/M checkpoints are controlled by p53-dependent and p53-independent pathways, both of which target the mitosis-promoting activity of the cyclinB/CDK1 complex. In the p53-dependent pathway, ATM/CHK2 or ATR/CHK1 phosphorylate and stabilize p53, which in turn upregulates p21 to suppress the activity of cyclinB/CDK1, leading to late G2 arrest. In the p53-independent pathway, CHK1/CHK2 control three parallel pathways: (i) phosphorylate and inhibit CDC25A, CDC25B and CDC25C; (ii) inhibit the activity of polo-like kinase 1 (PLK1), which is known to activate CDC25C and (iii) phosphorylate and upregulate the activity of WEE1 kinase, which catalyzes the inhibitory phosphorylation of CDK1. All these events block the activation of cyclinB/CDK1 in a concerted fashion, resulting in late G2 arrest (Fukasawa, 2007).
Effectors of DNA repair
NHEJ and HRR represent two major DSB repair pathways, which occur in different phases of the cell cycle: NHEJ in G0/G1-phase, HRR in late S/G2. Up to 90% of DSBs are repaired by NHEJ in G1-phase of the cell cycle. Seven core proteins required for NHEJ have been identified, Ku70, Ku80, DNA-PKcs, Artemis, XRCC4, XLF/Cernunnos and DNA ligase IV, which are assembled as two steps: the Ku heterodimer (Ku70/80) binds to DSB ends and recruits DNA-PKcs, and consequently to coordinate end processing with rejoining by recruiting XRCC4, Artemis, XLF and DNA ligase IV (Lieber, 2008).
DSBs can also be repaired by homologous recombination-mediated pathways. Repair is initiated by the CtIP-BRCA1 complex-mediated resection of a DSB to produce 3′ single-stranded DNA overhangs (Sartori et al., 2007) and followed by strand invasion and strand displacement, which is mediated by RAD52 and RAD51 paralogs (-A, -B, -C). DNA resynthesis of the broken portion with the undamaged sister chromatids serving as a template is then mediated by the RAD51-BRCA2 complex and RAD54 (San Filippo et al., 2008).
Effectors of apoptosis
If DNA damage cannot be repaired in a timely manner, apoptosis is initiated to remove these cells before the DNA lesions enlarge and lead to more serious consequences, such as cancers. DSBs are thought to be crucial apoptosis-triggering lesions, and p53 plays a critical role in the DSB-induced apoptosis. In response to DSBs, ATM/CHK2 and ATR/CHK1 activate and stabilize p53, which leads to transcriptional activation of pro-apoptotic factors, such as FAS, PUMA and BAX. To back up p53, CHK1/CHK2 can also activate E2F1 and p73, respectively, which in turn transcribe BAX, PUMA and NOXA. However, DNA damage-triggered signaling and the execution of apoptosis are cell type- and genotoxin-specific, depending on the p53 status and DNA repair capacity (Roos and Kaina, 2006).
miRNAs that regulate DSB-type damage response
Figure 2 also summarizes the reported miRNAs that regulate the expression of core protein components in the DSB DDR.
miR-24-H2AX
miR-24 is identified by miRNA arrays during post-mitotic differentiation of hematopoietic cell lines. Overexpression of miR-24 downregulates the histone variant H2AX, the initial sensor protein in the DSB response. miR-24-mediated suppression of H2AX renders hematopoietic cells hypersensitive to gamma-IR and genotoxic drugs, which might account for the reduced DNA repair capacity of terminally differentiated hematopoietic cells (Lal et al., 2009).
miR-421-ATM
By using a target prediction program, miR-421 is reported to suppress ATM expression by targeting the 3′ UTR of ATM transcripts. Ectopic expression of miR-421 results in a deficient S-phase cell-cycle checkpoint and an increased sensitivity to ionizing radiation. Blocking the interaction between miR-421 and ATM 3′ UTR with an antisense morpholino oligonucleotide rescues the defective phenotype caused by miR-421 overexpression, indicating that ATM mediates the effect of miR-421 on cell-cycle checkpoints and radiosensitivity. This is the first study to show that ATM, the chief transducer of DSB damage, is subject to miRNA regulation. The miR-421-ATM pathway most likely contributes to the DDR in a variety of ways given the many targets of transphosphorylation of ATM (Hu et al., 2010a).
miR-504-p53 and miR-125b-p53
Computational predictions suggest that several miRNAs are involved in the post-transcriptional regulation of p53. miR-504 downregulates human p53 through its direct binding to two sites in the p53 3′ UTR. Overexpression of miR-504 decreases p53 protein levels and regulates p53 transcriptional activity, p53-mediated apoptosis, and cell-cycle arrest in response to stress; it further promotes the tumorigenecity of cells in vivo (Hu et al., 2010b). miR-125b is another negative regulator of p53, depending on the binding of miR-125b to the 3′ UTR of p53 mRNA. Overexpression of miR-125b represses the endogenous level of p53 protein and suppresses apoptosis. Interestingly, miR-125b is downregulated following IR and is thought to mediate the increase in DNA damage-induced p53 protein levels and the subsequent p53-induced apoptosis during the stress response (Le et al., 2009).
miR-106b-p21
Overexpression of miR-106b promotes cell-cycle progression, whereas loss of function reverses this phenotype. The cyclin-dependent kinase inhibitor p21 is a direct target of miR-106b, and the miR-106b-mediated p21 downregulation overrides a doxorubicin-induced DNA damage checkpoint. Interestingly, miR-106b is overexpressed in multiple tumor types and this overexpression may contribute to tumor cell proliferation in part by suppressing the cell-cycle checkpoint (Ivanovska et al., 2008).
miR-21-CDC25A
miR-21 is induced by DNA damage, negatively regulating G1/S transition. It also participates in the DNA damage-induced G2/M checkpoint. This is achieved by downregulation of CDC25A, a cell-cycle regulator. miR-21 suppresses CDC25A expression through a defined sequence in 3′ UTR of CDC25A. Interestingly, miR-21 is underexpressed in a subset of CDC25A-overexpressing colon cancers. This study shows a role of miR-21 in modulating cell-cycle progression following stress, providing a molecular explanation of miR-21 in tumorigenesis and a potential therapeutic role for upregulation of miR-21 in colon cancer (Wang et al., 2009).
miR-210-RAD52 and miR-373-RAD52
Two miRNAs, miR-210 and miR-373, are upregulated in a hypoxia-inducible factor-1 alpha-dependent manner in hypoxic cells. Overexpression of miR-210 suppresses the levels of RAD52, which is a key factor in HRR while overexpression of miR-373 leads to a reduction in the nucleotide excision repair (NER) proteins, RAD23B and RAD52. Consistent with these results, both RAD52 and RAD23B are downregulated in hypoxia. These results indicate that hypoxia-inducible miR-210 and miR-373 play roles in modulating the expression levels of key proteins involved in the HRR and NER pathways, providing new mechanistic insight into the effects of hypoxia on DNA repair and genetic instability in cancer (Crosby et al., 2009).
There are other reported miRNAs that regulate the expression of core protein components of the DDR pathways, including miR-449a/b and miR-16, both targeting CDC25A (Pothof et al., 2009; Yang et al., 2009), miR-195 targeting WEE1 (Qi et al., 2009), miR-124a targeting CDK2 (Nakamachi et al., 2009) and miR-100 targeting PLK1 (Shi et al., 2010). Their roles in the DSB DDR need further study.
miRNAs in embryonic stem cells and induced pluripotent stem cells
Both embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are characterized by two fundamental properties: self-renewal and differentiation. Recent research indicates that miRNAs represent an important layer of regulation for stem cell self-renewal and differentiation (Gangaraju and Lin, 2009). Human iPSCs are morphologically indistinguishable from human ESCs; however, genome-wide expression analyses reveal differences by gene and miRNA expression signatures (Chin et al., 2009). Among the miRNA signature, miR-24, miR-421, miR-125b and miR-373 are shown to be significantly different between hESCs and hiPSCs, also confirmed by another study (Wilson et al., 2009). The four miRNAs described above are already known to regulate H2AX, ATM, p53 and RAD52 (Figure 2) and are all involved in the differentiated cell's DDR. This suggests that the DDR itself may distinguish iPSCs from ESCs. More interestingly, these four miRNAs are dramatically upregulated or downregulated after differentiation into somatic cells, suggesting that these miRNAs might influence the differentiation of ESCs and iPSCs through their controlled DDR or other unidentified protein targets.
ESCs have a very short G1-phase and lack the G1/S checkpoint. Recent experiments suggest that miRNAs play a central role in achieving this unique cell-cycle property (Marson et al., 2008; Wang et al., 2008). The miR-290 cluster (including miR-291-3p, miR-294 and miR-295) is highly expressed in ESCs and acts to suppress several well-known inhibitors of the G1/S transition (including p21, RBL2 and LATS2), thereby modulating the cell-cycle G1/S checkpoint of ESCs (Wang et al., 2008). Expression of the miR-290 cluster is downregulated as ESCs differentiate forward and consequently the G1-phase becomes longer and the G1/S checkpoint appears in the differentiated somatic cells (Wang et al., 2008). As ATM, p53 and p21 are all involved in the DNA damage-induced G1/S checkpoint control in differentiated somatic cells, whether and how the miR-421-ATM, miR-125b/miR-504-p53 and miR-106b-p21 pathways (Figure 2) contribute to the G1/S checkpoint of ESCs is an interesting open question and worthy of further investigation.
For ESCs to maintain genomic integrity so that they can retain the ability to differentiate into multiple cell types without propagating DNA errors, they would seem to require a more efficient repair system than differentiated somatic cells. In the ESCs, ATR rather than ATM is responsible for the DSB HRR, whereas in differentiated somatic cells, the HRR is mainly dependent on ATM (Adams et al., 2010). This relationship between ATR and ATM dynamically changes as cells differentiate. As miR-421 is highly expressed in ESCs and decreases with cell differentiation (Chin et al., 2009), it is intriguing to speculate that the miR-421-ATM pathway (Hu et al., 2010a) contributes to the switch from ATR in ESCs to ATM in differentiated somatic cells that are responsible for DSB HRR.
Cancer stem cells, radioresistance and miRNAs
Cancer stem cells (CSCs) represent a small subset of cells identified in a variety of tumors that are responsible for the origin and maintenance of tumors. Several pieces of evidence support the role of CSCs in the tumor radio/chemo-resistance model: (1) breast CSCs (CD44+/CD24−/low subpopulation) are more resistant than the non-CSCs to radiation treatment (Phillips et al., 2006); (2) this breast CSC subpopulation is elevated in patients treated with chemotherapy (Chang et al., 2005); (3) glioblastoma CSCs (CD133+ subpopulation) are also more radioresistant than the non-CSCs; the underlying mechanism is thought to be that CSCs have more efficient capacity of repairing DNA damage than non-CSCs (Bao et al., 2006). These findings suggest that radio/chemoresistance might be a general property of CSCs (Diehn and Clarke, 2006; Visvader and Lindeman, 2008). Recently, there is increasing evidence to show an association between miRNA expression in tumors and chemo- and radiosensitivity (Hummel et al., 2010). Application of these miRNA studies in differentiated cells to CSCs might provide new insights into the radio/chemoresistance development of cancers and offer a great promise for developing new cancer therapies with miRNAs as sensitizers to modulate the CSCs’ radio/chemosensitivity.
Conclusions and perspectives
It is now clear that miRNA expression is altered in response to DNA damage. miRNA microarray analysis will characterize the miRNA profiles for different types of cells and identify whether there is a common core miRNA signature for different cell types responding to DNA damage. DNA damage modulates miRNA expression by either inducing the transcription of miRNA genes or directly interacting with the processing and maturation machinery of miRNAs. Whether DNA damage affects the degradation or modification of miRNAs thereby to regulate the miRNA expression definitely deserves further investigation.
We have summarized the core protein components for the signal transduction pathways of DSB DDR. Only a few proteins to date are known to be regulated by miRNAs (Figure 2). It is estimated that 30% of human proteins are regulated by miRNAs. We envision that there will be additional proteins in the DDR pathways that are regulated by miRNAs. Studies from somatic cells indicate that cellular radiosensitivity reflects the efficiency of DNA repair and loss of the core components in the NHEJ and HRR pathways leads to the cellular phenotype of radiosensitivity of cells (Jeggo and Lavin, 2009). Therefore, the miRNA-mediated negative regulation of these core protein components provides new avenues to modulate cellular sensitivity to radiation or chemical therapeutic compounds.
The role of miRNA in regulating the self-renewal and differentiation of stem cells has been well documented. The new role of miRNA in regulating DDR is just emerging, and the role of miRNA in the DDR of stem cells needs to be scrutinized in details. Further study of the miRNA's role in DDR for CSCs will help us to better understand tumor radio/chemoresistance and offer us new opportunities for the use of miRNAs as new types of radio/chemotherapy sensitizers.
Funding
The research related to this work in the authors’ laboratory was supported by NIH U19AI067769-9001 pilot research grant (H.H.), California Breast Cancer Research Program 16IB-0016 (H.H.), NIH RO1NS052528 (R.A.G.) and the Ataxia-Telangiectasia Medical Research Foundation (Los Angeles, CA, (R.A.G.)).
Conflict of interest: none declared.
Acknowledgements
We apologize to those whose work was not cited here because of space limitations.
References
- Adams B.R., Golding S.E., Rao R.R., et al. Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS One. 2010;5:e10001. doi: 10.1371/journal.pone.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas C., Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bartek J., Bartkova J., Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C.J., Zhang X., Savelyeva I., et al. p53-responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell I.G., Kong Y.W., Johnston S.J., et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc. Natl. Acad. Sci. USA. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H.J., Shin S., Yoo H., et al. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int. J. Oncol. 2009;34:1661–1668. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- Chang J.C., Wooten E.C., Tsimelzon A., et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J. Clin. Oncol. 2005;23:1169–1177. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin M.H., Mason M.J., Xie W., et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby M.E., Kulshreshtha R., Ivan M., et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.K., Sokhi U.K., Bhutia S.K., et al. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl. Acad. Sci. USA. 2010;107:11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry W.B., Putzke A.P., Rothman J.H. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- Diehn M., Clarke M.F. Cancer stem cells and radiotherapy: new insights into tumor radioresistance. J. Natl. Cancer Inst. 2006;98:1755–1757. doi: 10.1093/jnci/djj505. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Gangaraju V.K., Lin H. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges S.A., Biery M.C., Kim S.Y., et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Du L., Nagabayashi G., et al. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc. Natl. Acad. Sci. USA. 2010a;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Chan C.S., Wu R., et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 2010b;38:689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem. Sci. 2010;35:101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel R., Hussey D.J., Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur. J. Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Ball A.S., Diaz R.L., et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P., Lavin M.F. Cellular radiosensitivity: how much better do we understand it? Int. J. Radiat. Biol. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- Josson S., Sung S.Y., Lao K., et al. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai Z.S., Pasquinelli A.E. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat. Struct. Mol. Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kato M., Paranjape T., Muller R.U., et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lal A., Pan Y., Navarro F., et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lavin M.F. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Le M.T., Teh C., Shyh-Chang N., et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Gromak N., et al. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamachi Y., Kawano S., Takenokuchi M., et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–1304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- Nudelman A.S., DiRocco D.P., Lambert T.J., et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., et al. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki J.M., Steitz J.A. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F., Visone R., Onelli M.R., et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Phillips T.M., McBride W.H., Pajonk F. The response of CD24−/low/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Pothof J., Verkaik N.S., van IJcken W., et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Yu J.Y., Shcherbata H.R., et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand T.A., Petersen S., Du F., et al. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J.L., et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W.P., Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Alajez N.M., Bastianutto C., et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int. J. Cancer. 2010;126:2036–2048. doi: 10.1002/ijc.24880. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shin S., Cha H.J., Lee E.M., et al. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int. J. Oncol. 2009;35:81–86. [PubMed] [Google Scholar]
- Siomi H., Siomi M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Smirnov D.A., Morley M., Shin E., et al. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H.I., Yamagata K., Sugimoto K., et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V., Jung P., Verdoodt B., et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Wagner-Ecker M., Schwager C., Wirkner U., et al. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat. Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zou F., Zhang X., et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.D., Venkatasubrahmanyam S., Jia F., et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Feng M., Jiang X., et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi P.T., Wang Y., Zhao S., et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]