SYNOPSIS
RNF4 family ubiquitin ligases are RING E3 ligases that regulate the homeostasis of SUMOylated proteins by promoting their ubiquitylation. Here we report that the RING domain of RNF4 forms a stable dimer, and that dimerization is required for ubiquitin transfer. Our data suggests that the stability of the E2~ubiquitin thioester bond is regulated by RING domain dimerization.
Keywords: Ubiquitin, E3 ligase, SUMO, protein-protein interaction, RING finger, protein domain
INTRODUCTION
RING (Really Interesting New Gene) domain ubiquitin ligases promote the transfer of ubiquitin from ubiquitin-conjugating enzymes (E2s) to lysine residues in target proteins [1]. Many RING E3 ligases also physically recruit substrate proteins through a distinct substrate recognition motif, and this interaction confers substrate specificity. By simultaneously recruiting both the substrate and an E2~ubiquitin conjugate, RING domain-containing proteins are thought to act as scaffolds that promote the direct transfer of ubiquitin from the E2 conjugating enzyme to the substrate. This mechanism is distinct from that used by the HECT domain E3 ligases, which form a covalent thioester intermediate with ubiquitin, and directly catalyze substrate ubiquitylation [2].
RNF4 (RING finger protein 4) is a RING E3 ligase that brings about the ubiquitylation of proteins that have been modified by addition of the ubiquitin-like protein SUMO (Small Ubiquitin-like MOdifier) [3–6]. RNF4 is distinguished from other RING E3 ligases by the presence of a tandem array of four SUMO interaction motifs (SIMs) near the N-terminus. SIMs are known to form a β-strand that selectively interacts with SUMO [6, 7]. This interaction allows RNF4 to select substrate proteins that are then targeted for ubiquitylation by the C-terminal RING domain. RNF4 is the functional fusion of the fission yeast proteins Rfp1/2 and Slx8 (Slx5/Hex3 in budding yeast) and together these proteins comprise the STUbL (SUMO-targeted ubiquitin ligase) family [4, 5, 8]. In yeast, this family of proteins has a critical role in maintaining genome stability, gene silencing and protein quality control [9–11]. In human cells, the STUbL activity of RNF4 mediates the arsenic trioxide (As2O3)-induced degradation of promyelocytic leukemia protein (PML) and contributes to the therapeutic effect of arsenic trioxide in treating acute promyelocytic leukemia [12, 13].
To understand the mechanism by which RNF4 promotes the ubiquitylation of substrate proteins we solved the structure of the RING domain and undertook biochemical studies to investigate the features that are important for activity. By utilizing a number of mutant proteins and a stable UbcH5b~ubiquitin oxyester conjugate we show that RING dimerization destabilizes the oxyester bond so that catalysis is favored. E2 recruitment is also required for activity, although binding of the E2 to the RING domain does not correlate with E3 ligase activity. Our complementation studies in yeast indicate that dimerization is functionally important and together these results suggest that RING dimerization is a critical activating event that is required for ubiquitin transfer.
EXPERIMENTAL
Protein expression and purification
Mouse RNF4 proteins (Swissprot Q9QZS2) were overexpressed in Escherichia coli (BL21) cells at 18 °C overnight following addition of 0.1 mM IPTG. Cells were recovered then lysed by sonication in 1×PBS (3.2 mM NaHPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, pH 7.4) containing 0.2% Tween-20, 1 mM dithiothreitol (DTT) and general protease inhibitors. Proteins were purified using glutathione affinity chromatography and the GST (glutathione-S-transferase) tag was removed using 3C protease fused to GST. Proteins were then purified by size exclusion chromatography using either a Superdex S75 or S200 column (GE Healthcare).
Crystallization and structure solution
Crystals of the rat RNF4 RING domain (Swissprot O88846) were grown at 18 °C using the vapor diffusion method (Note the sequence of the RING domain of rat and mouse RNF4 differ by only two residues and these are in the disordered N-terminus). Sitting drops (150 nL) were set up using the Mosquito robot (TTP LabTech Ltd.) and contained a 2:1 ratio of protein (22 mg/mL in PBS, pH 7.4) and crystallization buffer (0.2 M lithium sulfate, 0.1 M bis-Tris pH 6.2, 22% PEG 3350). Diffraction data were collected at beamline PX-2 at the Australian Synchrotron. Three datasets were collected around the zinc κ absorption edge, and the structure was solved using the multiple wavelength anomalous dispersion method (see Supplementary methods for details).
GST-pulldown, autoubiquitylation and discharge assays
To measure E3 ligase activity, reactions that contained 20 µM untagged ubiquitin, 5 µM UbcH5b, 50 nM E1 and ~20 µM RNF4 proteins, were incubated for 1.5 h at 37 °C in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM ATP, 2 mM MgCl2 and 2 mM DTT. Reactions were stopped by addition of 2× SDS-PAGE buffer and samples were resolved by 14.5% SDS-PAGE and detected by staining with Coomassie Blue.
For discharge assays, UbcH5b with a Cys to Ser mutation at the active site (C85S) was charged with ubiquitin in the presence of E1, ATP, creatine phosphate and phosphocreatine. Ubiquitin charged C85S-UbcH5b (referred to as UbcH5b~Ub conjugate) was purified by size-exclusion chromatography. To measure ubiquitin discharge, reactions containing ~10 µM UbcH5b~Ub and ~10 µM RNF4 proteins were incubated at 37 °C in 60 mM MES, pH 6.5. At the indicated time points reactions were stopped by addition of 2× SDS-PAGE buffer and samples were resolved by 16% SDS-PAGE and detected by staining with Coomassie Blue.
To evaluate the ability of the RNF4 proteins to bind to the UbcH5b~Ub conjugate we utilized GST-RING fusion proteins. Immobilized GST-RING proteins were incubated with UbcH5b~Ub conjugate in 1×PBS, pH 7.4, 0.2% Triton-X100 for 1 h at 4 °C, then washed twice with the same buffer. Following addition of 2× SDS-PAGE buffer and after separation by 14.5% PAGE, proteins were detected by staining with Coomassie Blue.
Yeast growth assay and plasmid shuffle
The growth and transformation of fission yeast Schizosaccharomyces pombe is based on Forsburg and Rhind [14]. Full length mouse RNF4 and its mutant forms were expressed in S. pombe using an nmt1 promoter-based vector pSGP73 [15]. To carry out plasmid shuffling, the RNF4 expression plasmids were transformed into a parental strain HSY443 (Δrfp1::his3+ Δrfp2::kanMX pSLF173-Rfp2-ura4+). Transformants were grown for 24 hr in EMM media containing uracil to allow the spontaneous loss of the pSLF173-Rfp2-ura4+ plasmid. The culture was then spotted in a serial dilution onto EMM plates containing 5-fluoroorotic acid (5-FOA) to eliminate cells still containing the Rfp2-ura4+ plasmid. Hydroxyurea (HU) (5 mM) was used to generate additional genotoxic stress in Δrfp1Δrfp2 cells that is relieved by expressing wild-type Rfp1, Rfp2, or RNF4. All assays were conducted in the presence of thiamine so that the nmt1 promoter activity was kept at a minimum.
SUMO-targeted ubiquitin ligase (STUbL) activity assay
The in vitro ubiquitin ligase assay was carried out as described before [5], except a Flag-tagged, diSUMO2-GST fusion protein was used as the substrate for detection of STUbL activity. For each reaction, 1 µg ubiquitin, 2 µg E1, 2 µg E2, and 0.4 µg Flag-diSUMO2-GST were mixed in 25 µl of reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT, and 5 mM ATP) and incubated for 1 h at 30 °C. The reaction was terminated by addition of 2× SDS-PAGE sample buffer and ubiquitylation was visualized by anti-Flag (M2, Sigma) immunoblotting.
RESULTS AND DISCUSSION
The RNF4 RING domain adopts a dimeric structure
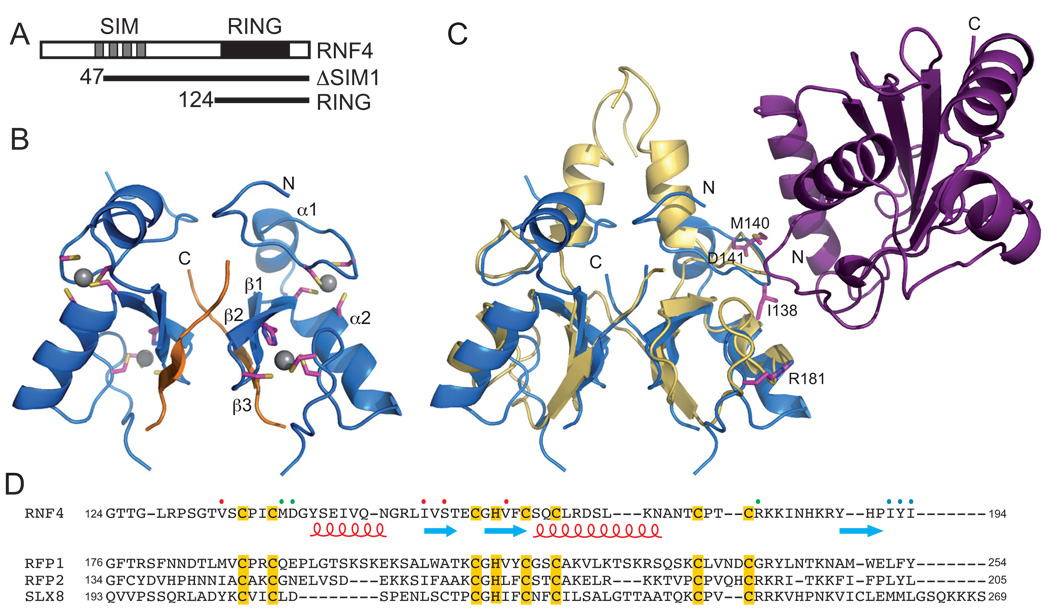
Isolated RING domains often retain the E3 ligase activity of the parent protein and allow activity to be evaluated. Therefore, to characterize ubiquitin transfer by RNF4 we initially identified the minimal domain that is able to promote autoubiquitylation of RNF4 (Figure 1A and Supplementary Figure S1). All deletion constructs studied promoted the efficient autoubiquitylation of RNF4 indicating that the purified RING domain (residues 124–194) was an active E3 ligase. We determined the crystal structure of the RING domain at 1.8 Å using the multiple anomalous dispersion method and scattering from the zinc ions (Figure 1B and Supplementary Table S1). The asymmetric unit contained a RING domain dimer, although the electron density maps indicated that the C-terminal residues of the RING domain were exchanged and that a crystallographic two-fold axis resulted in a domain-swapped tetramer (Supplementary Figures S2A and S2B). However, when analyzed at high protein concentrations, using a multi-angle laser light scattering (MALLS) detector coupled in-line to a size-exclusion column (Supplementary Figure S2C), only the dimer was observed indicating that the dimer predominates in solution and is the biologically relevant structure (Figure 1B).
Figure 1. Structural analysis of the RNF4 RING domain.
(A) Schematic representation of RNF4 constructs utilized in this study. (B) Ribbon diagram of the RNF4 RING dimer (residues 124–194) in blue with the domain-swapped C-terminal tail colored orange. Zinc ions are shown as grey spheres and zinc-chelating residues are shown as sticks. Elements of secondary structure are labeled. (C) Ribbon diagram showing the RNF4 RING domain structure (blue) overlaid on that of the cIAP2 RING (yellow):UbcH5b (pink) complex. Residues at the E2 interface in RNF4 that were mutated to alanine are indicated as sticks and are labeled. (D) Alignment showing the RNF4 sequence used for structural studies and the comparable regions from the yeast homologues Rfp1, Rfp2 and Slx8. Elements of secondary structure are indicated below the RNF4 sequence and residues mutated in this study are indicated with colored dots (green = E2 interface; red = dimer interface; blue = C-terminal tail).
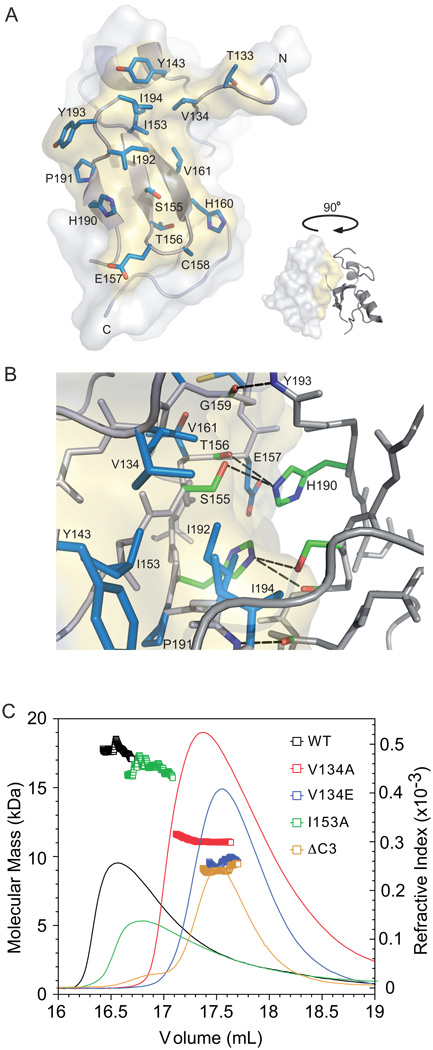
The RING domain of RNF4 adopts a typical RING domain fold that is stabilized by interaction with two zinc ions, and the two monomers in the dimer are almost identical (r.m.s.d. of 0.568 Å over 64 residues), with only small differences in the loop regions. The orientation of the two RING domains in the RNF4 dimer is similar to that observed for other C-terminal RING domain dimers, and the RNF4 structure overlays closely with the MDM2:MDMX RING heterodimer (r.m.s.d. 2.197 Å over 99 residues) (Supplementary Figure S3), and the cIAP2 RING homodimer (r.m.s.d. 2.018 Å over 78 residues) (Figure 1C) [16, 17]. The main difference between the three RING dimer structures is due to the distinct conformation of residues N-terminal to the RING domain. As a consequence, the dimer interface is considerably smaller in RNF4 (~550 Å2 compared to ~1000 Å2). The RNF4 dimer interface predominantly comprises residues in the first two β-strands (e.g. Ile153, Ser155 and Val161) that are part of the core RING domain, as well as residues in the third β-strand (e.g. His190 and Ile192) that is immediately C-terminal to the core RING domain (Figure 2A). In addition to hydrophobic contacts, the side chains of Ser155 and His190 are involved in a small network of hydrogen bonds (Figure 2B). Of the residues N-terminal to the RING domain, only Val134 packs closely. Despite differences in the size of the dimer interface, the minimal RING domain from RNF4 interacts in a similar manner to those of cIAP2 and MDM, suggesting that other RING domains that have a C-terminal location will form comparable dimers.
Figure 2. Characterization of the dimer interface.
(A) Surface representation of a RNF4 RING monomer showing the dimer interface as determined using PISA. The side chains of I153 and contact residues at the dimer interface are displayed as sticks (blue). (B) Close-up view of the dimer interface viewed from above relative to (A). In addition to the contact residues, those residues involved in the hydrogen-bonding network are shown as sticks (green). (C) Samples of the indicated RING-domain mutant proteins were separated on a Superdex S75 column that was coupled to a MALLS detector. The refractive index profile and the calculated mass are shown for each. The calculated mass of the RING domain monomer is 7.8 kDa.
The sequences of the RING domains from the yeast RNF4 homologues, Rfp1 and Rfp2 (Figure 1D), are similar to that of RNF4 and the C-terminal residues are conserved. As a consequence it is likely that the RING domains of these proteins adopt a similar structure to the RNF4 RING domain, with the C-terminal residues having an important role in dimer formation as suggested previously [5]. Nevertheless, their binding partner, Slx8 has a more extended C-terminus that is likely to be solvent accessible and the conformation adopted by these residues is uncertain.
RING dimerization is required for efficient ubiquitin transfer
To identify features of the RING domain that are important for ubiquitin transfer in RNF4 we generated a number of point mutants. First we mutated residues that were likely to be important for E2 recruitment. Residues in the RING domain of RNF4 that are predicted to be required for functional interaction with the E2, UbcH5b, were identified based on overlay of the RNF4 RING homodimer with the cIAP2 RING:UbcH5b complex structure (Figure 1C) [16]. Based on this comparison, Met140, Asp141 and Arg181 in the RING domain, which are predicted to contact UbcH5b, were mutated to Ala. The second set of mutants was focused on understanding the role of the C-terminal residues that we had previously shown were required for the biological activity of RNF4 [5]. We deleted three residues from the C-terminus (ΔC3) or mutated single residues in the C-terminal extension that lies beyond the RING domain (e.g. Y193A) because similar mutants in cIAP proteins abrogate activity [16]. Lastly, we also made mutations that we expected would disrupt dimerization but not affect the C-terminal tail (e.g. V161A, V134A/E, I153A/E, S155A/E) (Figure 2A and 2B). These mutants were generated because previous analysis of the MDM RING domains suggested that while the C-terminal residues were important for dimerization, they might also directly affect E3 ligase activity [16–18].
Initially we evaluated the ability of the dimer interface mutants to form a stable RING domain dimer. For some mutants we determined their oligomeric state by separating them on a size-exclusion column that was coupled to a MALLS detector (Figure 2C), while for others we compared their elution position relative to that of proteins that had been evaluated by MALLS (Supplementary Figure S4A and S4B). The theoretical mass of the RNF4 RING domain is 7.8 kDa; the wild-type protein had an average mass of 17.9 kDa, indicative of a dimer. Mutation of the aromatic residue at the C-terminus (Y193A) rendered the protein monomeric, as did deletion of the three C-terminal residues (ΔC3), with both mutants having an observed average mass of ~8–9 kDa. This indicated that the C-terminal residues are essential for dimer formation. Mutation of hydrophobic residues in the core had a variable effect on dimerization. While the V161A, I153E and V134E mutants behaved as monomers, the V134A mutant had an average mass of 11.1 kDa, slightly larger than the monomer, indicating it retained some ability to self-associate (Figure 2C). Moreover, when a comparable sample of the I153A mutant was analyzed it had an average mass of 15.9 kDa, suggesting that it was only slightly destabilized compared to the wild-type protein. Likewise, the S155A mutant behaved as a stable dimer, while the S155E mutant was monomeric. Thus mutation of the core dimer interface generated mutants of variable dimer stability. The oligomeric state of all mutants is summarized in Table 1.
Table 1. Summary of mutant protein data.
The oligomeric state from MALLS analysis (Mono/Dimer); the ability to bind the E2~Ub conjugate (Bind E2~Ub), and the results from autoubiquitylation (Auto), discharge (Discharge) and SUMO-targeted ubiquitin ligase (STUbL) assays; as well as yeast complementation (Yeast) assays are summarized.
| RING | ΔSIM1 | Full length | ||||
|---|---|---|---|---|---|---|
| Mono/Dimer | Bind E2~Ub1 |
Auto | Discharge | Yeast | STUbL | |
| WT | D | +++ | +++ | +++ | +++ | +++ |
| Dimer interface mutants | ||||||
| I153A2 | D* | +++ | +++ | +++ | +++ | n.d. |
| S155A | D | +++ | +++ | +++ | +++ | n.d. |
| V134A2 | M* | ++ | ++ | ++ | +++ | + |
| V134E | M | +++ | + | +/- | - | +/- |
| S155E | M | +/- | - | +/- | - | n.d. |
| V161A | M | - | - | +/- | - | - |
| I153E3 | M | - | n.d. | n.d. | - | n.d. |
| C-terminal tail mutants | ||||||
| Y193A | M | +/- | - | - | - | - |
| Y193M | n.d. | n.d. | n.d. | n.d. | - | - |
| ΔC3 | M | +/- | - | - | - | - |
| I192A | n.d. | n.d. | n.d. | n.d. | - | - |
| I194A | n.d. | n.d. | n.d. | n.d. | - | - |
| E2 interface mutants | ||||||
| D141A | n.d. | + | +++ | ++ | +++ | n.d. |
| M140A/R181A | D | - | - | - | - | n.d. |
n.d. not determined.
A weak interaction between E2~Ub and some mutants may not be detected because of the limits of the GST-pulldown assay utilized.
M* predominantly monomeric (monomer = 8–9 kDa, observed 11.1 kDa). D* predominantly dimeric (dimer = 17.9 kDa, observed 15.9 kDa).
Longer forms of purified I153E were unstable.
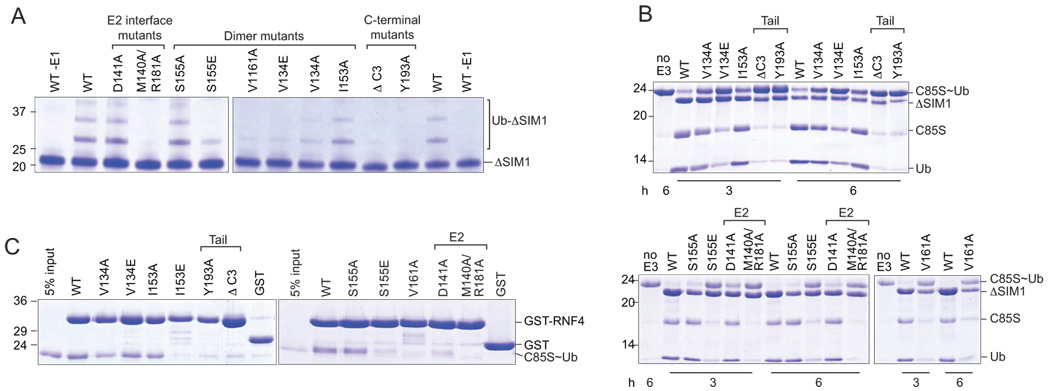
To assess the in vitro activity of these mutants, we carried out ubiquitin-ligase assays with an N-terminally truncated RNF4 protein (ΔSIM1, Figure 1A). In an autoubiquitylation assay the wild-type protein readily promoted the formation of a ubiquitin ladder that depended on the availability of E1 (Figure 3A). In contrast, the E2 interface mutant (M140A/R181A) that does not interact with the E2 (see Figure 3C) did not promote the formation of a ladder. Other mutants, such as I153A, V134A and S155A, retained significant E3 ligase activity and were comparable to the wild-type protein. In contrast the monomeric C-terminal tail mutants, Y193A and ΔC3, were inactive, while the monomeric dimer interface mutants, V161A, V134E and S155E had significantly reduced E3 ligase activity. These results showed that E2 recruitment and an intact C-terminal tail are required to support ubiquitin transfer, and suggested that high level activity depends on the RING domain retaining the capacity to dimerize.
Figure 3. Ubiquitin discharge and transfer depend on RING dimerization.
(A) Autoubiquitylation assays using ΔSIM1 (residues 47–194) mutant proteins and the E2, UbcH5b. Mutants are grouped by function. (B) Ubiquitin-discharge assay using the ΔSIM1 mutant proteins and the UbcH5b~Ub conjugate that was prepared using UbcH5b with an active-site mutation (C85S). Reactions were incubated in a MES buffer at 37 °C, and samples taken after 3 and 6 hours. Mutations within the C-terminal tail (Tail) or E2 interface (E2) are indicated. (C) Interaction of the UbcH5b~Ub conjugate with wild-type and mutant RNF4 RING domain proteins was assessed using a GST-pulldown assay. Samples were incubated in 1×PBS for 1 hour at 4 °C. Proteins in all panels were detected by staining with Coomassie Blue.
RING dimerization destabilizes the thioester bond in preparation for catalysis
In an effort to further dissect the role of the C-terminal tail and RING dimerization we utilized a ubiquitin discharge assay to assess the ability of the RNF4 mutant proteins to activate the E2~ubiquitin conjugate for transfer of ubiquitin to a target lysine. For this assay we utilized an oxyester E2~ubiquitin conjugate that was prepared using the C85S mutant of UbcH5b. The oxyester conjugate is stable in the absence of the RING domain, but, although activity is slowed, addition of active E3 ligases promotes hydrolysis of the oxyester bond as measured by the appearance of free E2 and ubiquitin (Figure 3B, lanes 1 and 2). Consistent with their ability to efficiently promote autoubiquitylation, the wild-type protein as well as the S155A, I153A and V134A mutants, which all had some ability to self-associate, resulted in appreciable discharge of ubiquitin (Figure 3B). The ability of the monomeric dimer interface mutant proteins (V134E, V161A and S155E) to promote ubiquitin discharge was diminished and paralleled their ability to support autoubiquitylation. In contrast, the C-terminal mutants and the disruptive E2 interface mutant (M140A/R181A) did not promote discharge, indicating that E2 recruitment and the C-terminal residues have an essential role. These results suggest that the RING domain influences the stability of the oxyester bond between the E2 and ubiquitin. Moreover, dimeric RING proteins have a destabilizing affect on the oxyester bond compared to their monomeric counterparts.
Binding of the E2~ubiquitin conjugate to the RING domain doesn’t correlate with activity
Because the monomeric dimer interface mutants (V134E, V161A and S155E) had a low level of activity in both the autoubiquitylation and ubiquitin discharge assays we wondered if the decreased activity of these mutants resulted from a diminished ability to bind to the UbcH5b~ubiquitin conjugate. To investigate the importance of E2 recruitment for activity we assessed the ability of the UbcH5b~ubiquitin conjugate to bind the mutant RNF4 proteins in a GST-pulldown assay under conditions where minimal discharge occurred (Figure 3C). An interaction between wild-type RNF4 and the conjugate was readily detected, while there was no detectable interaction with the disruptive E2 interface mutant (M140A/R181A) and binding to the D141A mutant was significantly reduced. All the highly active mutant proteins appeared to bind the conjugate as efficiently as the wild-type protein. In contrast, the monomeric dimer interface mutants exhibited a variable ability to interact with the conjugate. For example, the monomeric V134E mutant appeared to bind the UbcH5b~ubiquitin conjugate tightly, while the V161A and S155E mutants had diminished but detectable binding. Likewise, interaction of the E2~ubiquitin conjugate with the C-terminal tail mutants was reduced. These results suggest that E2~ubiquitin recruitment does not correlate well with activity. Notably, the D141A mutant is highly active but has a reduced affinity for the conjugate, while some inactive monomeric mutants still bind to the conjugate tightly.
RING dimerization is required for in vivo function
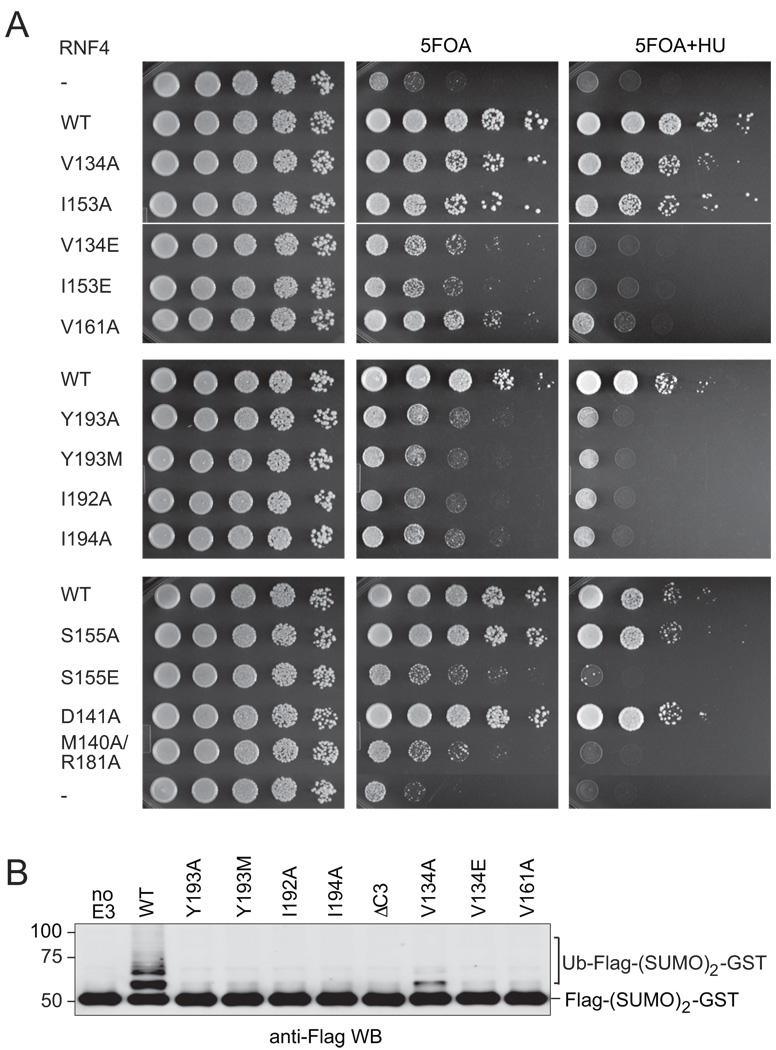
Together the in vitro experiments suggest that the ability to promote ubiquitin discharge and autoubiquitylation are highly correlated, and that both activities depend on the integrity of the E2 interface and C-terminal tail, with a stable RING-domain mediated dimer required for full activity. To determine whether the C-terminal residues or RING-mediated dimerization of RNF4 is important in a physiological paradigm, we took advantage of a fission yeast mutant strain lacking the RNF4 homologues Rfp1 and Rfp2. The Δrfp1Δrfp2 double mutant has a severe growth defect that can be compensated for by ectopic expression of Rfp1, Rfp2, or RNF4 alone [5]. We used a plasmid shuffle method to displace an Rfp2-expressing plasmid with a plasmid carrying RNF4 or its mutant forms, in the Δrfp1Δrfp2 background. The growth of the resulting strains (5-FOA-resistant colonies) reflected the activity of RNF4 in a biological context (Figure 4A). For example, as expected wild-type RNF4 supported growth but the inactive E2 interface mutant (M140A/R181A) did not. We predicted that if dimerization is essential for RNF4 function, the monomeric mutants should lack activity in vivo. Indeed, like wild-type the S155A, I153A and V134A mutants, which had significant ubiquitin discharge and E3-ligase activity (Figure 3) and retained the ability to self-associate, supported yeast growth even under conditions of genotoxic stress induced by growth in the presence of hydroxyurea (5-FOA+HU). In contrast, the C-terminal mutants (I192A, Y193A, Y193M, I194A) and the dimer interface mutants (S155E, V134E and V161A) that behaved as monomers and had no, or diminished in vitro E3 ligase activity (Figure 3A), functioned poorly in yeast under both normal growth conditions and in response to genotoxic stress (Figure 4A). These data suggest that although the monomeric mutants that have a wild-type C-terminus can support low level ubiquitin discharge and transfer in vitro, RING dimerization is required for RNF4 function in vivo.
Figure 4. RING dimerization is required for a functional RNF4 protein in vivo.
(A) RING domain mediated dimerization is necessary for RNF4 to compensate the simultaneous loss of Rfp1 and Rfp2 in the fission yeast S. pombe. Wild-type and mutant RNF4 were tested for their ability to rescue the growth defect of the Δrfp1Δrfp2 double mutant using a plasmid shuffle assay. In addition to the RNF4-expressing plasmid, the Δrfp1Δrfp2 double mutant strain also contained a plasmid expressing Rfp2 and ura4+; the latter plasmid supports normal growth regardless of the activity of the coexpressed RNF4 and is counter-selected by 5-fluoroorotic acid (5FOA). Cells were plated at increasing dilutions onto EMM plates, or EMM plates containing 5FOA, or 5FOA plus 5 mM hydroxyurea (HU). (B) STUbL assay using diSUMO2-GST and full-length RNF4 mutant proteins as indicated. The diSUMO2-GST was Flag-tagged and its ubiquitylation was detected using an anti-Flag immunoblot.
To investigate if the ability of RNF4 to function in a cellular context depends on its ability to target SUMO for ubiquitylation, we assessed the SUMO-targeted ubiquitin ligase (STUbL) activity of bacterially-expressed full-length RNF4 proteins [5]. This experiment clearly establishes a critical role for all of the C-terminal residues in ubiquitylation of SUMO-containing substrates (Figure 4B). Likewise, the monomeric mutants V134E and V161A are inactive, consistent with their inability to function in the yeast complementation assay. Only the V134A mutant that supports yeast growth and appeared to form dimers of modest affinity retained some ability to promote the ubiquitylation of a tandem (di-) SUMO repeat substrate (Figure 4B). We suggest that the E3 ligase activity of full-length V134A RNF4 may be further promoted in a cellular context by intracellular interactions, thus causing a nearly wild-type phenotype in the growth assay (Figure 4A). For example, in vivo a polySUMO chain or other protein, may provide a platform that leads to high local concentrations of RNF4 that favor dimerization.
Concluding remarks
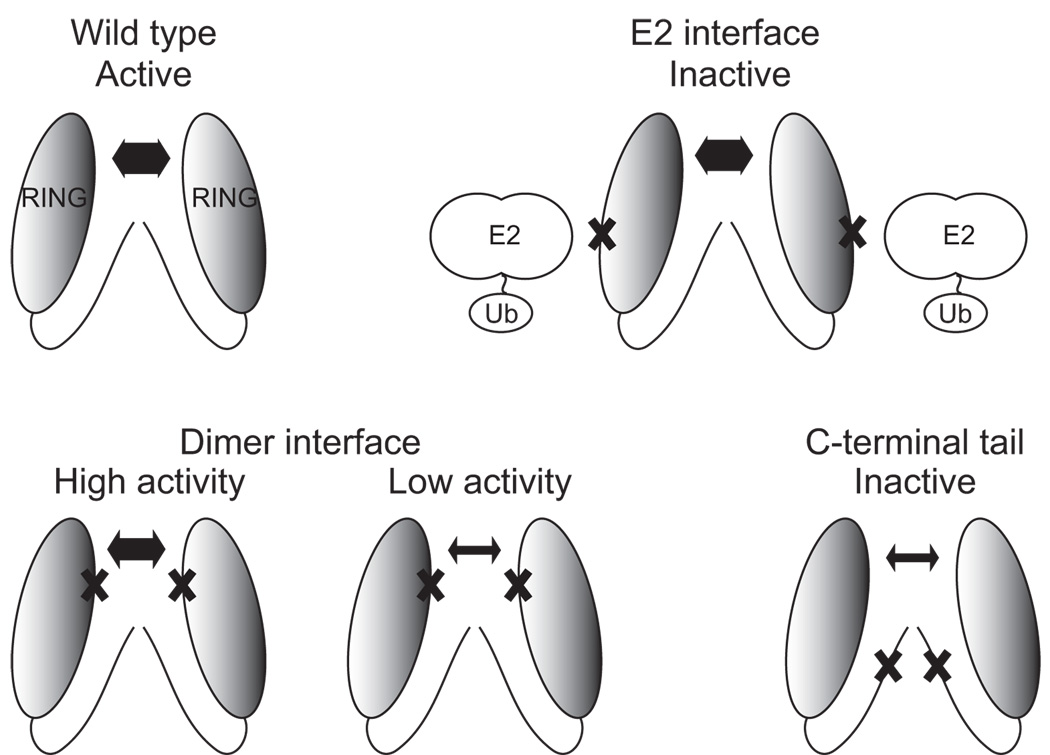
Our analysis of the structure and function of RNF4 indicates that RING dimerization and E2 recruitment are important for its E3-ligase activity and for the normal biochemical and physiological functions of RNF4 (summarized in Table 1). As a consequence disruption of either E2 recruitment or RING dimerization inactivates the E3 ligase activity of RNF4. Moreover, activity correlates with the apparent strength of the RING mediated dimer (see model in Figure 5). Our results also suggest that in addition to stabilizing RING dimerization, the C-terminal residues may have another role, because the monomeric mutants that disrupt the dimer interface retain some in vitro activity, but those that disrupt the C-terminal residues are inactive (Figure 5). Although a full understanding of the role played by the C-terminal residues remains uncertain, this observation is consistent with analysis of the RING domains from MDM2 and MDMX, which showed that mutation of the C-terminal residues disrupted ubiquitylation of the substrate, p53, but not MDM oligomerization [18].
Figure 5. The determinants of RNF4 E3 ligase activity.
Wild-type RNF4 forms a stable dimer and is an active E3 ligase. Disruption of E2 recruitment, RING dimerization or the C-terminal tail abrogates the E3 ligase activity. Predominantly monomeric mutant proteins are indicated by a thin double-headed arrow and largely dimeric proteins by a thick double-headed arrow. Crosses indicate the site of informative mutations.
Ubiquitin transfer from the E2 to the target lysine requires that the thioester bond is susceptible to attack by the incoming lysine. Our analysis of mutant RNF4 proteins suggests that RING dimerization destabilizes the E2~ubiquitin conjugate so that ubiquitin transfer is favored. This suggests that the RING domain actively contributes to ubiquitin transfer rather than just having a scaffolding role, whereby it increases transfer simply by simultaneously binding substrate and E2. Uncertainty has surrounded the role of RING domains in ubiquitin transfer because, as we observed, tighter E2 binding does not always correlate with increased activity (Figure 3) [1, 16, 19], and not all RING:E2 complexes promote transfer indicating that proximity of the E2~ubiquitin conjugate to the substrate is insufficient for activity [20, 21]. Here we suggest that although interaction with the E2~ubiquitin conjugate is required for activity, interaction between the RING domain and the E2 does not regulate the E3-ligase activity of the RNF4, instead the highly specific dimerization event regulates ubiquitin transfer by altering the stability of the thioester bond.
It remains uncertain how RING dimerization destabilizes the conjugate. Others have suggested that a RING-induced conformational change within the E2 is important for E3 ligase activity [1] and residues in the E2 that connect the E3 binding site with the catalytic center have been identified [22]. In addition, comparison of the cIAP2 RING structure in the presence and absence of the E2, UbcH5b, revealed small differences in the conformation of the C-terminal residues at the RING dimer interface [16]. However, the mechanism by which RING dimerization promotes changes in the E2 that trigger ubiquitin transfer remains unclear, and it is possible that the more rigid RING dimer stabilizes an E2~ubiquitin thioester conformation that favors catalysis. Further structural data will be required to distinguish these possibilities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter Mace and Yoshio Nakatani for help with collection and processing of x-ray data, Bodhi Bettjeman for excellent technical assistance and staff at the Australian synchrotron for assistance with data collection.
FUNDING
CLD is supported by the Marsden Fund (NZ). This work was also supported by USPHS grant CA80100 from the NCI to TH. TH is a Frank and Else Schilling American Cancer Society Professor.
Footnotes
AUTHOR CONTRIBUTION
CWL determined the crystal structure and carried out in vitro experiments; HS performed all yeast and STUbL assays; all authors interpreted the data and contributed to writing the manuscript.
Accession codes.
Protein Data Bank: Coordinates for the RNF4 RING structure have been deposited with the accession code 3NG2.
REFERENCES
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 4.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJP, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 8.Kosoy A, Calonge TM, Outwin EA, O'Connell MJ. Fission Yeast Rnf4 Homologs Are Required for DNA Repair. J. Biol. Chem. 2007;282:20388–20394. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- 9.Darst RP, Garcia SN, Koch MR, Pillus L. Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol Cell Biol. 2007;28:1361–1372. doi: 10.1128/MCB.01291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 13.Tatham M, Geoffroy M, Shen L, Plechanovova A, Hattersley N, Jaffray E, Palvimo J, Hay R. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 14.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siam R, Dolan WP, Forsburg SL. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 2004;33:189–198. doi: 10.1016/j.ymeth.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment. J. Biol. Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 17.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell. Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 18.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knipscheer P, Sixma TK. Protein-protein interactions regulate Ubl conjugation. Curr. Opin. Struct. Biol. 2007;17:665–673. doi: 10.1016/j.sbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 21.Huang A, Dejong R, Wienk H, Winkler G, Timmers H, Boelens R. E2–c-Cbl Recognition Is Necessary but not Sufficient for Ubiquitination Activity. J. Mol. Biol. 2009;385:507–519. doi: 10.1016/j.jmb.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 22.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.