Abstract
Iron deficiency is usually attributed to chronic blood loss or inadequate dietary intake. Here, we show that iron deficiency anemia refractory to oral iron therapy can be caused by germline mutations in TMPRSS6, which encodes a type II transmembrane serine protease produced by the liver that regulates the expression of the systemic iron regulatory hormone hepcidin. These findings demonstrate that TMPRSS6 is essential for normal systemic iron homeostasis in humans.
We and others have identified families with multiple individuals with iron deficiency anemia unresponsive to oral iron therapy but partially responsive to parenteral iron administration, suggesting that some cases of iron deficiency may be genetically determined1–6. We refer to this phenotype as iron-refractory iron deficiency anemia (IRIDA), the key features of which are: a congenital hypochromic, microcytic anemia, a very low mean corpuscular erythrocyte volume, a low transferrin saturation, abnormal iron absorption characterized by no hematological improvement following treatment with oral iron, and abnormal iron utilization characterized by a sluggish, incomplete response to parenteral iron (Table 1).
Table 1.
Clinical and biochemical parameters of IRIDA index cases with TMPRSS6 mutations
| Mutation 1 |
Mutation 2 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Ancestry | Consanguinity | Sex | Age at evaluation | RBC × 1012/l | Hb (g/dl) | MCV (fl) | Retics (%) | Transferrin saturation (%) | Nucleotide/amino acid change | Domain | Nucleotide/amino acid change | Domain |
| Multiplex kindreds | |||||||||||||
| A | Turkish | Yes | M | 6 y | 5.0 | 8.8 | 58 | n.d. | 2 | 1906_1907insGC (K636fs)a,b | Protease | 1906_1907insGC (K636fs)a,b | Protease |
| B | Northern European | No | F | 13 mo. | n.d. | 9.2 | 65 | 1.0 | 10 | 1813delG (A605fs)a | Protease | IVS13+1G>Aa | LDLRA |
| C | Nigerian | No | M | 17 mo. | 4.2 | 7.0 | 49 | n.d. | 5 | IVS16+1G>Ca,b | Protease | Not identified | n.a. |
| D | Northern European | No | F | 11 y | 4.9 | 8.2 | 56 | 1.6 | 3 | 1324G>A (G442R)a | CUB | 1561G>A (D521N)a | LDLRA |
| E | African American | No | M | 7 y | 5.1 | 7.5 | 49 | 0.6 | 4 | 2320C>T (R774C)b | Protease | Not identified | n.a. |
| Sporadic cases | |||||||||||||
| F | Nigerian | No | F | 3 y | 5.0 | 9.7 | 61 | 0.5 | 4 | IVS15-1G>Ca,b | Protease | IVS15-1G>Ca,b | Protease |
| G | African American | No | M | 15 mo. | 5.0 | 7.9 | 53 | 0.8 | 2 | 1065C>Ab (Y355X) | CUB | 1383delAb (E461fs) | LDLRA |
Kindreds A through E include an additional affected sibling who harbors the identical mutation(s) as the index case. Each mutation was present in heterozygous form in affected individuals, with the exception of the mutations present in the affected individuals from Kindreds A and F, which were present in homozygous form. Hematological parameters from kindreds B2,6, D5 and E4 have been previously reported. Transferrin saturation (%) was calculated by dividing the serum iron level by the total iron binding capacity and multiplying by 100. RBC, red blood cell count; Hb, hemoglobin; MCV, mean corpuscular volume; Retics, reticulocytes, CUB, complement factor C1r/C1s, urchin embryonic growth factor, and bone morphogenetic protein; LDLRA, LDL-receptor class A; n.d., not determined; n.a., not applicable. In all cases, Hb, MCV and transferrin saturation were below the respective reference ranges provided by the referring hospital laboratory.
Mutation not found in 100 control chromosomes from individuals of self-reported Caucasian ancestry.
Mutation not found in 100 control chromosomes from individuals of African-American ancestry.
To determine the genetic basis of IRIDA, we studied five multiplex kindreds. Acquired causes of iron deficiency and other inherited causes of microcytosis were rigorously excluded (Supplementary Note online). In all five families, recessive transmission was suggested by the absence of the phenotype in the parents of affected sibling pairs; one kindred was also notable for parental consanguinity (Table 1 and Supplementary Fig. 1 online). We excluded several genes involved in intestinal iron absorption and/or systemic iron utilization, including CYBRD1, HAMP, SLC11A2 and SLC40A1, as IRIDA candidates through haplotype analysis using flanking microsatellite markers and/or by sequencing coding regions and intron–exon boundaries (data not shown).
Other investigators recently described a Sardinian kindred with autosomal recessive IRIDA linked to chromosome 22q12–13 (R. Galanello, M. Cau, M.A. Melis, F. Deidda, A. Cao and M. Cazzola, unpublished data; M.A. Melis, M. Cau, R. Congiu, G. Sole, A. Cao and R. Galanello, unpublished data). The phenotype in the families we studied was similarly compatible with linkage to 22q12–13 (Supplementary Fig. 1 and Supplementary Methods online). TMPRSS6, located within the critical interval, encodes a type II transmembrane serine protease (also known as matriptase-2) that is expressed primarily in the liver7 (Supplementary Fig. 2 online), and we considered it an excellent positional candidate gene, as a recessive mutation in the mouse ortholog (Tmprss6) leads to anemia as a result of defective dietary iron uptake (E. Beutler, P. Lee, T. Gelbart, X. Du and B. Beutler, unpublished data).
We analyzed all TMPRSS6 coding regions and intron–exon boundaries and identified sequence variants in each of the five multiplex IRIDA kindreds (Table 1, Fig. 1, Supplementary Methods and Supplementary Table 1 online). Affected individuals harbored frame-shift mutations, splice junction mutations or missense mutations altering residues conserved in TMPRSS6 homologs from humans to fugu (Supplementary Fig. 3 online). In three of the four kindreds in which the phase of chromosomal segregation was known, we identified bialleic mutations. In the fourth family, we found a mutation only on the paternal allele; however, we did not exclude the presence of other types of mutations, such as large deletions, that would not be detectable by sequencing. Additionally, in the fifth kindred, for which DNA was available from only the affected individuals, we found a nonconservative missense mutation in both siblings. We also examined two individuals with sporadic IRIDA and found nonsense, frameshift or splice junction mutations in both (Table 1). None of the disease-associated variants were present in the NCBI and Ensembl SNP databases or in 100 control chromosomes (Table 1 and data not shown). These findings conclusively show that mutations in TMPRSS6 cause IRIDA.
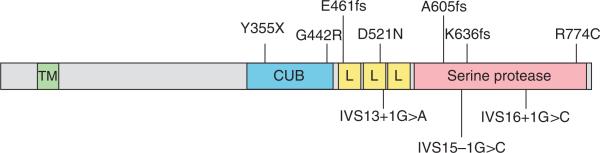
Figure 1.
Schematic representation of TMPRSS6 mutations and corresponding TMPRSS6 domains. The missense, nonsense, frameshift and splice junction mutations present in five familial and two sporadic cases of IRIDA are diagrammed adjacent to the affected TMPRSS6 domain (Table 1). The transmembrane (TM), complement factor C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein (CUB), LDL-receptor class A (L) and trypsin-like serine protease domains are shown in green, blue, yellow and red, respectively.
All of the TMPRSS6 mutations that we identified in individuals with IRIDA lie distal to exon 8, in regions that encode several conserved structural domains, most notably a trypsin-like serine protease domain (Fig. 1). This catalytic domain is highly similar to that of other type II transmembrane serine proteases, particularly the S1 family of trypsin-like serine proteases. Whether it is the putative catalytic activity of TMPRSS6 that is essential for normal systemic iron homeostasis, rather than another function of the molecule, is uncertain. Nonetheless, the finding of individuals with IRIDA harboring homozygous frameshift mutations predicted to disrupt only the catalytic domain suggests that this portion of the molecule is important for iron homeostasis. Of note, the Tmprss6 mouse mutant also results from a splice site mutation that disrupts the catalytic domain (E. Beutler, P. Lee, T. Gelbart, X. Du and B. Beutler, unpublished data).
To gain insight into the pathophysiology of IRIDA, we determined levels of hepcidin, a hormone produced by the liver that regulates intestinal iron absorption and macrophage iron release8. Under normal circumstances, hepcidin is induced by iron overload and repressed by iron deficiency and anemia. Although urinary hepcidin levels are typically undetectable in individuals with iron deficiency9, in the five affected individuals from three IRIDA kindreds we examined, urinary hepcidin/creatinine ratios were either within or above the normal range (Supplementary Note and Supplementary Table 2 online). The finding of inappropriately elevated urinary hepcidin levels in individuals with IRIDA provides insight into the pathophysiology of the disorder, as it may explain the failure to absorb dietary iron despite systemic iron deficiency, as well as the coexistent failure to respond to parenteral iron administered as iron-dextran, which must be processed and exported by macrophages before utilization for erythropoiesis.
How TMPRSS6 mutations lead to inappropriately elevated hepcidin levels remains unclear. The simplest explanation would be that TMPRSS6 normally cleaves a protein that acts in or on hepatocytes to negatively regulate hepcidin production, secretion or clearance. Studies in the Tmprss6 mouse mutant suggest that TMPRSS6 is a negative regulator of hepcidin transcription (E. Beutler, P. Lee, T. Gelbart, X. Du and B. Beutler, unpublished data). This transcriptional effect might be achieved by cleaving a protein that upregulates a pathway that normally represses hepcidin transcription, or that down-regulates a pathway that normally activates hepcidin transcription.
The identification of TMPRSS6 mutations in individuals with IRIDA has broad implications for clinical disorders of iron metabolism. Mutations or polymorphisms in TMPRSS6 may contribute to iron deficiency anemia in individuals with or without other predisposing factors. Furthermore, the finding that TMPRSS6 regulates hepcidin levels in humans may have potential applications for treatment of iron disorders. For example, inhibition of the putative protease function of TMPRSS6 might be a potential treatment for disorders in which hepcidin is inappropriately low, such as primary hemochromatosis and iron loading anemias. Similarly, treatment with agonists or with the endogenous substrate of TMPRSS6 might be employed in the anemia of chronic disease, in which hepcidin is inappropriately high.
Supplementary Material
ACKNOWLEDGMENTS
We thank the families for their invaluable contribution to this study. We are indebted to E. Neufeld for ongoing mentorship. We thank C. Trenor, A. Donovan, I. Rubio-Aliaga and other members of the Andrews laboratory for their contributions to the early stages of this project. We thank A.J. Iafrate and J. Miller for technical advice and assistance. K.E.F. was supported by T32 CA009216 awarded to the Department of Pathology, Massachusetts General Hospital. This work was also supported by R01 DK080011 (M.D.F.), K12 HL087164 (M.M.H.), R01 DK066373 (N.C.A.) and DK053813 (N.C.A.).
Footnotes
AUTHOR CONTRIBUTIONS
K.E.F. obtained institutional review board approval and consents, designed, conducted and interpreted results from the segregation studies and sequence analysis, and prepared the manuscript. M.M.H. obtained institutional review board approval and consents and coordinated clinical sample acquisition and clinical data analysis. D.R.C. conducted and interpreted results from the sequencing analysis and assisted with the segregation studies and other technical aspects of the project. Y.A., H.A.P., K.R.H., M.M.M. S.M.S. and J.J.S. were clinical collaborators who provided samples from affected individuals, phenotypic information and results of laboratory testing. K.M. interpreted results from the segregation studies and sequencing analysis. N.C.A. and M.D.F. obtained institutional review board approval and consents, supervised the design of experiments and data interpretation, and prepared the manuscript. N.C.A. also provided samples from affected individuals, phenotypic information and results of laboratory testing.
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Andrews NC. Yale J. Biol. Med. 1997;70:219–226. [PMC free article] [PubMed] [Google Scholar]
- 2.Brown AC, et al. Am. J. Hematol. 1988;27:1–6. doi: 10.1002/ajh.2830270102. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan GR, Sheehan RG. J. Pediatr. 1981;98:723–728. doi: 10.1016/s0022-3476(81)80831-1. [DOI] [PubMed] [Google Scholar]
- 4.Hartman KR, Barker JA. Am. J. Hematol. 1996;51:269–275. doi: 10.1002/(SICI)1096-8652(199604)51:4<269::AID-AJH4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Mayo MM, Samuel SM. Clin. Lab. Sci. 2001;14:135–138. [PubMed] [Google Scholar]
- 6.Pearson HA, Lukens JN. J. Pediatr. Hematol. Oncol. 1999;21:412–417. doi: 10.1097/00043426-199909000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. J. Biol. Chem. 2002;277:37637–37646. doi: 10.1074/jbc.M203007200. [DOI] [PubMed] [Google Scholar]
- 8.Nemeth E, Ganz T. Annu. Rev. Nutr. 2006;26:323–342. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- 9.Kemna EH, Tjalsma H, Podust VN, Swinkels DW. Clin. Chem. 2007;53:620–628. doi: 10.1373/clinchem.2006.079186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.