Abstract
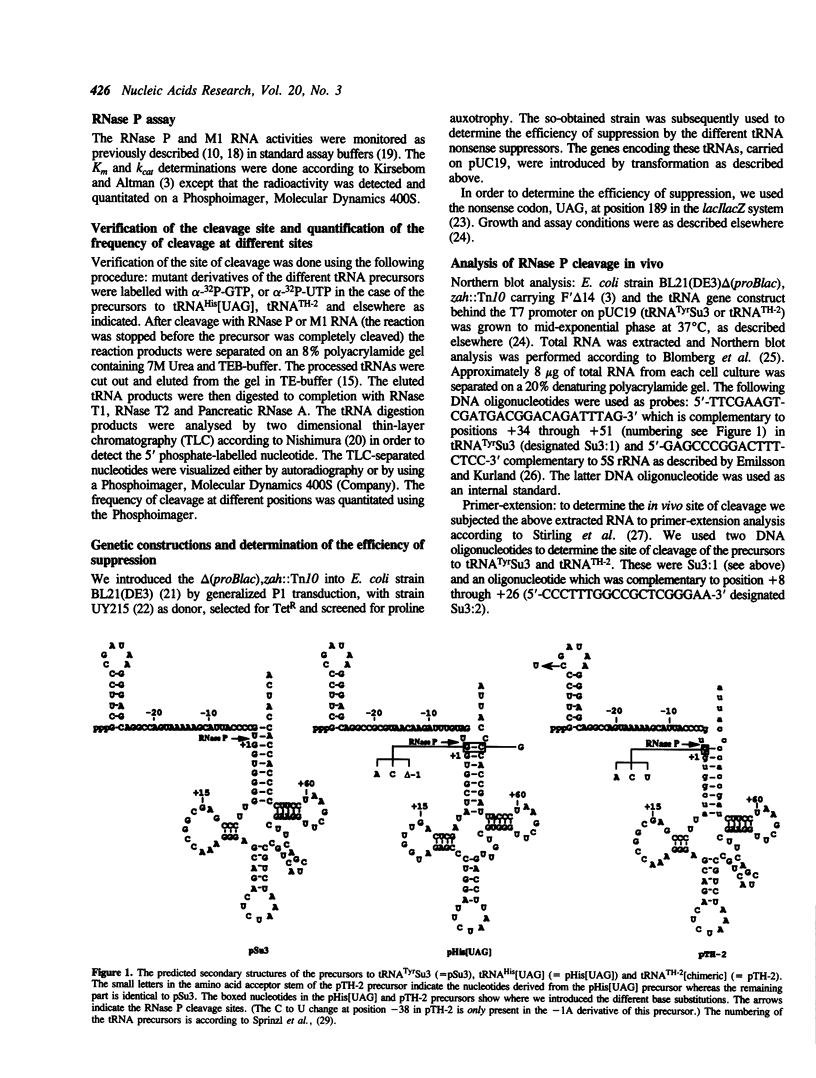
Cleavage by RNase P of the tRNA(His precursor yields a mature tRNA with an 8 base pair amino acid acceptor stem instead of the usual 7 base pair stem. Here we show, both in vivo and in vitro, that this is mainly dependent on the primary structure and length of the acceptor stem in the precursor. Furthermore, the tRNA(His) precursor used in this study was processed with a change in both kinetic constants, Km and kcat, in comparison to the kinetics of cleavage of the precursor to tRNA(Tyr)Su3. Cleavage of a chimeric tRNA precursor showed that these altered kinetics were due to a difference in the primary structure and in the length of the acceptor stems of these two tRNA precursors. We also studied the cleavage reaction as a function of base substitutions at positions -1 and/or +73 in the precursor to tRNA(His). Our results suggest that the nucleotide at position +73 in tRNA(His) plays a significant role in the kinetics of cleavage of its precursor, possibly in product release. In addition, it appears that the C5 protein of RNase P is involved in the interaction between the enzyme and its substrate in a substrate-dependent manner, as previously suggested.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Smith J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971 Sep 8;233(36):35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- Blomberg P., Wagner E. G., Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990 Jul;9(7):2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard U., Söll D. The unusually long amino acid acceptor stem of Escherichia coli selenocysteine tRNA results from abnormal cleavage by RNase P. Nucleic Acids Res. 1988 Dec 23;16(24):11617–11624. doi: 10.1093/nar/16.24.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard U., Willis I., Söll D. Processing of histidine transfer RNA precursors. Abnormal cleavage site for RNase P. J Biol Chem. 1988 Feb 15;263(5):2447–2451. [PubMed] [Google Scholar]
- Emilsson V., Kurland C. G. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 1990 Dec;9(13):4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8655–8659. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Structural requirements for processing of synthetic tRNAHis precursors by the catalytic RNA component of RNase P. J Biol Chem. 1988 Jan 15;263(2):652–657. [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., van Belkum A., Pleij C. W., Altman S. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988 Apr 22;53(2):267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989 Oct 11;17(19):7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. M., Klee H. J., Zagorski J., Fournier M. J. Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs. J Bacteriol. 1984 Jun;158(3):934–942. doi: 10.1128/jb.158.3.934-942.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Altman S. Reaction in vitro of some mutants of RNase P with wild-type and temperature-sensitive substrates. J Mol Biol. 1989 Jun 20;207(4):837–840. doi: 10.1016/0022-2836(89)90250-7. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Baer M. F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J Mol Biol. 1988 Dec 20;204(4):879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Isaksson L. A. Functional interactions in vivo between suppressor tRNA and mutationally altered ribosomal protein S4. Mol Gen Genet. 1986 Nov;205(2):240–247. doi: 10.1007/BF00430434. [DOI] [PubMed] [Google Scholar]
- Krupp G., Kahle D., Vogt T., Char S. Sequence changes in both flanking sequences of a pre-tRNA influence the cleavage specificity of RNase P. J Mol Biol. 1991 Feb 20;217(4):637–648. doi: 10.1016/0022-2836(91)90522-8. [DOI] [PubMed] [Google Scholar]
- Mans R. M., Guerrier-Takada C., Altman S., Pleij C. W. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990 Jun 25;18(12):3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Farabaugh P. J. Correlation of nonsense sites in the lacI gene with specific codons in the nucleotide sequence. Nature. 1978 Aug 24;274(5673):770–775. doi: 10.1038/274770a0. [DOI] [PubMed] [Google Scholar]
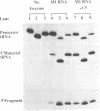
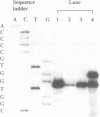
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols M., Söll D., Willis I. Yeast RNase P: catalytic activity and substrate binding are separate functions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1379–1383. doi: 10.1073/pnas.85.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana O., Cooley L., Söll D. The additional guanylate at the 5' terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol. 1986 Feb;6(2):525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck-Miller K. A., Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991 Sep 5;221(1):1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- Reich C., Olsen G. J., Pace B., Pace N. R. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988 Jan 8;239(4836):178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. A., Hulton C. S., Waddell L., Park S. F., Stewart G. S., Booth I. R., Higgins C. F. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989 Aug;3(8):1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol. 1988 Aug 20;202(4):835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]