Abstract
The objective of this study was to characterize the lipidome and electron transport chain activities in purified non-synaptic (NS) and synaptic (Syn) mitochondria from C57BL/6J mouse cerebral cortex. Contamination from subcellular membranes, especially myelin, has hindered past attempts to accurately characterize the lipid composition of brain mitochondria. An improved Ficoll and sucrose discontinuous gradient method was employed that yielded highly enriched mitochondrial populations free of myelin contamination. The activities of Complexes I, II, III, and II/III were lower in Syn than in NS mitochondria, while Complexes I/III and IV activities were similar in both populations. Shotgun lipidomics showed that levels of cardiolipin (Ptd2Gro) were lower, whereas levels of ceramide and phosphatidylserine were higher in Syn than in NS mitochondria. Coenzyme Q9 and Q10 was also lower in Syn than in NS mitochondria. Gangliosides, phosphatidic acid, sulfatides, and cerebrosides were undetectable in brain mitochondria. The distribution of Ptd2Gro molecular species was similar in both populations and formed a unique pattern, consisting of seven major molecular species groups, when arranged according to mass to charge ratios. Remodeling involving choline and ethanolamine phosphoglycerides could explain Ptd2Gro heterogeneity. NS and Syn mitochondrial lipidomic heterogeneity could influence energy metabolism, which may contribute to metabolic compartmentation of the brain.
Keywords: cardiolipin, lipidome, myelin, non-synaptic, shotgun lipidomics, synaptic
Lipids of the mitochondrial membrane can influence numerous mitochondrial functions to include electron transport chain (ETC) activities, nucleotide transport, mitochondrial protein import, membrane fluidity/permeability properties, and ATP synthesis (Petrushka et al. 1959; Daum 1985; Hoch 1992; Brand et al. 2003; Shinzawa-Itoh et al. 2007). Brain contains at least two major populations of mitochondria, based on cellular localization (Lai and Clark 1989). These include the non-synaptic (NS) mitochondria, which originate from neuronal and glial cell bodies, and the synaptic (Syn) mitochondria, which primarily originate from the Syn bouton of neurons. These mitochondrial populations express differences in specific ETC activities, believed to contribute to the metabolic compartmentation of the brain (Lai et al. 1977; Battino et al. 1991). The origin of this metabolic compartmentation could arise from differences in lipid composition of the mitochondrial membrane. However, contamination from subcellular membranes to include myelin has hindered past attempts to accurately characterize the lipid composition of brain mitochondria (Cuzner and Davison 1968; Lai and Clark 1989; Villa et al. 1989b; Bangur et al. 1995).
Shotgun lipidomics by electrospray ionization mass spectrometry (ESI/MS) can provide a rapid and detailed analysis of lipids in biological membranes (Han and Gross 2005a). This approach recently identified nearly 100 molecular species of brain cardiolipin (1,3-diphosphatidyl-sn-glycerol, Ptd2Gro), a mitochondrial specific lipid (Cheng et al., unpublished observation). The origin of brain Ptd2Gro heterogeneity is not well defined, but may be related to the unique synthesis of Ptd2Gro (Hauff and Hatch 2006; Schlame and Ren 2006). Ptd2Gro is synthesized as immature Ptd2Gro through the condensation of phosphatidylglycerol (PtdGro) and CDP-diacylglycerol. Ptd2Gro is then deacylated to monolysocardiolipin (MLCL) and reacylated to form mature Ptd2Gro (Hauff and Hatch 2006). This Ptd2Gro remodeling process uses fatty acyl groups from the sn-2 position of donor choline (ChoGpl) and ethanolamine (EtnGpl) glycerophospholipids (Xu et al. 2003; Schlame and Ren 2006). No prior studies have evaluated Ptd2Gro heterogeneity in purified NS and Syn mitochondrial populations.
The objective of this study was to analyze the lipid composition of NS and Syn mitochondria. We designed a modified Ficoll and sucrose discontinuous gradient system for the isolation and purification of mitochondria from mouse brain free of contaminating myelin or organelle and plasma membranes (Lai et al. 1977; Dagani et al. 1983). Shotgun lipidomics and HPLC connected to an electrochemical detector were used to determine if quantitative or qualitative differences in lipid composition existed between these two brain mitochondrial populations. Moreover, ours is the first comprehensive examination of the brain mitochondrial lipidome, including molecular species and mass content of ceramide (Cer), PtdGro, and lysophosphatidylcholine (LysoPtdCho). ETC activities were measured to determine if differences in enzyme activities correlated with differences in the lipid composition of mitochondrial populations. Our results provide new insight into the importance of brain mitochondrial lipids, which could contribute to metabolic compartmentation of the brain.
Materials and methods
Animals
C57BL/6J (B6) mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). All mice were propagated at the Boston College Animal Facility and were housed in plastic cages with filter tops containing Sani-Chip bedding (P. J. Murphy Forest Products Corp., Montville, NJ, USA). The room was maintained at 22°C on a 12 h light/dark cycle. Food (Prolab RMH 3000; PMI LabDiet, Richmond, IN, USA) and water were provided ad libitum. This study was conducted with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and was approved by the Institutional Animal Care Committee.
Non-synaptic and synaptic brain mitochondrial isolation
C57BL/6J mice (4 months of age) were killed by cervical dislocation and the cerebral cortex was dissected. Mitochondria were isolated in a cold room (4°C) and all reagents were kept on ice. The isolation procedure employed a combination of gradients and strategies as previously described (Lai and Clark 1976; Lai et al. 1977; Mena et al. 1980; Dagani et al. 1983; Rendon and Masmoudi 1985; Battino et al. 1991; Brown et al. 2006) (Fig. 1). The cerebral cortexes (a pool of six per sample) were diced on an ice cold metal plate and then placed in 12 mL of mitochondria isolation buffer (MIB; 0.32 M sucrose, 10 mM Tris–HCl, and 1 mM EDTA-K, pH 7.4). The pooled cerebral cortexes were homogenized using a Potter Elvehjem homogenizer with a Teflon coated pestle attached to a hand-held drill (Wheaton, Millville, NJ, USA). Samples were homogenized using 15 up- and downstrokes at 500 rpm. The homogenate was centrifuged at 1000 g for 5 min. The supernatant was collected and the pellet was washed twice by centrifugation at 1000 g for 5 min, collecting the supernatants each time. The supernatants were pooled and centrifuged at 1000 g for 5 min. The collected supernatant was then spun at 14 000 g for 15 min. The supernatant was discarded and the pellet, which contained primarily NS mitochondria, synaptosomes, and myelin, was resuspended in 12 mL MIB and was layered on a 7.5/12% discontinuous Ficoll gradient. Each Ficoll gradient layer contained 12 mL for a total volume of 36 mL. The Ficoll gradients were made from a 20% Ficoll stock with MIB. The gradient was centrifuged at 73 000 g for 36 min (4°C) in a Sorvall SW 28 rotor with slow acceleration and deceleration (Optima L-90K Ultracentrifuge, Beckman Coulter, Fullerton, CA, USA). The centrifugation time used permitted sufficient acceleration and deceleration to achieve maximum g force (Battino et al. 1991) and to prevent synaptosomal contamination of the mitochondrial fraction below the 12% Ficoll layer. Crude myelin collected at the MIB/7.5% Ficoll interface was discarded. Synaptosomes were collected at the 7.5/12% interface and were resuspended in MIB and centrifuged at 16 000 g for 15 min. The Ficoll gradient purified NS mitochondria (FM) were collected as a pellet below 12% Ficoll.
Fig. 1.
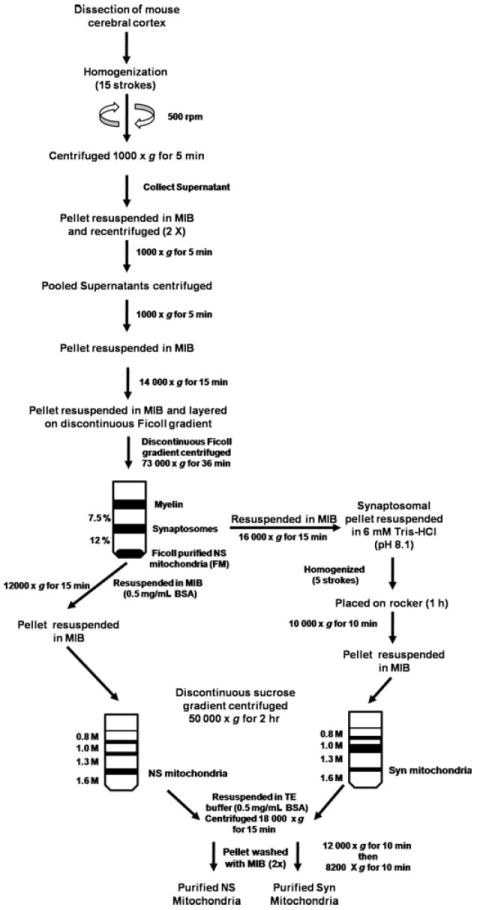
Procedure used for the isolation and purification of NS and Syn mitochondria from mouse cerebral cortex.
Purification of non-synaptic mitochondria
The FM pellet, containing NS mitochondria, was resuspended in MIB containing 0.5 mg/mL bovine serum albumin (BSA) and was centrifuged at 12 000 g for 15 min. The resulting pellet was collected and resuspended in 6 mL of MIB. The resuspended FM pellet was layered on a discontinuous sucrose gradient containing 0.8/1.0/1.3/1.6 M sucrose. The volumes for the sucrose gradient were 6/6/10/8 mL, respectively. The gradients were made from a 1.6 M sucrose stock containing 1 mM EDTA-K and 10 mM Tris–HCl, pH 7.4. The discontinuous sucrose gradient was centrifuged at 50 000 g for 2 h (4°C) in a Sorvall SW 28 rotor using slow acceleration and deceleration to prevent disruption of the gradient. Purified NS mitochondria were collected at the interface of 1.3 and 1.6 M sucrose. NS mitochondria were collected and resuspended in (1 : 3, v/v) Tris-EDTA buffer (1 mM EDTA-K and 10 mM Tris–HCl, pH 7.4) containing 0.5 mg/mL BSA and centrifuged at 18 000 g for 15 min. The pellet was then resuspended in MIB and centrifuged at 12 000 g for 10 min. The pellet was again resuspended in MIB and centrifuged at 8200 g for 10 min.
Purification of synaptic mitochondria
Synaptosomes were burst by homogenization in 6 mM Tris–HCl, pH 8.1, using five up- and downstrokes. The homogenized synaptosomes were transferred to a 15 mL conical tube and then placed on a rocker for 1 h (4°C). The burst synaptosomes were centrifuged at 10 000 g for 10 min. The pellet was resuspended in 6 mL of MIB. The resuspended pellet was layered on a discontinuous sucrose gradient and centrifuged following the same procedure as described above for NS mitochondria.
Western blot analysis of subcellular fractions
Protein concentration of isolated subcellular fractions was determined by the Dc Protein Assay using BSA standards (Bio-Rad, Hercules, CA, USA). Total protein (2–20 μg) from fractions were loaded on 4–12% NuPage Bis–Tris gradient gels using MES sodium dodecyl sulfate running buffer (Invitrogen, Carlsbad, CA, USA) and electrophoresed. Proteins were transferred to an immobilon TM-P membrane (Millipore, Bedford, MA, USA) for 2 h at 80 V at 4°C. The TM-P membrane was then washed with Tris- or phosphate-buffered saline (PBS), depending on manufacturers’ instructions for primary antibody and then blocked in 5% non-fat powdered milk in Tris-buffered saline or PBS with Tween 20, pH 7.6, for 1 h or overnight. Primary antibodies were used at the dilutions suggested by the manufacturers’ protocols and included Complex IV, subunit IV (1 : 1250) (clone 20E8; Molecular Probes, Eugene, OR, USA), monoamine oxidase-A (1 : 200) (clone T-19; Santa Cruz Biotechnology, Santa Cruz, CA, USA), SNAP25 (1 : 10 000) (clone, SMI 81; Sternberger monoclonal, Lutherville, MD, USA), β-actin (1 : 7000) (AC-15; Abcam, Cambridge, UK), tuberin (1 : 400) (clone C-20; Santa Cruz Biotechnology), proliferating cell nuclear antigen (1 : 100) (clone PC10; Dako, Carpinteria, CA, USA), calnexin (1 : 500) (clone C8.B6; Cell Signaling, Beverly, MA, USA), and proteolipid protein (1 : 200) (clone AA3). Also, proteins representative of individual ETC Complexes (I–V) were evaluated using the Rodent Total OXPHOS Complexes Detection Kit (1 : 500) (MS604; Mitosciences, Eugene, OR, USA). The kit consisted of antibodies to Complex I (ND6), Complex II (FeS 30 kDa), Complex III (Core 2 subunit), Complex IV (subunit I), and Complex V (α subunit). Blots were incubated with corresponding secondary antibody conjugated with horseradish peroxidase. Bands were visualized using the Pierce chemiluminescence detection system (Pierce, Rockford, IL, USA). Band densities were quantified using a densitometer (Molecular Dynamics, Sunnyvale, CA, USA) and IQ Mac software (version 1.2, GE Healthcare, Piscataway, NJ, USA). Protein was loaded in incremental amounts to confirm that the signal detected was not saturated.
Ganglioside GM1a analysis using cholera toxin b immunostaining
Gangliosides were isolated and purified from subcellular fractions as we previously described (Hauser et al. 2004). The ganglioside fraction was desalted by LH-20 Sephadex column chromatography (Varian, Harbor City, CA, USA). Purified gangliosides were spotted on a plastic backed Nagel PolyGram Sil G TLC plate (Macherey-Nagel, Germany) based on 100 μg protein, as previously described (Hirabayashi et al. 1990). Immunostaining for GM1a was performed as previously described (Brigande et al. 1998). Approximately 1.6 ng of purified GM1a was spotted as a standard. The concentration of GM1a standard used was calculated based its level of immunoreactivity with cholera toxin b. The TLC plate was developed in one ascending run for 45 min in CHCl3/CH3OH/0.2% CaCl2 (55 : 45 : 10). The plate was thoroughly dried after development and was then blocked in PBS containing 1% BSA for 2 h at 37°C on a rocker. The plate was then washed with PBS for 5 min. Next, the plate was incubated with the β-subunit of cholera toxin/peroxidase conjugated (1 μg/mL) (List Biological Laboratories Inc., Campbell, CA, USA) in PBS containing 1% BSA for 2 h at 37°C. Post-incubation, the plate was washed five times (5 min each time) with PBS and was then developed using 10 mL PBS, 2 mL 4-chloro-1-napthol (3 mg/mL in CH3OH), and 20 μL 30% H2O2 at 22°C for 15 min.
Materials for mass spectrometry
Synthetic phospholipids including 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (14:1-14:1 PtdCho), 1,2-dipalmitoleoyl-sn-glycero-3-phosphoethanolamine (16:1-16:1 PtdEtn), 1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol (15:0-15:0 PtdGro), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (14:0-14:0 phosphatidylserine; PtdSer), N-lauroryl sphingomyelin (N12:0 CerPCho), 1,1′,2,2′-tetramyristoyl cardiolipin (T14:0 Ptd2Gro), heptadecanoyl ceramide (N17:0 Cer), and 1-heptadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine (17:0 LysoPtdCho) were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA). It should be noted that the prefix ‘N’ denotes the amide-linked acyl chain. Deuterated cholesterol was purchased from Cambridge Isotope Laboratories Inc. (Cambridge, MA, USA). All the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Burdick and Jackson, Muskegon, MI, USA). All other chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA).
Sample preparation for mass spectrometric analysis
An aliquot of the mitochondrial preparation was transferred to a disposable culture borosilicate glass tube (16 × 100 mm). Internal standards were added based on protein concentration and included 16:1-16:1 PtdEtn (100 nmol/mg protein), 14:1-14:1 PtdCho (45 nmol/mg protein), T14:0 Ptd2Gro (3 nmol/mg protein), 15:0-15:0 PtdGro (7.5 nmol/mg protein), 14:0-14:0 PtdSer (20 nmol/mg protein), 17:0 LysoPtdCho (1.5 nmol/mg protein), N12:0 CerPCho (20 nmol/mg protein), and N17:0 Cer (5 nmol/mg protein). This allowed the final quantified lipid content to be normalized to the protein content and eliminated potential loss from the incomplete recovery. The molecular species of internal standards were selected because they represent < 0.1% of the endogenous cellular lipid mass as demonstrated by ESI/MS lipid analysis.
A modified Bligh and Dyer procedure was used to extract lipids from each mitochondrial preparation as previously described (Cheng et al. 2006). Each lipid extract was reconstituted with a volume of 500 μL/mg protein (which was based on the original protein content of the samples as determined from protein measurement) in CHCl3/MeOH (1 : 1, v/v). The lipid extracts were flushed with nitrogen, capped, and stored at −20°C for ESI/MS analysis. Each lipid solution was diluted approximately 50-fold immediately prior to infusion and lipid analysis.
Instrumentation and mass spectrometry
A triple-quadrupole mass spectrometer (Thermo Scientific TSQ Quantum Ultra, Plus, San Jose, CA, USA), equipped with an electrospray ion source and Xcalibur system software, was utilized as previously described (Han et al. 2004). The first and third quadrupoles serve as independent mass analyzers using a mass resolution setting of peak width 0.7 Th while the second quadrupole serves as a collision cell for tandem MS. The diluted lipid extract was directly infused into the ESI source at a flow rate of 4 μL/min with a syringe pump. Typically, a 2-min period of signal averaging in the profile mode was employed for each mass spectrum. For tandem MS, a collision gas pressure was set at 1.0 mTorr, but the collision energy varied with the classes of lipids as described previously (Han et al. 2004; Han and Gross 2005b). Typically, a 2- to 5-min period of signal averaging in the profile mode was employed for each tandem MS spectrum. All the mass spectra and tandem MS spectra were automatically acquired by a customized sequence subroutine operated under Xcalibur software. Data processing of 2D MS analyses including ion peak selection, data transferring, peak intensity comparison, and quantitation was conducted using self-programmed MicroSoft Excel macros (Han et al. 2004).
Mathematical computation for predicting the distribution of cardiolipin molecular species
A PERL script was developed for predicting the distribution of molecular species in mouse brain Ptd2Gro. This program calculated the expected concentration F × c1 × c2 × c3 × c4 for each Ptd2Gro species, given the concentration of each fatty acid found in Ptd2Gro; c represents a given concentration of fatty acids in any one of the four possible fatty acid tails of Ptd2Gro. F was calculated for each possible Ptd2Gro molecular species using an enumerative approach, with trivial computation time.
Isolation and quantification of coenzyme Q
Aliquots of NS and Syn mitochondria were subjected to lipid extraction and analyzed for Q content as previously described (Jonassen et al. 2002). Q6 was used to gauge coenzyme Q recovery in the lipid extracts and was added to all standards and samples. Lipids were extracted by adding 0.05 mL water, 0.9 mL methanol, and 0.6 mL petroleum ether to each sample. The mixtures were vortexed for 1 min, centrifuged at 1422 g, and the top layer of petroleum ether was removed from each mixture and saved in a separate vial. Fresh petroleum ether (0.6 mL) was added to each vial containing the aqueous phase and vortexed for 1 min. The vials were subjected to centrifugation as before and the second petroleum ether layer removed. The process was repeated once more, and the three pooled petroleum ether fractions were dried under nitrogen and resuspended in 150 μL methanol. The Q6 and Q10 standards were from Sigma-Aldrich, and Q9 was a gift from Dr Youssef Hatefi.
The coenzyme Q present in extracted standards and samples were separated and quantified by HPLC connected to an electrochemical detector as previously described (Jonassen et al. 2002), with the following exceptions: the pre-column electrode was the only electrode used and was set at +650 mV to oxidize all coenzyme Q, and a Gilson 118 UV/Vis detector (Gilson, Middletown, WI, USA) was utilized to detect coenzyme Q as they eluted from the column. The amounts of Q9 and Q10 in the standards and samples were normalized to the amount of Q6 recovered in the individual lipid extracts.
Electron transport chain enzyme activities
Purified mitochondrial samples were freeze-thawed three times before use in enzyme analysis to give substrate access to the inner mitochondrial membrane. All assays were performed on a temperature controlled SpectraMax M5 plate reader (Molecular Devices, Palo Alto, CA, USA) and were performed in triplicate. Specific enzyme activities were calculated using ETC complex inhibitors to subtract background activities.
Complex I
Complex I (NADH-ubiquinone oxidoreductase) activity was determined by measuring the decrease in the concentration of NADH at 340 nm as previously described (Birch-Machin and Turnbull 2001; Ellis et al. 2005). The assay was performed in buffer containing 50 mM potassium phosphate, pH 7.4, 2 mM KCN, 5 mM MgCl2, 2.5 mg/mL BSA, 2 μM antimycin, 100 μM decylubiquinone, and 0.3 mM K2NADH. The reaction was initiated by adding purified mitochondria (20 μg). The enzyme activity was measured for 5 min and values were recorded 30 s after the initiation of the reaction. Specific activities were determined by calculating the slope of the reaction in the linear range in the presence or absence of 1 μM rotenone (Complex I inhibitor).
Complex II
Complex II (succinate decylubiquinone 2,6-dichloroindophenol (DCIP) oxidoreductase) activity was determined by measuring the reduction of DCIP at 600 nm as previously described (King 1967; Ellis et al. 2005). The Complex II assay was performed in buffer containing 25 mM potassium phosphate, pH 7.4, 20 mM succinate, 2 mM KCN, 50 μM DCIP, 2 μg/mL rotenone, and 2 μg/mL antimycin. Purified mitochondria (10 μg) were added prior to initiation of the reaction. The reaction was initiated by adding 56 μM decylubiquinone. Specific activities were determined by calculating the slope of the reaction in the linear range in the presence or absence of 0.5 mM thenoyltrifluoroacetone (Complex II inhibitor).
Complex III
Complex III (ubiquinol-cytochrome c reductase) activity was determined by measuring the reduction of cytochrome c at 550 nm. The Complex III assay was performed in buffer containing (25 mM potassium phosphate, pH 7.4, 1 mM EDTA, 1 mM KCN, 0.6 mM dodecyl maltoside, and 32 μM oxidized cytochome c) using purified mitochondria (2.5 μg). The reaction was initiated by adding 35 μM decylubiquinol. The reaction was measured following the linear slope for 1 min in the presence or absence of 2 μM antimycin (Complex III inhibitor). Decylubiquinol was made by dissolving decylubiquinone (10 mg) in 2 mL acidified ethanol, pH 2, and using sodium dithionite as a reducing agent. Decylubiquinol was further purified by cyclohexane (Birch-Machin and Turnbull 2001; Degli Esposti 2001; Ellis et al. 2005).
Complex IV
Complex IV (cytochrome c oxidase) activity was determined by measuring the oxidation of ferrocytochrome c at 550 nm. The Complex IV assay was performed in buffer containing (10 mM Tris–HCl and 120 mM KCl, pH 7.0) using purified mitochondria (5 μg). The reaction was initiated by adding 11 μM reduced ferrocytochrome c and monitoring the slope for 30 s in the presence or absence of 2.2 mM KCN (Complex IV inhibitor) (Yonetan 1967; Ellis et al. 2005). Cytochrome c oxidase was additionally measured by a multiplexing assay (MS446; Mitosciences).
Complex I/III
Complex I/III (NADH cytochrome c reductase) activity was determined by measuring the reduction of cytochrome c at 550 nm. The Complex I/III assay was performed in buffer (50 mM potassium phosphate, pH 7.4, 1 mM EDTA, 2 mM KCN, 32 μM oxidized cytochrome c, and 105 μM K2NADH) and was initiated by adding purified mitochondria (10 μg). The reaction was measured for 30 s with a linear slope in the presence or absence of 1 μM rotenone and 2 μM antimycin (Complexes I and III inhibitors) (Birch-Machin and Turnbull 2001; Degli Esposti 2001; Ellis et al. 2005).
Complex II/III
Complex II/III (succinate cytochrome c reductase) activity was measured following the reduction of cytochrome c at 550 nm. The Complex II/III assay was performed in buffer (25 mM potassium phosphate, pH 7.4, 20 mM succinate, 2 mM KCN, 2 μg/mL rotenone) using purified mitochondria (10 μg). The reaction was initiated by adding 40 μM oxidized cytochrome c in the presence or absence of 2 μM antimycin (Complex III inhibitor) (Birch-Machin and Turnbull 2001; Ellis et al. 2005).
Results
Analysis of mitochondrial purity
Ficoll as well as sucrose discontinuous gradients were used to purify NS and Syn mitochondria (Fig. 1). As FM contained markers for cytoskeletal (β-actin) and membrane (SNAP25, PCNA, tuberin, PLP, and calnexin) contamination, we further purified mitochondria using a discontinuous sucrose gradient. None of these markers were present in the Ficoll and sucrose discontinuous gradient purified mitochondria, which contained only mitochondrial-enriched markers representing the inner mitochondrial membrane (Complex IV, subunit IV) and the outer mitochondrial membrane (monoamine oxidase-A) (Fig. 2a). Cholera toxin b immunostaining is a sensitive procedure for detecting gangliosides with the GM1a structure in cells and tissues (Brigande et al. 1998). The toxin can have slight cross-reactivity with GD1a. GM1a and a low amount of GD1a was found in the total homogenate, crude myelin, and FM fractions, indicating the presence of myelin and microsomal membranes in these subcellular fractions (Fig. 2b). Only a trace amount of GM1a was detected in the NS mitochondria and no GM1a was detected in Syn mitochondria. These finding attest to the high degree of mitochondrial purity achieved with the isolation procedure and also indicate that GM1a is not present in B6 mouse brain mitochondria.
Fig. 2.
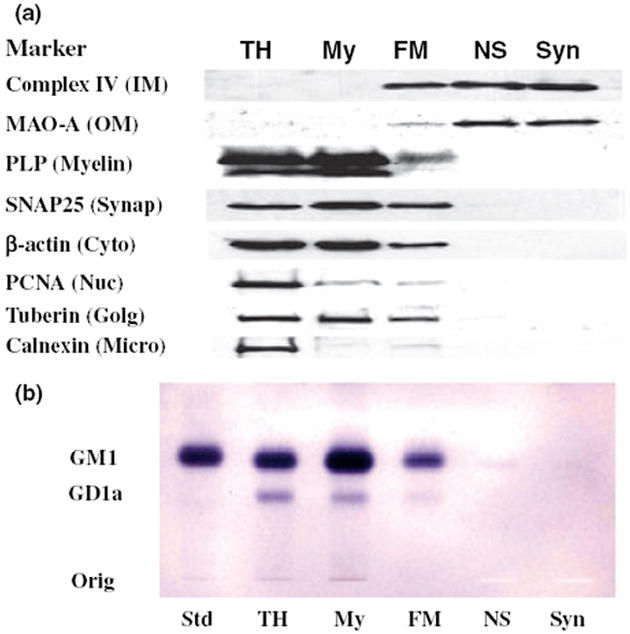
Distribution of protein markers on western blots (a) and of gangliosides on TLC (b) in subcellular fractions from mouse cerebral cortex. Subcellular fractions included total homogenate (TH), crude myelin (My), Ficoll gradient purified NS mitochondria (FM), Ficoll and sucrose gradient purified non-synaptic mitochondria (NS), and Ficoll and sucrose gradient purified synaptic mitochondria (Syn). Western blots were performed to determine the distribution of specific protein markers for the inner mitochondria membrane (Complex IV, subunit IV), outer mitochondrial membrane (monoamine oxidase-A), myelin (proteolipid protein), synaptosomal membrane (SNAP25), cytoskeleton (β-actin), nuclear membrane (proliferating cell nuclear antigen), Golgi membrane (tuberin), and microsomal membrane (calnexin). GM1a was visualized on TLC plates with cholera toxin b immunostaining as described in Materials and methods. Std, is GM1a.
Lipid composition of C57BL/6J mouse brain mitochondria
We used shotgun lipidomics to evaluate lipid content and distribution of fatty acid molecular species in the NS and Syn mitochondria. The lipid classes were listed according to their relative abundance (Table 1). Although the content of most lipids was similar in the NS and Syn mitochondria, the content of Ptd2Gro was lower whereas the content of PtdSer and Cer were higher in the Syn mitochondria than in the NS mitochondria. The myelin-enriched lipids, sulfatides and cerebrosides, were not detected in either NS or Syn mitochondria.
Table 1.
Lipid composition of C57BL/6J mouse brain mitochondria
| Lipid | Non-synaptic | Synaptic |
|---|---|---|
| Ethanolamine glycerophospholipids | 187.4 ± 12.1 | 211.7 ± 21.3 |
| Phosphatidylethanolamine | 164.9 ± 10.0 | 184.6 ± 20.3 |
| Plasmenylethanolamine | 22.5 ± 2.2 | 27.0 ± 1.0 |
| Choline glycerophospholipids | 129.9 ± 7.7 | 156.3 ± 26.1 |
| Phosphatidylcholine | 119.6 ± 5.3 | 137.4 ± 17.2 |
| Plasmenylcholine | 1.2 ± 0.1 | 2.4 ± 1.1 |
| Plasmanylcholine | 9.1 ± 3.2 | 16.5 ± 8.5 |
| Cholesterol | 139.0 ± 46.7 | 126.7 ± 31.2 |
| Cardiolipin | 52.7 ± 4.5 | 39.9 ± 3.4* |
| Phosphatidylinositol | 9.4 ± 0.8 | 10.2 ± 0.9 |
| Phosphatidylglycerol | 7.1 ± 0.5 | 6.4 ± 0.7 |
| Sphingomyelin | 5.3 ± 1.2 | 6.5 ± 0.6 |
| Phosphatidylserine | 4.6 ± 1.5 | 14.1 ± 3.0* |
| Lysophosphatidylcholine | 2.7 ± 0.6 | 3.3 ± 0.4 |
| Ceramide | 0.7 ± 0.2 | 1.6 ± 0.2** |
Values are expressed as mean nmol/mg protein ± SD (n = 3). Significantly different values from NS mitochondria at
p < 0.02;
p < 0.005 as determined from the two-tailed t-test.
The distribution of fatty acid molecular species within each major lipid of the NS and the Syn mitochondria is shown in Figs 3 and 4 and in supplementary Tables S1–S9. The individual phospholipids were arranged according to the number of molecular species with phosphatidylinositol having the fewest and Ptd2Gro having the greatest number. Phosphatidylinositol contained predominantly 18:0-20:4, with lower amounts of 16:0-20:4 (Fig. 3a and Table S4). CerPCho contained predominantly N20:0 and N18:0 (Fig. 3b and Table S6). PtdSer contained predominantly 18:0-22:6 (Fig. 3c and Table S7). PtdGro contained predominantly 16:0-18:1 (Fig. 3d and Table S5). LysoPtdCho consisted of a range of fatty acids, but predominantly 16:0, 18:1, 18:0, and 22:6 (Fig. 3e and Table S8). ChoGpl contained predominantly D16:0-18:1, D18:2-18:2/D16:0-20:4, D16:0-22:6/D18:2-20:4, D18:2-20:2/D18:0-20:4, D16:0-16:0, and A16:0-20:0 (Fig. 3f and Table S2). EtnGpl contained predominantly D18:0-20:4/D16:0-22:4 and D18:0-22:6/D18:1-22:5 (Fig. 3g and Table S1). Cer consisted primarily of N18:0 (Table S9).
Fig. 3.
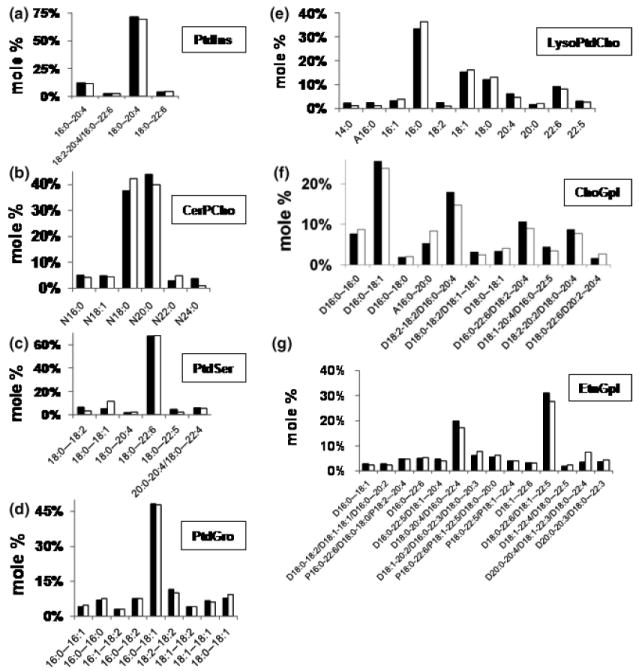
Distribution of fatty acid molecular species in NS (black bar) and Syn (white bar) mitochondrial phospholipids. Phospholipids included (a) phosphatidylinositol (PtdIns), (b) sphingomyelin (CerPCho), (c) phosphatidylserine (PtdSer), (d) phosphatidylglycerol (PtdGro), (e) lysophosphatidylcholine (LysoPtdCho), (f) choline glycerophospholipids (ChoGpl), and (g) ethanolamine glycerophospholipids (EtnGpl). Molar percentages less than 2% of the total lipid are not shown. Glycerolipid subclasses include D (phosphatidyl), P (plasmenyl), and A (plasmanyl). The total molecular species content of each lipid class is presented in supplementary Tables S1–S9. All values are expressed as the mean of three independent samples (n = 3), where six cortexes were pooled for each sample.
Fig. 4.
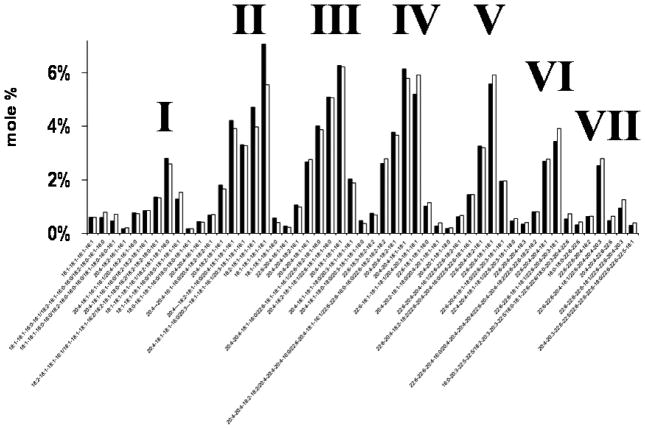
Distribution of Ptd2Gro molecular species in NS (black bar) and Syn (white bar) mitochondria. Ptd2Gro molecular species were arranged according to the mass to charge ratio based on percentage distribution. Ptd2Gro molecular species were subdivided into seven groups, which contained a predominance of oleic, arachidonic, and/or docosahexanoic fatty acids in varying concentrations. Corresponding mass content of molecular species in NS and Syn mitochondria can be found in supplementary Table S3. All values are expressed as the mean of three independent samples (n = 3), where six cortexes were pooled for each sample.
Cardiolipin composition in non-synaptic and synaptic mitochondria
Despite a lower amount of Ptd2Gro in the Syn than in the NS mitochondria, the mole percentage of Ptd2Gro molecular species was remarkably similar for NS and Syn mitochondria (Fig. 4 and Table S3). Both NS and Syn mitochondria contained Ptd2Gro species enriched in 18:1, 20:4, 22:6, 18:2, and 16:1 fatty acids (Table S3). The predominant molecular species of Ptd2Gro were 18:1-18:1-18:1-18:1, 20:4-18:1-18:1-18:1, 20:4-20:4-18:1-18:1, 22:6-20:4-18:1-18:1, 22:6-18:1-18:1-18:1/20:4-20:3-18:1-18:1, and 20:4-18:2-18:1-18:1/22:6-18: 1-18:1-16:0 (Fig. 4). A unique pattern was discovered comparing mole percentage based on the mass to charge ratios. The unique pattern consisted of seven major molecular species groups, with group I having the shortest chains and the least unsaturation, and group VII having the longest chains and the greatest unsaturation.
Mathematical model for the distribution of mouse brain cardiolipin molecular species
We hypothesized that the unique distribution of Ptd2Gro molecular species detected with shotgun lipidomics could be explained by a mathematical model in which Ptd2Gro fatty acids are at equilibrium with respect to the available quantities of each fatty acid chain. In other words, we expected that the concentration c of a given species of Ptd2Gro (specified by its four fatty acids) would be proportional to the product of the concentrations of its four fatty acids (c1 × c2 × c3 × c4). As there are multiple permutations in which fatty acids can be arranged in the four Ptd2Gro fatty acid positions, c should also be proportional to a combinatorial pre-factor F. For example, if there are four distinct fatty acids attached to the Ptd2Gro, there are F = 4! = 24 possible permutations in which these fatty acids can be arranged. However, if the four fatty acids are identical, there is only one way to arrange them, i.e. F = 1. The F × c1 × c2 × c3 × c4 data were summed in groups of Ptd2Gro species with identical numbers of carbons and double bonds. This grouping allows the predictions of the model to be compared directly with the observed data from shotgun lipidomic analysis. Our results show that the distribution of Ptd2Gro fatty acid molecular species predicted by the mathematical model fell into seven major groups that corresponded closely with data obtained from shotgun lipidomic analysis (Fig. 5). Data were condensed from about 1800 possible Ptd2Gro molecular species to 54 species that represented greater than 99% of the total Ptd2Gro species. Subtle differences between the theoretical and observed distribution of Ptd2Gro molecular species, as demonstrated in the molecular species between 72:7 and 72:4, could be the result of a slight difference in the rate of Ptd2Gro synthesis or remodeling. Our analysis showed that the unique pattern of Ptd2Gro molecular species could arise from a simple process of random fatty acid incorporation.
Fig. 5.
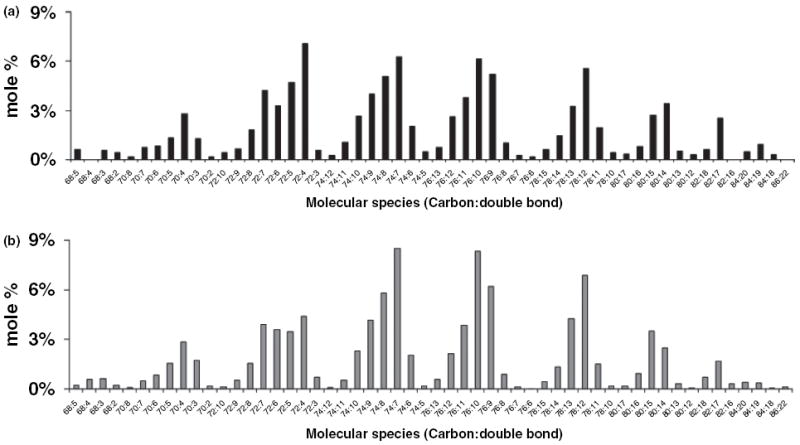
Mathematical prediction of the distribution of brain cardiolipin molecular species. (a) Distribution of molecular species of NS brain Ptd2Gro as determined by shotgun lipidomics. (b) Distribution of molecular species of Ptd2Gro as determined by mathematical modeling as described in Materials and methods.
Coenzyme Q content in non-synaptic and synaptic mitochondria
Coenzyme Q content was measured as this is a lipid constituent and involved in mitochondrial ETC activities (Lenaz 2001). The relative amounts of Q9 and Q10 were approximately 24% lower in the Syn mitochondria than in the NS mitochondria. The Q9 : Q10 ratio was about 3 : 1 in both the NS and Syn mitochondria (Fig. 6).
Fig. 6.
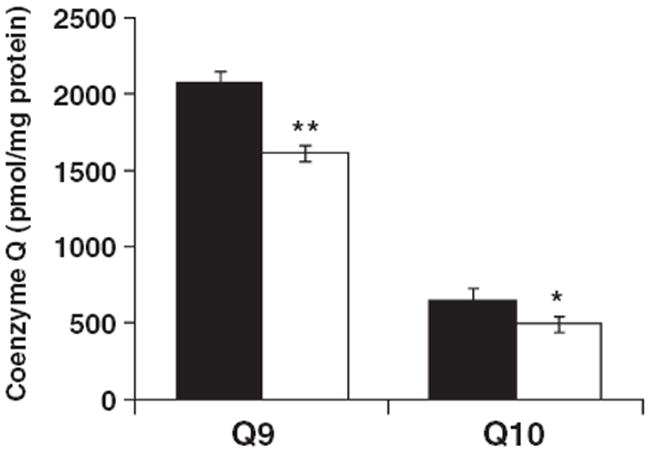
Coenzyme Q9 and Q10 content in NS (black bar) and Syn (white bar) mitochondria. Coenzyme Q content was measured by HPLC connected to an electrochemical detector as described in Materials and methods. The content of Q9 and Q10 were significantly lower in Syn mitochondria compared with NS mitochondria. Values are expressed as the mean pmol/mg protein ± SD (n = 3), where six cortexes were pooled for each sample. Asterisks indicate that the Q9 and Q10 values were lower in the Syn mitochondria than in the NS mitochondria at *p < 0.02 and **p < 0.001.
Electron transport chain activities of brain mitochondrial complexes
Electron transport chain activities have been used to characterize the metabolic properties of NS and Syn mitochondrial populations (Lai and Clark 1976; Lai et al. 1977; Gorini et al. 1989; Hevner and Wong-Riley 1989; Villa et al. 1989a; Battino et al. 1991). We examined the activities of the individual ETC complexes (Complexes I, II, III, and IV) as well as those of the linked enzyme complexes (Complexes I/III and II/III) (Table 2). The activities of Complexes I, II, III, and II/III were 55%, 53%, 63%, and 69%, respectively, in Syn mitochondria than those observed in NS mitochondria (Table 2). The activities of Complexes IV and I/III were similar in the NS and the Syn mitochondria. To determine whether differences in ETC enzyme activities might be associated with differences in protein content of complexes, we evaluated protein markers representative for ETC complexes using the Rodent OXPHOS Complexes Detection Kit. No differences were found for the expression of ETC protein markers for each of the complexes (Fig. 7). These data indicate that the differences found between the NS and Syn mitochondria for ETC enzyme activities were not observed for the content of individual ETC protein markers.
Table 2.
Electron transport chain activities of C57BL/6J mouse brain mitochondria
| Non-synaptic | Synaptic | |
|---|---|---|
| Complex I | 975 ± 64 | 533 ± 37*** |
| NADH ubiquinone oxidoreductase | ||
| Complex II | 292 ± 18 | 177 ± 27** |
| Succinate decylubiquinone DCIP oxidoreductase | ||
| Complex III | 437 ± 71 | 235 ± 47* |
| Ubiquinol cytochrome c oxidoreductase | ||
| Complex IV | 179 ± 17 | 184 ± 50 |
| Cytochrome c oxidase | ||
| Complex I/III | 269 ± 59 | 244 ± 40 |
| NADH cytochrome c oxidoreductase | ||
| Complex II/III | 309 ± 30 | 212 ± 22* |
| Succinate cytochrome c reductase |
Enzyme activities are expressed as (nmol/min/mg protein) ± SD (n = 3). Significantly different values from NS mitochondria at
p < 0.025;
p < 0.01;
p < 0.005 as determined from the two-tailed t-test. DCIP, 2,6-dichloroindophenol.
Fig. 7.
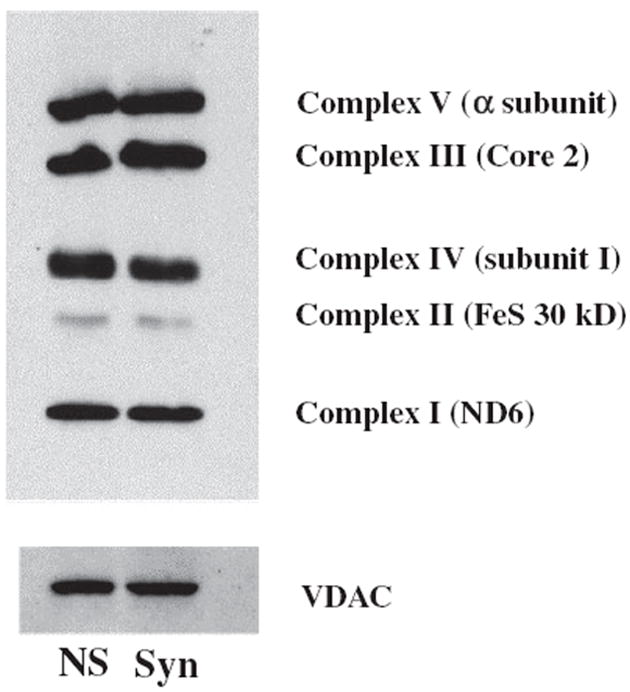
Expression of ETC protein markers in NS and Syn mitochondria. Representative protein markers for each of the five ETC complexes were evaluated by western blot as described in Materials and methods. This pattern is representative of expression found from three separate isolations, where six cortexes were pooled for each sample. ETC protein expression was evaluated at least three times to confirm accuracy of data. Voltage-dependent anion channel (VDAC) was used as a loading control for mitochondrial protein.
Discussion
The objective of this study was to characterize the lipidome and ETC activities in highly purified populations of NS and Syn mitochondria from B6 mice. Information on the lipid composition of NS and Syn mitochondria can provide insight into the metabolic compartmentation in brain (Sonnewald et al. 2004). However, subcellular contamination or technical limitations have hindered past attempts to accurately characterize the lipidome of brain mitochondria (Villa et al. 1989b; Ruggiero et al. 1992; Bangur et al. 1995). This has been due in large part to contamination from myelin, a lipidrich membrane that is difficult to separate from mitochondria (Seminario et al. 1964; Villa et al. 1989b). The failure to remove contaminating membranes or differences in experimental procedures could account for the variability in brain mitochondrial lipid composition found in previous studies (Ruggiero et al. 1992; Bangur et al. 1995). We used multiple discontinuous gradients to obtain highly enriched brain mitochondrial populations free from detectable contamination including that from myelin and cytoskeletal proteins. As the primary goal of this investigation was to isolate highly enriched NS and Syn mitochondria free from any detectable level of contamination, the length as well as choice of discontinuous gradients employed was designed solely for the purpose of mitochondrial lipid analysis as well as assessment of ETC enzyme activities by standard biochemical procedures. GM1a content of the isolated fractions was also evaluated as an independent marker for membrane contamination, as GM1a is enriched in myelin (Seyfried et al. 1979). Our data show that the ganglioside GM1a, sulfatides, cerebrosides, and cytoskeletal proteins do not exist in neural cell mitochondria. We suggest that the presence of glycosphingolipids in brain mitochondria would indicate contamination from non-mitochondrial subcellular membranes or a physiological environment different from that which exists normally in mouse cerebral cortex.
We used mouse cerebral cortex in this study as previous studies in rats showed that the relative differences between NS and Syn mitochondria for ETC enzyme activities and coenzyme Q content are generally similar across brain regions (Leong et al. 1984; Battino et al. 1991, 1995). It is therefore likely that our findings for mitochondrial lipids and ETC activities in the cerebral cortex would be similar for other brain regions. We chose to examine B6 mice because many existing neurological mutations that alter CNS function and behavior are expressed on this genetic background (Bedell et al. 1997a,b). Consequently, our study will provide useful information on brain mitochondrial function to a broad range of neurological and neurodegenerative diseases expressed in B6 mice.
Membrane lipids can influence the activity of mitochondrial enzymes to include those of the ETC (Daum 1985; Di Paola et al. 2000; Chicco and Sparagna 2007). We found that the levels of Ptd2Gro, coenzyme Q9, and Q10 were lower and the levels of Cer and PtdSer were higher in the Syn mitochondria than in the NS mitochondria. Additionally, we found that the ETC activities of Complexes I, II, III, and II/III were lower in the Syn mitochondria than in the NS mitochondria. As enzyme activities from individual (unlinked) complexes (Complexes I, II, and III) could include activities of the combined (linked) measurements (Complexes I/III and II/III), it is possible that these differences arise from the ETC existing in two states, a random collisions state or a solid electron channeling state (Lenaz and Genova 2007). Linked ETC activities can represent the combined activity of individual complexes or the activity of the supercomplex (Schagger and Pfeiffer 2000). Recently, it was shown that the Complex I/III linked activity in the α-synuclein knockout mouse brain was lower than that of normal mouse brain without corresponding changes in either the Complexes I or III individual ETC activities (Ellis et al. 2005). This suggests that the linked activity corresponded to the measurement of the ETC complexes in the supercomplex state. Hence, differences in individual ETC activities may not always correspond to differences in linked ETC activities.
No differences were found between NS and Syn mitochondria for the content of major protein markers for the individual ETC complexes. This suggests that other factors, such as differences in lipid content, may account the differences found between NS and Syn mitochondria for linked ETC activities. As Ptd2Gro is required for the enzymatic activities of Complexes I and III (Fry and Green 1981; Gohil et al. 2004; Chicco and Sparagna 2007), a lower Ptd2Gro content in Syn than in NS mitochondria could account for the lower activities of Complexes I and III in Syn mitochondria. We suggest that the differences found in ETC complex activities between the NS and Syn mitochondria might be due in part to quantitative differences in their lipid composition, which could differentially affect unlinked or linked enzyme activities.
It is interesting that Ptd2Gro content is lower, while PtdSer content is higher in Syn than NS mitochondria. These lipid differences might be related to the greater susceptibility of Syn mitochondria to calcium overload and permeability transition than non-synaptic mitochondria (Brown et al. 2006; Naga et al. 2007). Although Ptd2Gro and PtdSer are both anionic lipids that can buffer calcium, PtdSer is less prone than Ptd2Gro to calcium-induced peroxidation, which would occur during intense Syn activity (Grijalba et al. 1999; Brown et al. 2006). Differences in Ptd2Gro content between Syn and NS mitochondria might also influence susceptibility to apoptosis, cytochrome c retention, or lipid peroxidation (Kagan et al. 2004). We do not exclude the possibility that the difference between NS and Syn mitochondria for PtdSer content is related to differences in lipid synthesis, transport protein activity, or cellular heterogeneity related to Syn activity (Zborowski et al. 1983; Daum and Vance 1997).
Plasmalogen glycerophospholipids contain a vinyl-ether linkage in the sn-1 position and can function as antioxidants and facilitators of membrane fusion (Gross 1984; Zoeller et al. 1988; Farooqui and Horrocks 2001). The ESI/MS procedures we used can easily measure the content and composition of plasmalogens (Han et al. 2001; Han and Gross 2005a). Only brain and heart mitochondria contain significant quantities of choline and ethanolamine plasmalogen glycerophospholipids suggesting a unique role for plasmalogens in these tissues (Eichberg et al. 1964; Getz et al. 1968). We found that the content of plasmenylethanolamine and plasmenylcholine was similar in the NS and the Syn mitochondria. Hence, the role of plasmalogens is likely similar in the NS and Syn mitochondria. This role could involve management of free radicals or regulation of unique fusogenic properties of brain mitochondrial membranes.
Almost 100 molecular species of Ptd2Gro were detected in the NS and Syn mitochondria of B6 mice. Moreover, these molecular species formed a unique pattern consisting of seven major groups when arranged according to mass to charge ratios. Interestingly, this Ptd2Gro pattern in brain was also observed in human, rat, and rabbit, suggesting conservation across some mammalian species (Cheng et al., unpublished observation). Molecular species heterogeneity was greater in the most abundant groups (groups II–VI) than in the least abundant groups (groups I and VII). This Ptd2Gro species pattern in brain differs markedly from those found previously in non-neural tissues, which contain predominantly tetralinoleic Ptd2Gro (18:2-18:2-18:2-18:2) (Schlame et al. 2005). Surprisingly, no tetralinoleic Ptd2Gro was found in brain mitochondria. Hence, Ptd2Gro composition from brain differs markedly from the Ptd2Gro composition found in non-neural tissues.
We suggest that the unique brain Ptd2Gro pattern arises from the remodeling activities of phospholipases and acyl transferases, which remove and transfer, respectively, fatty acid species from the sn-2 position of the ChoGpl and EtnGpl to form mature Ptd2Gro from MLCL. This suggestion comes from finding an abundance of fatty acids in mature Ptd2Gro that match those expressed in the predicted sn-2 position of ChoGpl and EtnGpl. Our findings also indicate that only a small percentage of mature Ptd2Gro contains fatty acids originating from the condensation of PtdGro and CDP-diacylglycerol, which contain predominantly shorter chain fatty acids.
The mechanism by which the acyl transferases select the sn-2 fatty acids from either ChoGpl or EtnGpl remains unclear. Taffazin is the only known Ptd2Gro acyltransferase (AT) that is highly expressed in brain (Bione et al. 1993; Ma et al. 1999; Lu et al. 2004). Although taffazin may have specificity for linoleic acid in non-neural tissues (Xu et al. 2003), linoleic acid is a minor fatty acid in mature Ptd2Gro of brain. Our findings together with those of other groups indicate that brain Ptd2Gro contains predominantly oleic, arachidonic, and docosahexaenoic fatty acids (Yamaoka et al. 1988; Nakahara et al. 1992; Bayir et al. 2007). Although other enzymes have been implicated in Ptd2Gro remodeling to include scramblase 3, acyl-CoA:lysocardiolipin AT, MLCL AT, and acylglycerol-3-phosphate ATs, the ATs involved in the complex remodeling of brain Ptd2Gro have yet to be determined (Hauff and Hatch 2006). Nevertheless, our mathematical model indicates that the unique Ptd2Gro pattern for normal brain can arise from a random incorporation of selected fatty acids during the Ptd2Gro remodeling process.
The functional significance of the unique brain Ptd2Gro molecular species pattern remains unclear. A greater ratio of PUFA to saturated fatty acids could cause Ptd2Gro to be more prone to form a non-bilayer HII hexagonal phase lipid (Sankaram et al. 1989; Kraffe et al. 2002). This conformational change can create regions of small curvature within the membrane, which could affect cristae structure (Hoch 1992; Stuart et al. 1998). Increased docosahexaenoic containing Ptd2Gro results in decreased Complex IV activity, but increases F1 ATPase activities (Yamaoka et al. 1988). This might explain the lower Complex IV activities that we found in the NS and Syn mitochondria relative to those found in non-neural tissues (Benard et al. 2006). Long-chain PUFA Ptd2Gro species could also enhance membrane phase transformations thus allowing optimal activity of oxidative phosphorylation in response to environmental changes (Kraffe et al. 2002). The prevalence of oleic fatty acids in Ptd2Gro could also enhance mitochondrial respiratory chain ratios and degree of coupling (Brand et al. 2003). The unique Ptd2Gro species pattern of brain might also be interesting with regard to the ‘respirasome’ formation, as Ptd2Gro can ‘glue’ the complexes together in an organized supercomplex to increase efficiency of ATP production (Zhang et al. 2002; Pfeiffer et al. 2003). Further studies are needed to resolve the role of the unique pattern of Ptd2Gro molecular species in brain mitochondrial function.
Alterations in brain energy metabolism have been implicated in variety of neurological and neurodegenerative disorders to include Parkinson’s disease, Alzheimer’s disease, Huntington’s disease, epilepsy, brain tumors, storage diseases, bipolar disorder, schizophrenia, and autism to name a few (Bowling and Beal 1995; Lombard 1998; Blass 2002; Seyfried and Mukherjee 2005). Abnormalities in NS and Syn mitochondrial lipids, at the quantitative as well as the molecular species level, may occur in these disorders as either a cause or an effect of disease pathogenesis. Although differences in lipid content as well as ETC activities exist between NS and Syn mitochondria, these differences likely represent metabolic compartmentation in the brain. However, these populations of brain mitochondria might be affected differently in disease states. We suggest that the integrity of the mitochondrial lipidome is critical for efficient ATP production and that alterations in the lipidome will impair oxidative phosphorylation thus compromising normal neurological function.
Supplementary Material
Acknowledgments
We would like to thank Susan Bothwell, Christopher Ellis, Mary F. Roberts, Purna Mukherjee, Rena Baek, and John Mantis for technical assistance and helpful discussions.We would also like to thank Steven Pfeiffer and Cara-Lynne Schengrund for the gifts of the PLP and SNAP25 antibodies, respectively. This work was supported by Grants from NIH (HD39722), NCI (CA102135), and NIA (AG23168).
Abbreviations used
- A
plasmanyl
- AT
acyltransferase
- B6
C57BL/6J mice
- BSA
bovine serum albumin
- Cer
ceramide
- CerPCho
sphingomyelin
- ChoGpl
choline glycerophospholipids
- D
phosphatidyl
- DCIP
2,6-dichloroindophenol
- ESI/MS
electrospray ionization mass spectrometry
- ETC
electron transport chain
- EtnGpl
ethanolamine glycerophospholipids
- FM
Ficoll purified NS mitochondria
- LysoPtdCho
lysophosphatidylcholine
- MIB
mitochondrial isolation buffer
- MLCL
monolysocardiolipin
- NS
non-synaptic
- P
plasmenyl
- PBS
phosphate-buffered saline
- Ptd2Gro
cardiolipin
- PtdCho
phosphatidylcholine
- PtdCho
phosphatidylcholine
- PtdGro
phosphatidylglycerol
- PtdSer
phosphatidylserine
- Syn
synaptic
Footnotes
The following supplementary material is available for this article:
Table S1 Mass content of ethanolamine glycerophospholipid molecular species in mouse brain mitochondria.
Table S2 Mass content of choline glycerophospholipid molecular species in mouse brain mitochondria.
Table S3 Mass content of cardiolipin molecular species in brain mitochondria.
Table S4 Mass content of phosphatidylinositol molecular species in mouse brain mitochondria.
Table S5 Mass content of phosphatidylglycerol molecular species in mouse brain mitochondria.
Table S6 Mass content of sphingomyelin molecular species in mouse brain mitochondria.
Table S7 Mass content of phosphatidylserine molecular species in mouse brain mitochondria.
Table S8 Mass content of lysophosphatidyicholine molecular species in mouse brain mitochondria.
Table S9 Mass content of ceramide molecular species in mouse brain mitochondria.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1471-4159.2008.05383.x. (This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bangur CS, Howland JL, Katyare SS. Thyroid hormone treatment alters phospholipid composition and membrane fluidity of rat brain mitochondria. Biochem J. 1995;305:29–32. doi: 10.1042/bj3050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battino M, Bertoli E, Formiggini G, Sassi S, Gorini A, Villa RF, Lenaz G. Structural and functional aspects of the respiratory chain of synaptic and nonsynaptic mitochondria derived from selected brain regions. J Bioenerg Biomembr. 1991;23:345–363. doi: 10.1007/BF00762227. [DOI] [PubMed] [Google Scholar]
- Battino M, Gorini A, Villa RF, Genova ML, Bovina C, Sassi S, Littarru GP, Lenaz G. Coenzyme Q content in synaptic and non-synaptic mitochondria from different brain regions in the ageing rat. Mech Ageing Dev. 1995;78:173–187. doi: 10.1016/0047-6374(94)01535-t. [DOI] [PubMed] [Google Scholar]
- Bayir H, Tyurin VA, Tyurina YY, et al. Selective early cardiolipin peroxidation after traumatic brain injury: an oxidative lipidomics analysis. Ann Neurol. 2007;62:154–169. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Jenkins NA, Copeland NG. Mouse models of human disease Part I: techniques and resources for genetic analysis in mice. Genes Dev. 1997a;11:1–10. doi: 10.1101/gad.11.1.1. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Largaespada DA, Jenkins NA, Copeland NG. Mouse models of human disease Part II: recent progress and future directions. Genes Dev. 1997b;11:11–43. doi: 10.1101/gad.11.1.11. [DOI] [PubMed] [Google Scholar]
- Benard G, Faustin B, Passerieux E, et al. Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol. 2006;291:C1172–C1182. doi: 10.1152/ajpcell.00195.2006. [DOI] [PubMed] [Google Scholar]
- Bione S, Tamanini F, Maestrini E, Tribioli C, Poustka A, Torri G, Rivella S, Toniolo D. Transcriptional organization of a 450-kb region of the human X chromosome in Xq28. Proc Natl Acad Sci USA. 1993;90:10977–10981. doi: 10.1073/pnas.90.23.10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- Blass JP. Glucose/mitochondria in neurological conditions. Int Rev Neurobiol. 2002;51:325–376. doi: 10.1016/s0074-7742(02)51010-2. [DOI] [PubMed] [Google Scholar]
- Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 1995;56:1151–1171. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- Brand MD, Turner N, Ocloo A, Else PL, Hulbert AJ. Proton conductance and fatty acyl composition of liver mitochondria correlates with body mass in birds. Biochem J. 2003;376:741–748. doi: 10.1042/BJ20030984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigande JV, Platt FM, Seyfried TN. Inhibition of glycosphingolipid biosynthesis does not impair growth or morphogenesis of the postimplantation mouse embryo. J Neurochem. 1998;70:871–882. doi: 10.1046/j.1471-4159.1998.70020871.x. [DOI] [PubMed] [Google Scholar]
- Brown MR, Sullivan PG, Geddes JW. Synaptic mitochondria are more susceptible to Ca2+ overload than nonsynaptic mitochondria. J Biol Chem. 2006;281:11658–11668. doi: 10.1074/jbc.M510303200. [DOI] [PubMed] [Google Scholar]
- Cheng H, Guan S, Han X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: implications for the role of altered triacylglycerols in diabetic neuropathy. J Neurochem. 2006;97:1288–1300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- Cuzner ML, Davison AN. The lipid composition of rat brain myelin and subcellular fractions during development. Biochem J. 1968;106:29–34. doi: 10.1042/bj1060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagani F, Gorini A, Polgatti M, Villa RF, Benzi G. Synaptic and non-synaptic mitochondria from rat cerebral cortex. Characterization and effect of pharmacological treatment on some enzyme activities related to energy transduction. Farmaco [Sci] 1983;38:584–594. [PubMed] [Google Scholar]
- Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- Daum G, Vance JE. Import of lipids into mitochondria. Prog Lipid Res. 1997;36:103–130. doi: 10.1016/s0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- Degli Esposti M. Assessing functional integrity of mitochondria in vitro and in vivo. Methods Cell Biol. 2001;65:75–96. doi: 10.1016/s0091-679x(01)65005-2. [DOI] [PubMed] [Google Scholar]
- Di Paola M, Cocco T, Lorusso M. Ceramide interaction with the respiratory chain of heart mitochondria. Biochemistry. 2000;39:6660–6668. doi: 10.1021/bi9924415. [DOI] [PubMed] [Google Scholar]
- Eichberg J, Whittaker VP, Dawson RM. Distribution of lipids in subcellular particles of guinea-pig brain. Biochem J. 1964;92:91–100. doi: 10.1042/bj0920091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CE, Murphy EJ, Mitchell DC, Golovko MY, Scaglia F, Barcelo-Coblijn GC, Nussbaum RL. Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking alpha-synuclein. Mol Cell Biol. 2005;25:10190–10201. doi: 10.1128/MCB.25.22.10190-10201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui AA, Horrocks LA. Plasmalogens: workhorse lipids of membranes in normal and injured neurons and glia. Neuroscientist. 2001;7:232–245. doi: 10.1177/107385840100700308. [DOI] [PubMed] [Google Scholar]
- Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J Biol Chem. 1981;256:1874–1880. [PubMed] [Google Scholar]
- Getz GS, Bartley W, Lurie D, Notton BM. The phospholipids of various sheep organs, rat liver and of their subcellular fractions. Biochim Biophys Acta. 1968;152:325–339. doi: 10.1016/0005-2760(68)90040-4. [DOI] [PubMed] [Google Scholar]
- Gohil VM, Hayes P, Matsuyama S, Schagger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J Biol Chem. 2004;279:42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
- Gorini A, Arnaboldi R, Ghigini B, Villa RF. Brain cytochrome oxidase activity of synaptic and nonsynaptic mitochondria during aging. Basic Appl Histochem. 1989;33:139–145. [PubMed] [Google Scholar]
- Grijalba MT, Vercesi AE, Schreier S. Ca2+-induced increased lipid packing and domain formation in submitochondrial particles. A possible early step in the mechanism of Ca2+-stimulated generation of reactive oxygen species by the respiratory chain. Biochemistry. 1999;38:13279–13287. doi: 10.1021/bi9828674. [DOI] [PubMed] [Google Scholar]
- Gross RW. High plasmalogen and arachidonic acid content of canine myocardial sarcolemma: a fast atom bombardment mass spectroscopic and gas chromatography-mass spectroscopic characterization. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005a;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Exp Rev Proteomics. 2005b;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- Han X, Holtzman DM, McKeel DW., Jr Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Hauff KD, Hatch GM. Cardiolipin metabolism and Barth Syndrome. Prog Lipid Res. 2006;45:91–101. doi: 10.1016/j.plipres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Hauser EC, Kasperzyk JL, d’Azzo A, Seyfried TN. Inheritance of lysosomal acid beta-galactosidase activity and gangliosides in crosses of DBA/2J and knockout mice. Biochem Genet. 2004;42:241–257. doi: 10.1023/b:bigi.0000034429.55418.71. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MT. Brain cytochrome oxidase: purification, antibody production, and immunohistochemical/histochemical correlations in the CNS. J Neurosci. 1989;9:3884–3898. doi: 10.1523/JNEUROSCI.09-11-03884.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Hyogo A, Nakao T, Tsuchiya K, Suzuki Y, Matsumoto M, Kon K, Ando S. Isolation and characterization of extremely minor gangliosides, GM1b and GD1 alpha, in adult bovine brains as developmentally regulated antigens. J Biol Chem. 1990;265:8144–8151. [PubMed] [Google Scholar]
- Hoch FL. Cardiolipins and biomembrane function. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Marbois BN, Faull KF, Clarke CF, Larsen PL. Development and fertility in Caenorhabditis elegans clk-1 mutants depend upon transport of dietary coenzyme Q8 to mitochondria. J Biol Chem. 2002;277:45020–45027. doi: 10.1074/jbc.M204758200. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, Fujii Y. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic Biol Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- King TE. Preparation of succinate dehydrogenase and reconstitution of succinate oxidase. In: Estabrook RW, Pullman ME, editors. Methods Enzymol. Academic Press; New York: 1967. pp. 322–331. [Google Scholar]
- Kraffe E, Soudant P, Marty Y, Kervarec N, Jehan P. Evidence of a tetradocosahexaenoic cardiolipin in some marine bivalves. Lipids. 2002;37:507–514. doi: 10.1007/s11745-002-0925-z. [DOI] [PubMed] [Google Scholar]
- Lai JC, Clark JB. Preparation and properties of mitochondria derived from synaptosomes. Biochem J. 1976;154:423–432. doi: 10.1042/bj1540423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JCK, Clark JB. Isolation and characterization of synaptic and nonsynaptic mitochondria from mammalian brain. In: Boulton AA, Baker GB, Butterworth RF, editors. Carbohydrates and Energy Metabolism. Vol. 11. The Humana Press Inc.; Clifton, NJ: 1989. pp. 43–98. [Google Scholar]
- Lai JC, Walsh JM, Dennis SC, Clark JB. Synaptic and non-synaptic mitochondria from rat brain: isolation and characterization. J Neurochem. 1977;28:625–631. doi: 10.1111/j.1471-4159.1977.tb10434.x. [DOI] [PubMed] [Google Scholar]
- Lenaz G. A critical appraisal of the mitochondrial coenzyme Q pool. FEBS Lett. 2001;509:151–155. doi: 10.1016/s0014-5793(01)03172-6. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Kinetics of integrated electron transfer in the mitochondrial respiratory chain: random collisions vs. solid state electron channeling. Am J Physiol Cell Physiol. 2007;292:C1221–C1239. doi: 10.1152/ajpcell.00263.2006. [DOI] [PubMed] [Google Scholar]
- Leong SF, Lai JC, Lim L, Clark JB. The activities of some energy-metabolising enzymes in nonsynaptic (free) and synaptic mitochondria derived from selected brain regions. J Neurochem. 1984;42:1306–1312. doi: 10.1111/j.1471-4159.1984.tb02788.x. [DOI] [PubMed] [Google Scholar]
- Lombard J. Autism: a mitochondrial disorder? Med Hypotheses. 1998;50:497–500. doi: 10.1016/s0306-9877(98)90270-5. [DOI] [PubMed] [Google Scholar]
- Lu B, Kelher MR, Lee DP, Lewin TM, Coleman RA, Choy PC, Hatch GM. Complex expression pattern of the Barth syndrome gene product tafazzin in human cell lines and murine tissues. Biochem Cell Biol. 2004;82:569–576. doi: 10.1139/o04-055. [DOI] [PubMed] [Google Scholar]
- Ma BJ, Taylor WA, Dolinsky VW, Hatch GM. Acylation of monolysocardiolipin in rat heart. J Lipid Res. 1999;40:1837–1845. [PubMed] [Google Scholar]
- Mena EE, Hoeser CA, Moore BW. An improved method of preparing rat brain synaptic membranes. Elimination of a contaminating membrane containing 2′,3′-cyclic nucleotide 3′-phosphohydrolase activity. Brain Res. 1980;188:207–231. doi: 10.1016/0006-8993(80)90569-7. [DOI] [PubMed] [Google Scholar]
- Naga KK, Sullivan PG, Geddes JW. High cyclophilin D content of synaptic mitochondria results in increased vulnerability to permeability transition. J Neurosci. 2007;27:7469–7475. doi: 10.1523/JNEUROSCI.0646-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara I, Kikuchi H, Taki W, Nishi S, Kito M, Yonekawa Y, Goto Y, Ogata N. Changes in major phospholipids of mitochondria during postischemic reperfusion in rat brain. J Neurosurg. 1992;76:244–250. doi: 10.3171/jns.1992.76.2.0244. [DOI] [PubMed] [Google Scholar]
- Petrushka E, Quastel JH, Scholefield PG. Role of phospholipids in oxidative phosphorylation and mitochondrial structure. Can J Biochem Physiol. 1959;37:989–998. [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Rendon A, Masmoudi A. Purification of non-synaptic and synaptic mitochondria and plasma membranes from rat brain by a rapid Percoll gradient procedure. J Neurosci Methods. 1985;14:41–51. doi: 10.1016/0165-0270(85)90113-x. [DOI] [PubMed] [Google Scholar]
- Ruggiero FM, Cafagna F, Petruzzella V, Gadaleta MN, Quagliariello E. Lipid composition in synaptic and non-synaptic mitochondria from rat brains and effect of aging. J Neurochem. 1992;59:487–491. doi: 10.1111/j.1471-4159.1992.tb09396.x. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Powell GL, Marsh D. Effect of acyl chain composition on salt-induced lamellar to inverted hexagonal phase transitions in cardiolipin. Biochim Biophys Acta. 1989;980:389–392. doi: 10.1016/0005-2736(89)90331-3. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys Lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Seminario LM, Hren N, Gomez CJ. Lipid distribution in subcellular fractions of the rat brain. J Neurochem. 1964;11:197–209. doi: 10.1111/j.1471-4159.1964.tb06129.x. [DOI] [PubMed] [Google Scholar]
- Seyfried TN, Mukherjee P. Targeting energy metabolism in brain cancer: review and hypothesis. Nutr Metab (Lond) 2005;2:30. doi: 10.1186/1743-7075-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried TN, Glaser GH, Yu RK. Genetic variability for regional brain gangliosides in five strains of young mice. Biochem Genet. 1979;17:43–55. doi: 10.1007/BF00484473. [DOI] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Schousboe A, Qu H, Waagepetersen HS. Intracellular metabolic compartmentation assessed by 13C magnetic resonance spectroscopy. Neurochem Int. 2004;45:305–310. doi: 10.1016/j.neuint.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Gillis TE, Ballantyne JS. Remodeling of phospholipid fatty acids in mitochondrial membranes of estivating snails. Lipids. 1998;33:787–793. doi: 10.1007/s11745-998-0271-1. [DOI] [PubMed] [Google Scholar]
- Villa RF, Gorini A, Geroldi D, Lo Faro A, Dell’Orbo C. Enzyme activities in perikaryal and synaptic mitochondrial fractions from rat hippocampus during development. Mech Ageing Dev. 1989a;49:211–225. doi: 10.1016/0047-6374(89)90072-9. [DOI] [PubMed] [Google Scholar]
- Villa RF, Gorini A, Lo Faro A, Dell’Orbo C. A critique on the preparation and enzymatic characterization of synaptic and nonsynaptic mitochondria from hippocampus. Cell Mol Neurobiol. 1989b;9:247–262. doi: 10.1007/BF00713032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J Biol Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Urade R, Kito M. Mitochondrial function in rats is affected by modification of membrane phospholipids with dietary sardine oil. J Nutr. 1988;118:290–296. doi: 10.1093/jn/118.3.290. [DOI] [PubMed] [Google Scholar]
- Yonetan T. Cytochrome oxidase: beef heart. In: Estabrook RW, Pullman ME, editors. Methods Enzymol. Academic Press; New York: 1967. pp. 332–335. [Google Scholar]
- Zborowski J, Dygas A, Wojtczak L. Phosphatidylserine decarboxylase is located on the external side of the inner mitochondrial membrane. FEBS Lett. 1983;157:179–182. doi: 10.1016/0014-5793(83)81141-7. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- Zoeller RA, Morand OH, Raetz CR. A possible role for plasmalogens in protecting animal cells against photosensitized killing. J Biol Chem. 1988;263:11590–11596. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.