Summary
Coenzyme Qn is a fully substituted benzoquinone containing a polyisoprene tail of distinct numbers (n) of isoprene groups. Caenorhabditis elegans fed Escherichia coli devoid of Q8 have a significant lifespan extension when compared to C. elegans fed a standard ‘Q-replete’ E. coli diet. Here we examine possible mechanisms for the lifespan extension caused by the Q-less E. coli diet. A bioassay for Q uptake shows that a water-soluble formulation of Q10 is effectively taken up by both clk-1 mutant and wild-type nematodes, but does not reverse lifespan extension mediated by the Q-less E. coli diet, indicating that lifespan extension is not due to the absence of dietary Q per se. The enhanced longevity mediated by the Q-less E. coli diet cannot be attributed to dietary restriction, different Qn isoforms, reduced pathogenesis or slowed growth of the Q-less E. coli, and in fact requires E. coli viability. Q-less E. coli have defects in respiratory metabolism. C. elegans fed Q-replete E. coli mutants with similarly impaired respiratory metabolism due to defects in complex V also show a pronounced lifespan extension, although not as dramatic as those fed the respiratory deficient Q-less E. coli diet. The data suggest that feeding respiratory incompetent E. coli, whether Q-less or Q-replete, produces a robust life extension in wild-type C. elegans. We believe that the fermentation-based metabolism of the E. coli diet is an important parameter of C. elegans longevity.
Keywords: aging, Caenorhabditis elegans, clk-1, coenzyme Q or ubiquinone, dietary restriction, respiratory defective Escherichia coli
Introduction
Ubiquinone (coenzyme Q or Q) is found in both prokaryotes and eukaryotes where it functions as an essential component of the respiratory electron transport chain (Gennis & Stewart, 1996; Dutton et al., 2000). In addition to its role in respiration, Q is also found widely distributed throughout cellular membranes where in its reduced form it scavenges lipid peroxyl radicals (Bentinger et al., 2007). Q may also play a part in cell signaling as it acts as a pro-oxidant generating the superoxide free radical, which in turn undergoes dismutation and forms the important signaling molecule, hydrogen peroxide (Linnane et al., 2007). Based on its importance in respiration and as an antioxidant, Q is a popular dietary supplement and has shown promise as a therapeutic agent in treating neurodegenerative and cardiovascular disease (Galpern & Cudkowicz, 2007; Pepe et al., 2007).
Q is composed of a benzoquinone ring with a polyisoprenoid side chain of varying length. The length of the isoprenoid side chain varies among organisms. For example, Escherichia coli produce Q8, Caenorhabditis elegans produce Q9, and humans produce Q10 (Olson & Rudney, 1983). The length of the isoprenoid side chain in Q is determined by the specific long-chain polyisoprenyl diphosphate synthase in each organism (Kawamukai, 2002). The eukaryotic biosynthetic pathway has been elucidated primarily in Saccharomyces cerevisiae. In this organism, nine genes designated COQ1–COQ9 are required for production of Q6 (Tran & Clarke, 2007). Homologues of COQ1–COQ8 have been identified in C. elegans and humans based on sequence homology. The yeast coq mutant strains fail to grow on media containing a nonfermentable carbon source, and this lack of growth is rescued by provision of exogenous Q6 in the medium (Jonassen et al., 1998; Do et al., 2001; James et al., 2005). Restoration of growth of yeast coq mutants on medium containing a nonfermentable carbon source by supplementation with exogenous Q6 constitutes strong evidence that the growth defect is caused solely by the absence of Q.
As would be expected from its essential role in respiratory metabolism, Q is required for normal development. The C. elegans clk-1 mutant has a defect in Q9 biosynthesis, accumulates the intermediate demethoxyubiquinone-9, and relies on the Q supplied via a diet of E. coli for development to adulthood and fertility (Jonassen et al., 2001; Miyadera et al., 2001; Burgess et al., 2003; Hihi et al., 2003). clk-1 mutants fed E. coli strains lacking Q from the time of hatching stop growing during the L2 larval stage (Jonassen et al., 2001), and after about 1 week will eventually develop into sterile adults (Burgess et al., 2003). Similarly, clk-1 dauer larvae fed the Q-less diet recover to adulthood but are completely sterile (Jonassen et al., 2001). The growth arrest and sterility phenotypes of the clk-1 mutants fed Q-less E. coli are very reproducible and can be assessed quantitatively over a period of several days (Jonassen et al., 2001; Burgess et al., 2003). The clk-1 mutants also experience growth arrest in axenic media (Braeckman et al., 1999). Provision of Q-replete E. coli rescues the growth on axenic media and also restores fertility in clk-1 mutants developing from dauer larvae. Mice homozygous for a clk-1 null mutation show embryonic lethality, lack Q9, and accumulate demethoxyubiquinone-9 and are not rescued by dietary Q supplementation (Levavasseur et al., 2001; Nakai et al., 2001).
Caenorhabditis elegans serves as a powerful model system to identify genetic and environmental factors that influence aging (Houthoofd & Vanfleteren, 2007). For example, worms harboring mutations in the daf-2 gene of the insulin/IGF-like signaling pathway exhibit a profound life extension and stress-resistant phenotypes that are dependent on the product of the daf-16 gene, a FOXO-family transcription factor (Kenyon, 2005). Many other classes of genes modulating aging have been identified in C. elegans by RNAi screens (Hamilton et al., 2005). Dietary restriction extends C. elegans lifespan (Walker et al., 2005), as does treatment with a variety of drugs and natural products such as vitamin E (Collins et al., 2006), and plant extracts such as blueberry polyphenols (Wilson et al., 2006).
Several lines of evidence suggest that Q has a conserved function in aging in organisms ranging from worms to mammals. clk-1 mutant worms fed the standard Q-replete OP50 E. coli live longer than wild-type animals (Lakowski & Hekimi, 1996; Ewbank et al., 1997) and the Q acquired from their diet is present at decreased levels compared to wild-type worms (Jonassen et al., 2001). Wild-type worms fed an E. coli diet devoid of Q have an extended adult lifespan (Larsen & Clarke, 2002). The long-lived phenotype of worms fed the Q-less E. coli diet is independent of daf-16 and augments the long life of daf-2 mutants, daf-2;daf-12 double mutants and even clk-1 mutants, provided the Q-less diet is administered after the clk-1 animals have reached the L4 larval stage of development (Larsen & Clarke, 2002). Heterozygous mice harboring one clk-1 null allele show normal growth and reproduction, and although Q content in young mice is normal, a significant decline in liver Q content is observed with age (Liu et al., 2005). Intriguingly, clk-1 heterozygous mice have a longer lifespan than their wild-type littermate controls (Liu et al., 2005). These observations led to the hypothesis that decreased intake of dietary Q in nematodes, or decreased de novo biosynthesis of Q in worms and mice, can cause lifespan extension. The mechanisms responsible for this lifespan extension are not known.
Here we describe a bioassay for the uptake and assimilation of added Q. Supplementation of plate or liquid growth media with NovaSOL® Q10 restores fertility of the clk-1 mutants fed Q-less E. coli. We show both clk-1 mutant and N2 wild-type nematodes transport this exogenous Q10 into their mitochondria. We then use this water-soluble Q formulation to test whether the lifespan extension produced by feeding the Q-less E. coli diet can be reversed. Surprisingly, supplementation of the Q-less E. coli diet with NovaSOL Q10 does not reverse the lifespan extension, indicating that other attributes of the E. coli ‘Q-less’ diet are responsible for the lifespan extension. The results presented here suggest that the respiratory deficiency of the E. coli diet can produce lifespan extension of wild-type C. elegans.
Results
Development of a bioassay for Q uptake
To evaluate the role of Q in modulating nematode lifespan, it is crucial to ascertain whether the Q provided exogenously is assimilated. C. elegans clk-1 mutants fed Q-less E. coli (GD1; Table 1) show arrested or greatly slowed development and are sterile (Jonassen et al., 2001; Burgess et al., 2003). GD1 E. coli harbor a deletion of the ubiG gene, encoding an O-methyl-transferase in the coenzyme Q biosynthetic pathway, and fail to synthesize Q8 (Hsu et al., 1996). Provision of Q-replete E. coli (e.g. OP50 or GD1/pAHG, harboring a plasmid expressing ubiG) rescues growth arrest and restores fertility. However, our initial attempts to rescue clk-1 mutant sterility by adding either purified Q10 or Q9 to plate or liquid media (from 0.5 to 50 μM final concentration added as a suspension in ethanol) failed to rescue sterility of either the clk-1(qm30) or the clk-1(e2519) mutants fed GD1. Various additives (present at the indicated final concentrations) were tested during the preparation of plate and liquid growth media to help solubilize or disperse the added Q, including Triton X-100 (1%), dimethylsulfoxide (2%), ethanol (1%), glycerol (1%), or Cremophor EL (1%) (Sigma-Aldrich, St Louis, MO, USA). In addition, a suspension of Q10 in Tween 80 was prepared as described (Ishii et al., 2004). While none of these additives appeared to harm development of N2 wild-type C. elegans, only a few progeny were sporadically produced by the clk-1 mutants in response to these Q supplementations (data not shown).
Table 1.
Genotype and source of bacterial strains
| Strain | Genotype | Reference or source |
|---|---|---|
| OP50 | ura | Sulston & Hodgkin (1988) |
| GD1 | HW272 ubiG::Kanr | Hsu et al. (1996) |
| KO229:pMN18 (produce Q7) | FS1576 ispB::Cmr pMN18 plasmid harboring H. influenzae ispB homologue | Okada et al. (1997) |
| KO229:pKA3 (produce Q8) | FS1576, ispB::Cmr, pKA3 plasmid harboring E. coli ispB | Okada et al. (1997) |
| KO229:pSN18 (produce Q9) | FS1576 ispB::Cmr pSN18 plasmid harboring Synechocystis sp. strain PCC6803 ispB homologue | Okada et al. (1997) |
| KO229:pLD23 (produce Q10) | FS1576 ispB::Cmr pLD23 plasmid harboring G. suboxydans ispB homologue | Okada et al. (1998) |
| AN180 | argE3 thi-1 strr | Butlin et al. (1971) |
| AN120 | argE3 thi-1 strr atpA401 | Butlin et al. (1971) |
| 1100 | bglR thi-1 rel-1 HfrP01 | Humbert et al. (1983) |
| 1100 Δbc | 1100 deleted for atpBEFHAGDC | Klionsky et al. (1983), Stack & Cain (1994) |
It seemed possible that the lack of rescue might lie with the efficiency of the uptake of the exogenously provided Q. Micelles of the hydrophobic compound, vitamin E, significantly increased its short-term bioavailability in healthy human patients (Back et al., 2006). In addition, a recent study showed that the degree of in vitro dissolution of exogenous coenzyme Q presented in various formulations correlated well with the level of uptake by Caco-2 human intestinal cells (Bhagavan et al., 2007). Thus, nematode growth media (NGM) were supplemented with NovaSOL Q, a water-soluble formulation containing Q10 as a neutral lipid core component packaged in 30-nm-diameter micelles. The clk-1 mutants fed Q-less E. coli on media supplemented with Q10 in this manner grew to adulthood and were fertile (Fig. 1). Although the NovaSOL Q supplementation provides only a partial rescue of fertility, the brood size of the clk-1(e2519) and clk-1(qm30) mutants was similar to those of animals fed the standard Q8-replete OP50 E. coli diet (Jonassen et al., 2001; Hihi et al., 2003). Plate media containing identical amounts of micelle particles but with medium-chain triglycerides as the neutral core component in place of Q10 (NovaSOL vehicle) failed to rescue growth arrest or restore fertility (Fig. 1). In order to determine the minimum amount of Q10 required to rescue the clk-1 mutants from sterility, the growth medium was supplemented with decreasing amounts of NovaSOL Q. Supplementation with Q10 at concentrations as low as 10 μg mL−1 (11.6 μM) sustained the growth and fertility of clk-1(e2519) animals fed Q-less E. coli (data not shown).
Fig. 1.
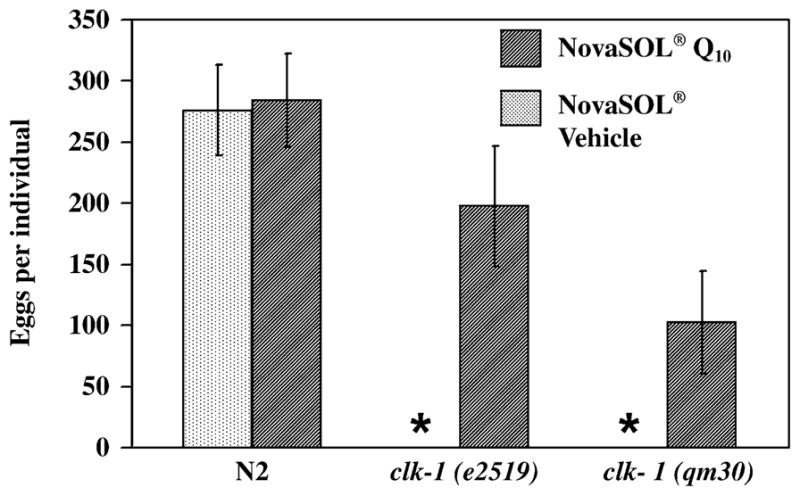
Supplementation with NovaSOL Q restores fertility of Caenorhabditis elegans clk-1 mutants fed Q-less Escherichia coli. The number of eggs laid per individual were determined for N2, clk-1 (e2519), and clk-1 (qm30) worms grown from the time of hatching on plates supplemented with NovaSOL Q (150 μg Q10 mL−1 plate media; dark cross-hatched bars), or with the NovaSOL vehicle control (light stippled bar). A lawn of the Q-less E. coli strain, GD1, was present on all plates. clk-1 mutants on nematode growth medium (NGM) plates with vehicle control arrested as L2 larvae for the duration of the brood size determination (*, 9 days). n = 12 for all conditions except for N2 worms grown on NGM with vehicle control where n = 10.
Q10 supplementation fails to rescue the Q-less E. coli mutant
It was possible that the water-soluble formulation of Q10 also rescued the respiratory growth defect of the GD1 E. coli. In this event, the exogenous Q may rescue the clk-1 mutants not necessarily by providing Q to the worms, but rather because the altered Q-less metabolism of the E. coli had been repaired. To determine whether this was the case serial dilutions of GD1 E. coli were plated onto succinate defined medium (SDM) plates containing succinate as a carbon source, where Q-dependent respiration is needed for growth. These plates were supplemented with either NovaSOL® Q or NovaSOL® vehicle control. As shown in Fig. 2(C and D), GD1 E. coli fail to grow on SDM under any of the conditions tested, indicating that the presence of the Q10 supplement fails to restore respiratory metabolism in the GD1 E. coli mutant. Because addition of Q6 to liquid media with vigorous aeration restores growth of yeast coq mutants (Jonassen et al., 1998; Do et al., 2001; James et al., 2005), we tested whether NovaSOL Q or addition of Q4, Q6, Q9, or Q10 would rescue growth of GD1 E. coli in liquid SDM. However, no rescue was observed (data not shown). In contrast, clk-1 mutants fed AN120, a respiratory defective E. coli strain lacking complex V but with normal levels of Q8 (Butlin et al., 1971), showed normal development and fertility (data not shown). These results identify Q as the essential component required for growth and fertility of the clk-1 mutant, and indicate that defective respiration or altered metabolites in the Q-less E. coli diet are not responsible for the clk-1 mutant growth arrest.
Fig. 2.
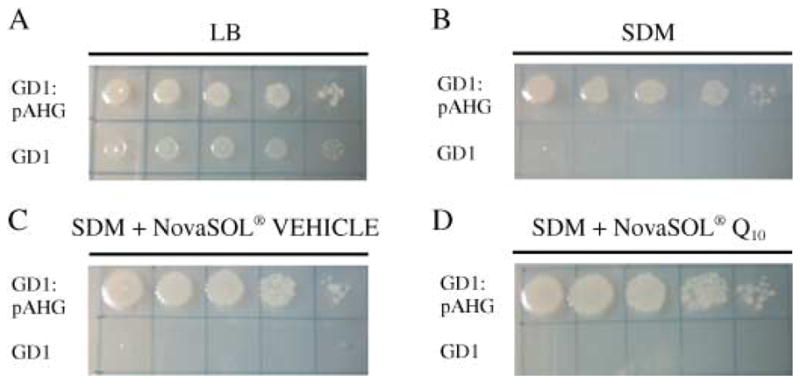
NovaSOL Q fails to restore growth of the Q-less Escherichia coli strain, GD1, on succinate defined medium (SDM). Serial dilutions of GD1 and the rescued strain, GD1:pAHG were applied to the following plate media: (A) Luria-Bertani, (B) SDM, (C) SDM supplemented with NovaSOL vehicle control, and (D) SDM supplemented with NovaSOL Q. Each plate was incubated overnight at 37 °C.
Caenorhabditis elegans mitochondria contain the exogenously provided Q10
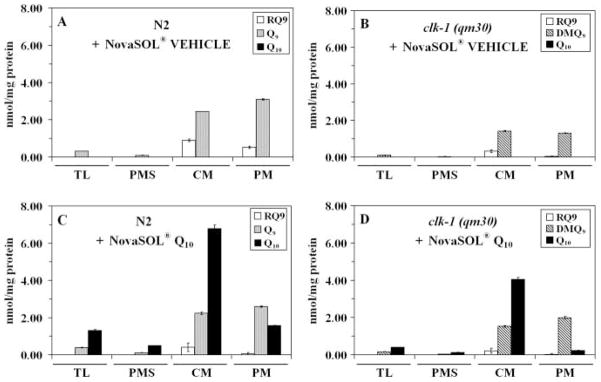
To determine the extent of uptake of the exogenously supplied Q10, mitochondria were isolated as described in the Experimental procedures from N2 and clk-1(qm30) nematodes fed the Q-less E. coli supplemented with either NovaSOL Q or NovaSOL vehicle control. The clk-1(qm30) mutants cultured in liquid media with vehicle control remain sterile but eventually reach adulthood after 6–7 days in culture. Lipids were extracted from worm lysates and isolated mitochondria, and the quinone content was determined by reverse phase high-performance liquid chromatography (HPLC) with an electrochemical detector. Significant amounts of Q10 were present in samples of worm lysate and mitochondria prepared from animals grown in medium supplemented with Q10, but were absent in mitochondria isolated from animals grown in medium supplemented with vehicle control (Fig. 3). While the majority of the exogenously supplied Q10 is found in the ‘crude mitochondria’ fraction, a small yet significant portion is present in the gradient purified mitochondria. Thus, both clk-1 mutant and N2 wild-type nematodes are able to transport this exogenous Q10 into their mitochondria.
Fig. 3.
Coenzyme Q10 is transported from the growth medium into both N2 and clk-1 (qm30) worm mitochondria. Measurement of rhodoquinone-9 (RQ9), demethoxyubiquinone-9 (DMQ9), ubiquinone-9 (Q9), and ubiquinone-10 (Q10) content in lipid extracts from worm total lysate (TL), post-mitochondrial supernatant (PMS), crude mitochondria (CM), and pure mitochondria (PM). N2 and clk-1 (qm30) worms were grown from starved L1 larvae to young adults in S medium supplemented with either NovaSOL vehicle control (A and B, respectively) or in S medium supplemented with NovaSOL Q (C and D, respectively). The Q-less Escherichia coli strain, GD1, was the bacterial food source in all cases. Data shown are representative of two independent experiments.
Q10 supplementation does not decrease lifespan of C. elegans fed a Q-less E. coli diet
After establishing the uptake of exogenous Q, we decided to use this water-soluble formulation to test whether the lifespan extension produced by feeding wild-type C. elegans the GD1 Q-less E. coli diet can be reversed. Adult C. elegans fed a Q-less E. coli diet show significant lifespan extension compared to worms fed a Q8-replete E. coli diet (Larsen & Clarke, 2002). This result implies that the supply of external Q8 from E. coli shortens worm lifespan. If Q is indeed the critical component affecting nematode lifespan, then supplementation of the GD1 Q-less E. coli diet with exogenously added Q would be expected to restore the short lifespan. To identify whether Q is the missing component responsible for the lifespan extension observed with the Q-less diet, we assayed lifespan with adult N2 worms fed the GD1 Q-less diet on NGM plates supplemented with 150 μg NovaSOL Q10 per milliliter. N2 worms were fed OP50 or GD1 with or without addition of NovaSOL Q10. A vehicle control with a neutral core containing medium-chain triglycerides instead of Q10 was also tested. Surprisingly, the NovaSOL Q10 supplement failed to restore the short lifespan (Fig. 4 and Table 2). In fact, both NovaSOL Q10 and the vehicle control supplement produced a slight but significant lifespan extension. Results were similar when N2 worms were fed the vehicle or Q10-supplemented diets for either one or two generations (data not shown). The data indicate that the lack of Q is not the critical factor responsible for lifespan extension mediated by the E. coli GD1 diet.
Fig. 4.
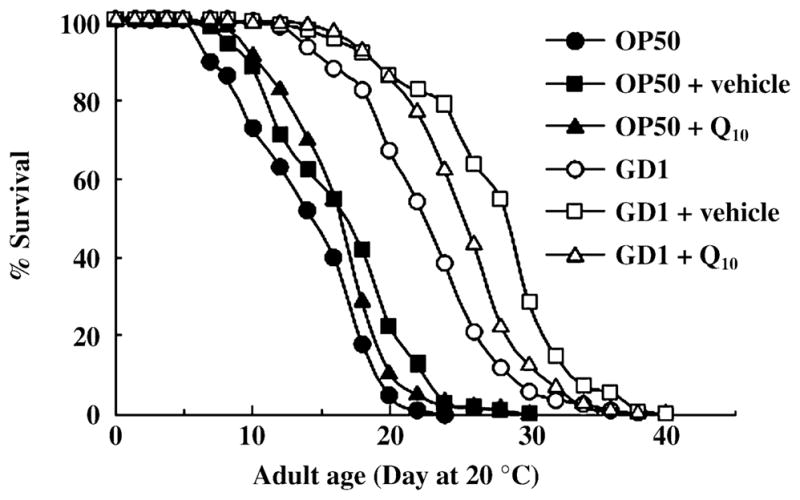
Adult Caenorhabditis elegans lifespan extension mediated by the GD1 Escherichia coli diet is not decreased by supplementation with NovaSOL Q10. Survival curves are shown for N2 worms cultured at 20 °C and fed either Q8-replete OP50 (black symbols) or Q-less GD1 (white symbols) E. coli. Worms were cultured for two generations on unsupplemented nematode growth medium (●,○), medium supplemented with NovaSOL Q10 (150 μg Q10 mL−1, ▲, △), or vehicle control (■, □). The vehicle control contains medium chain triglycerides as the neutral lipid core in place of Q10. Similar results were obtained for N2 worms fed the each of the stated diet for one generation.
Table 2.
Effects of dietary variables on Caenorhabditis elegans lifespan. Independent experiments are in groups
| Condition | Number of deaths (n)† | Mean lifespan (days ± SE)‡ | Maximum (days) | Percentage of mean on OP50 |
|---|---|---|---|---|
| OP50 | 135 | 14.48 ± 0.39 | 22 | – |
| OP50 + vehicle | 125 | 16.77 ± 0.46** | 28 | 115.9 |
| OP50 + Q10 | 131 | 16.75 ± 0.35** | 28 | 115.7 |
| GD1 | 119 | 23.29 ± 0.47** | 36 | 160.8 |
| GD1 + vehicle | 115 | 27.91 ± 0.53** | 38 | 192.7 |
| GD1 + Q10 | 123 | 25.87 ± 0.41** | 36 | 178.7 |
| OP50 | 152 | 18.60 ± 0.41 | 27 | – |
| GD1 | 160 | 25.02 ± 0.35** | 33 | 134.5 |
| KO229/Q7 | 158 | 20.63 ± 0.42** | 33 | 110.9 |
| KO229/Q8 | 157 | 19.34 ± 0.38 | 27 | 104.0 |
| KO229/Q9 | 156 | 20.13 ± 0.31 | 27 | 108.2 |
| KO229/Q10 | 159 | 19.37 ± 0.37 | 29 | 104.1 |
| OP50 | 163 | 22.20 ± 0.39 | 32 | – |
| OP50 + Amp | 136 | 27.43 ± 0.43** | 38 | 123.6 |
| OP50 | 178 | 19.33 ± 0.35 | 32 | – |
| GD1 | 174 | 26.25 ± 0.29** | 36 | 135.8 |
| OP50:pUG6 + Amp | 165 | 19.10 ± 0.37 | 30 | 98.8 |
| GD1:pUG6 + Amp | 140 | 24.82 ± 0.39** | 40 | 128.4 |
| GD1 + Amp (non-proliferating) | 154 | 30.53 ± 0.45** | 46 | 157.9 |
| OP50 | 140 | 17.73 ± 0.38 | 31 | – |
| GD1 | 162 | 26.46 ± 0.39** | 39 | 149.2 |
| OP50 + UV | 138 | 20.99 ± 0.29** | 37 | 118.4 |
| GD1 + UV | 85 | 22.81 ± 0.37** | 33 | 128.7 |
| OP50 | 167 | 20.20 ± 0.32 | 30 | – |
| GD1 | 123 | 31.15 ± 0.47** | 44 | 154.2 |
| AN180 | 146 | 21.11 ± 0.35* | 28 | 104.5 |
| AN120 (atpA−) | 143 | 27.06 ± 0.47** | 40 | 134.0 |
| 1100 | 160 | 21.01 ± 0.29 | 30 | 104.0 |
| 1100 Δbc (atpA-H−) | 115 | 25.70 ± 0.44* | 36 | 127.2 |
Animals that crawled away, had internally hatched larvae, or had eviscerated gonads were excluded.
Log rank test from survival analysis P-values:
0.001 < P < 0.05;
P < 0.001.
Diets containing Q8, Q9, Q10 isoforms do not influence worm lifespan
Differences in polyisoprenoid tail length of Qn isoforms among various species are well known. C. elegans produce Q9 while E. coli synthesize Q8. It is important to determine whether the endogenous Q8 isoform produced by E. coli might be the dietary component detrimental to lifespan. To determine this we measured the adult lifespan of N2 worms fed E. coli engineered to produce Q7, Q8, Q9 or Q10 (Table 1). The designated Qn isoform is the predominant species in each of these E. coli strains (Okada et al., 1996, 1997, 1998; Jonassen et al., 2003). In order to determine whether the Q isoforms produced might be altered following the extended incubations required for lifespan determination, we determined the quinone content for each of these E. coli strains recovered from NGM plates following incubation conditions that simulate a lifespan experiment. As shown in Table 3, the predominance of the major Q isoforms was not affected. N2 worms fed Q8-, Q9-, or Q10-producing E. coli showed no significant difference in lifespan when compared to worms fed the standard OP50 diet (Fig. 5 and Table 2). A modest but significant increase in lifespan was observed in N2 worms fed Q7. These results indicate that the shorter lifespan of N2 worms fed OP50 cannot be attributed to the ‘unnatural’ Q8 isoform provided by their E. coli diet.
Table 3.
Determination of Q isoform content in Escherichia coli diets provided on nematode growth medium (NGM) plate media
| pmol Qn isoform per milligram E. coli protein
| |||||
|---|---|---|---|---|---|
| Strain | Q6 | Q7 | Q8 | Q9 | Q10 |
| OP50 (Q8) | ND | 192.9 ± 14.6 | 2751.3 ± 135.2 | ND | ND |
| KO229:pMN18 (Q7) | 165.2 ± 2.8 | 1219.2 ± 7.3 | 143.9 ± 1.6 | ND | ND |
| KO229:pKA3 (Q8) | ND | 103.6 ± 3.6 | 1640.1 ± 48.9 | < 10 | ND |
| KO229:pSN18 (Q9) | ND | < 10 | 372.1 ± 3.1 | 1371.9 ± 11.6 | ND |
| KO229:PLD23 (Q10) | ND | < 10 | 26.7 ± 0.7 | 174.3 ± 3.4 | 1182.7 ± 6.0 |
Lipid extracts were prepared from the indicated E. coli strains seeded on NGM plate media as described in the Experimental procedures. ND, not detected.
Fig. 5.
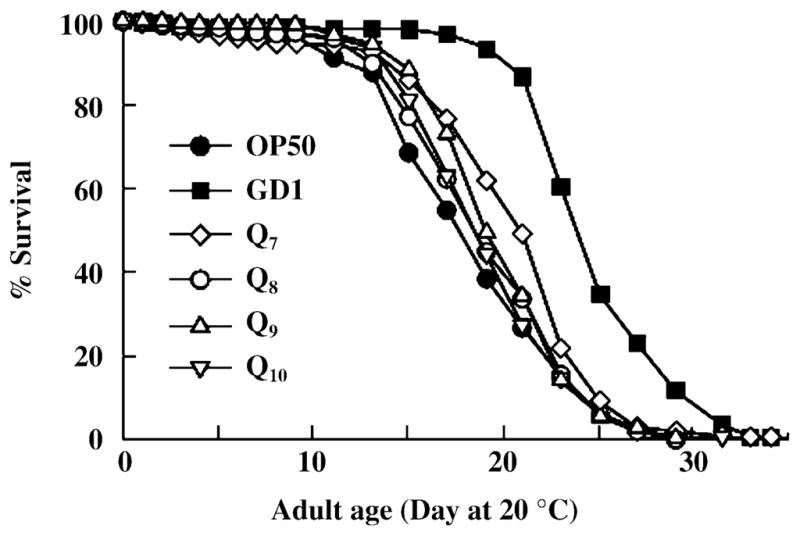
Caenorhabditis elegans fed Q8-, Q9-, or Q10-producing Escherichia coli have similar lifespans. N2 worms were cultured at 20 °C and fed the standard Q8-replete OP50 E. coli diet (●). Alternatively, worms were reared from hatching until the L4 larval stage on OP50 E. coli and then were transferred to the designated diets: GD1 Q-less E. coli (■), or E. coli engineered to produce predominantly Q7 (◇), Q8 (○), Q9 (△), or Q10 (▽). Results are representative of two experiments.
Caenorhabditis elegans consume Q-replete OP50 and Q-less GD1 E. coli at identical rates
To investigate whether C. elegans fed the E. coli GD1 diet experience dietary restriction, we measured the rate of consumption of OP50 or GD1 E. coli by N2 adult worms. There was no discernable difference in the rates of bacteria consumed by N2 worms (Fig. 6), suggesting that differences in the food consumption rate do not account for the effects on worm lifespan. Previous studies have shown that lifespan extension afforded by dietary restriction in nematodes is associated with low brood size (Klass, 1977; Bishop & Guarente, 2007). Since the brood size of N2 worms fed either the GD1 or OP50 E. coli diets are not significantly different (Jonassen et al., 2001), it seems unlikely that the worms fed the GD1 E. coli diet are experiencing a dietary restriction.
Fig. 6.
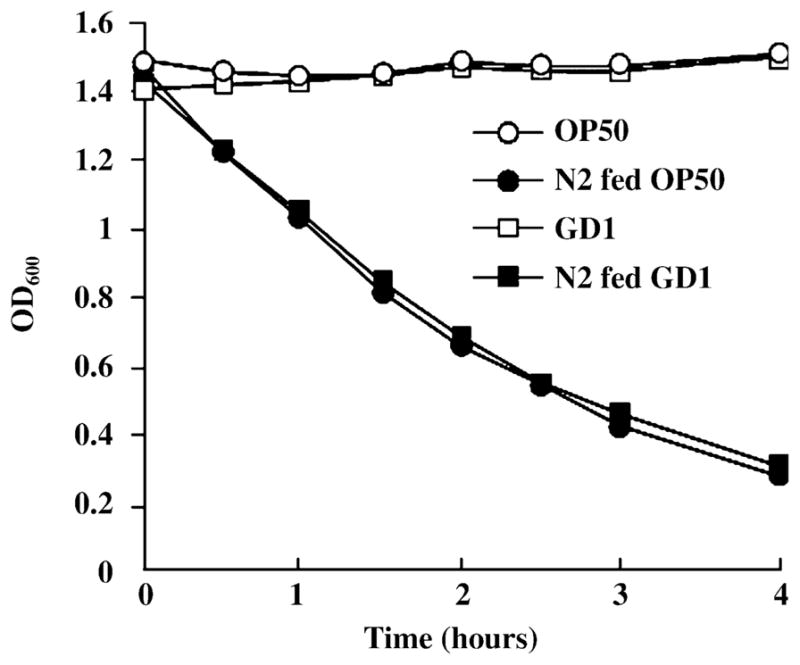
Adult Caenorhabditis elegans (N2) animals consume the Q-less (GD1) and Q-replete (OP50) Escherichia coli at identical rates. The rate of bacterial consumption was measured as previously described (Jones et al., 1996). Young adult worms were cultured in S medium containing equivalent amounts of either GD1 or OP50. Aliquots were taken from these cultures at 30-min intervals, and the OD600 of each aliquot was measured after briefly allowing the worms to settle on ice.
A diet of nonproliferating E. coli produces a lifespan extension greater than that observed with a diet of proliferating E. coli, whether Q-replete or Q-less
The Q-less GD1 E. coli grow more slowly than either Q-replete OP50 or the rescued strain, GD1/pAHG (Fig. 2). It was previously reported that N2 worms fed nonproliferating OP50 treated with antibiotics live longer than worms fed proliferating OP50 (Garigan et al., 2002; Walker et al., 2005). Indeed, N2 worms fed OP50 E. coli treated with ampicillin (at bacteriostatic concentrations), showed 23% life extension compared to worms fed the standard OP50 diet (Fig. 7A and Table 2). Based on these results, we hypothesized that the longer lifespan resulting from the GD1 diet might be attributed to the slow growth of the Q-less mutant strain. However, N2 worms fed ampicillin-treated GD1 E. coli showed approximately 16% lifespan extension over worms fed either untreated GD1, or GD1 E. coli harboring an ampicillin resistance plasmid (Fig. 7B and Table 2). These results suggest that it is unlikely that the lifespan extension can be attributed solely to the slower growth of the GD1 E. coli, or is due to differences in bacterial cell density provided on the plate growth media.
Fig. 7.
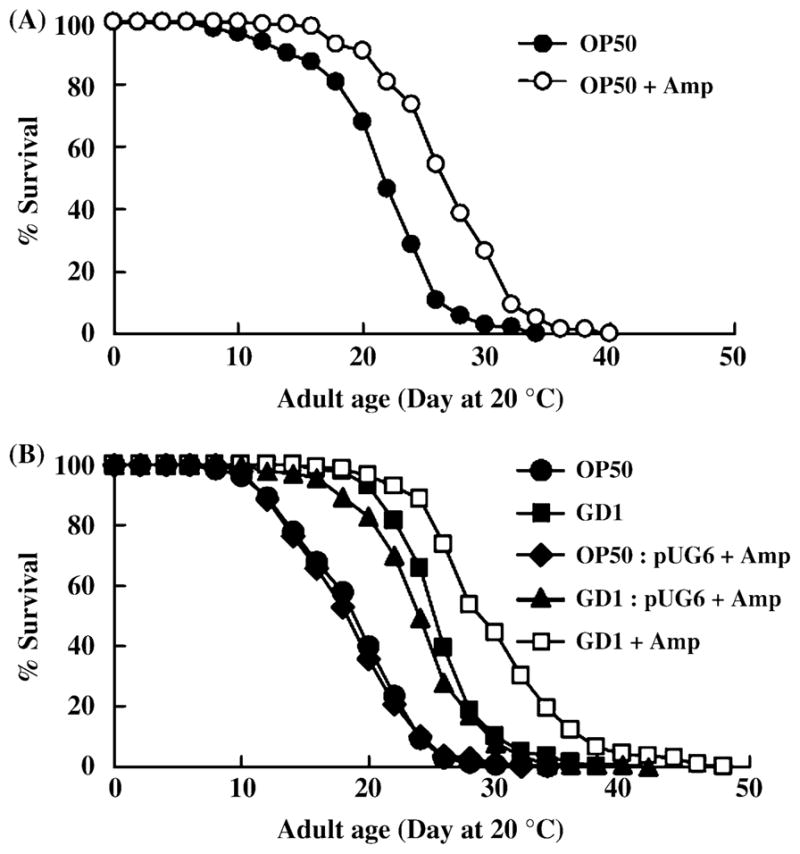
A diet of nonproliferating GD1 causes further lifespan extension relative to a diet of proliferating GD1. (A) Adult N2 worms were cultured at 20 °C and fed OP50 on nematode growth medium (NGM) plates with (○) or without ampicillin (●). Worms were cultured on regular NGM plates with OP50 until the L4 larval stage and subsequently transferred to the plates containing either proliferating or nonproliferating bacteria. (B) L4 stage worms were cultured on proliferating OP50 (●,◆), proliferating GD1 (■,▼) or nonproliferating GD1 (□). Bacteria harboring pUG6, an Escherichia coli vector conferring ampicillin resistance, are able to grow on NGM + ampicillin plates.
Ultraviolet irradiation destroys the GD1-mediated lifespan extension
Because N2 worms fed nondividing OP50 showed a lifespan extension (Gems & Riddle, 2000; Garigan et al., 2002; Walker et al., 2005), we anticipated that a bactericidal treatment such as ultraviolet (UV) irradiation would also enhance lifespan. However, N2 worms fed UV-treated GD1 showed significantly shorter lifespan than the worms fed untreated GD1 (Fig. 8 and Table 2). The UV treatment completely prevents bacteria from forming colonies when transferred to a new plate, whereas the ampicillin treatment inhibits bacterial growth but allows recovery upon transfer to a media without antibiotic. This indicated that the UV treatment might be destroying a beneficial component present in the GD1 E. coli. Additionally, factors produced by metabolically active GD1 E. coli may be important for the lifespan extension mediated by the GD1 diet.
Fig. 8.
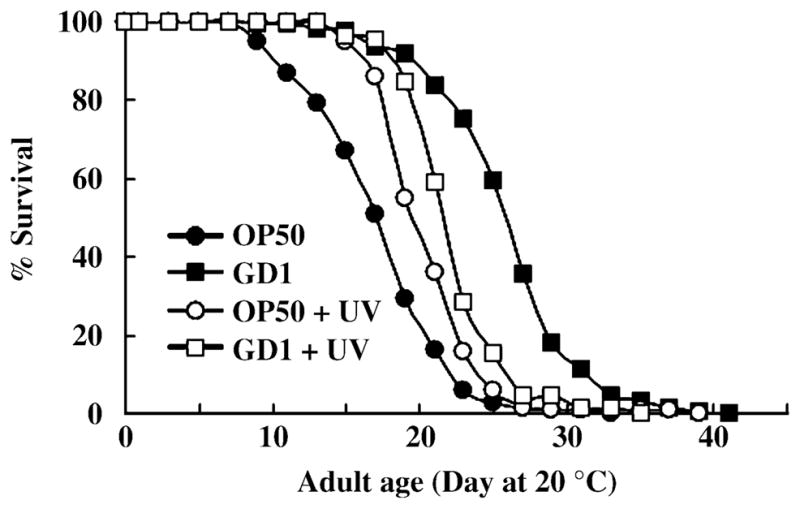
Ultraviolet (UV) irradiation of OP50 and GD1 E. coli produces opposing effects on worm lifespan. N2 worms were cultured at 20 °C and at the L4 larval stage transferred to plates containing either proliferating bacteria (closed symbols) or UV irradiated bacteria (open symbols). Where indicated, each nematode growth medium (NGM) plate with bacteria was irradiated with UV light by a DNA Transfer Lamp for 5 min. N2 worms were fed either UV irradiated OP50 (○), untreated OP50 (●), UV irradiated GD1 (□), or untreated GD1 (■).
N2 worms fed ATP synthase deficient bacteria have extended lifespan
An obvious metabolic difference between GD1 and OP50 E. coli is the severely compromised respiratory metabolism in GD1 due to the lack of Q8. For example, the GD1 E. coli fail to grow on minimal media containing succinate as sole carbon source, and this trait is not rescued by the provision of NovaSOL Q10 (Fig. 2). To elucidate whether the respiratory defective metabolism of E. coli modulates worm lifespan, we decided to test whether providing another strain of E. coli unable to grow on succinate might extend worm lifespan. The respiratory electron transport chain of E. coli has multiple oxidoreductases that use Q as electron acceptor and multiple terminal electron acceptors that utilize QH2 as electron donor (Gennis & Stewart, 1996). Because of the versatile respiratory metabolism in E. coli, it is difficult to ‘knock out’ respiration per se. Thus, we decided to assay lifespan in worms fed an ATP synthase (complex V) E. coli mutant. Although the respiratory chain is intact in this mutant, it cannot utilize the proton motive force generated by respiration and hence is unable to grow on succinate (Butlin et al., 1971). Therefore, we fed two independently derived ATP synthase mutants: AN120, which harbors an S373F mutation in atpA (uncA) encoding the ATP synthase alpha-subunit (Maggio et al., 1987); and the 1100 Δbc strain, in which the entire ATP synthase operon has been deleted (Klionsky et al., 1983; Stack & Cain, 1994). Importantly, the AN120 mutant has been shown to have similar levels of Q8 as compared to its parental strain, AN180 (Butlin et al., 1971). These ATP synthase defective mutants, or the corresponding parental strains were provided as E. coli diets. N2 worms fed the AN120 atpA mutant showed 34.9% and 29.7% longer lifespan than N2 fed OP50, or AN180, respectively (Fig. 9 and Table 2). Brood sizes of N2 fed OP50, AN120, or AN180 are not significantly different (306 ± 52, 327 ± 37, and 301 ± 29, respectively; n = 12 for each condition), indicating that this effect is probably not due to dietary restriction. Similarly, N2 worms fed the 1100 Δbc E. coli mutant also showed longer lifespans than the worms fed OP50 or 1100, the parental strain of 1100 Δbc (Table 2). Although the lifespan of worms fed either E. coli ATP synthase mutant is significantly longer, the degree of the lifespan extension is smaller than that produced by the GD1 diet. This result suggests that although the impaired respiratory metabolism of GD1 appears to be an important factor, additional attributes may be required for the lifespan extension observed with the GD1 diet.
Fig. 9.
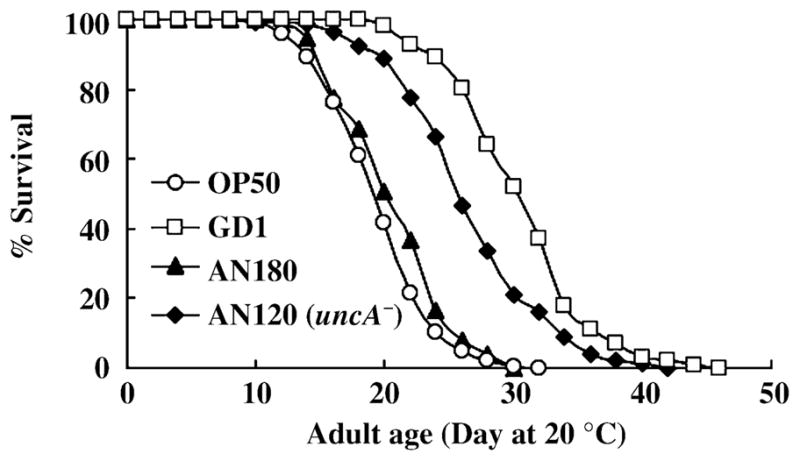
Impaired respiratory metabolism is an important factor contributing to the GD1 lifespan extension. N2 worms were reared on OP50 from hatching and at the L4 larval stage N2 worms were transferred to one of the following Escherichia coli strains: OP50 (○); GD1 (□); AN120 (◆), harboring a point mutation in the atpA gene encoding ATP synthase alpha subunit; or AN180 (▲), the ATP synthase parental strain.
Discussion
A water-soluble formulation of Q10 is assimilated by nematodes but not E. coli
This study shows that a water-soluble formulation of Q10, NovaSOL Q, is able to rescue the growth defect and restore fertility of the C. elegans clk-1 mutant. NovaSOL Q contains a neutral lipid core of Q10 encapsulated in 30-nm-diameter micelles that readily disperse in aqueous solutions. This enhanced dispersion into micelles is most likely responsible for the cellular uptake of Q10 observed in C. elegans cultured on medium containing NovaSOL Q, because addition of purified Q to media with a variety of non-ionic detergents such as Triton X-100 failed to rescue the clk-1 mutants. Micelles of Triton X-100 are about 8.8 nm in diameter (Kumbhakar et al., 2004), and may accommodate Q10 as a neutral lipid core component less efficiently. Although the NovaSOL Q was able to rescue the sterility and growth arrest of clk-1 nematodes, it did not restore growth of the Q-less GD1 E. coli mutant on media containing succinate. This result identifies Q as the essential component required for growth and fertility of the clk-1 mutant, and indicates that defective respiration or altered metabolites in the Q-less E. coli diet are not responsible for the clk-1 mutant growth arrest.
Relatively small amounts of exogenous coenzyme Q10 in the clk-1 mitochondria allowed rescue of growth arrest and sterility normally observed with a Q-less E. coli diet. Previously, it was shown that the small amounts of coenzyme Q8 assimilated by clk-1 animals fed the standard OP50 E. coli diet are sufficient to allow for growth and reproduction (Jonassen et al., 2002). The maternal effect rescue of clk-1 phenotypes is attributed to the small contribution of maternally supplied Q in the egg (Burgess et al., 2003). In addition, tRNA suppressors present in the clk-1(e2519) genetic background supplied only a low level of Q9 biosynthesis, yet allowed these animals to have brood sizes similar to wild type, and many of the suppressed mutants also had normal lifespans (Branicky et al., 2006). Thus, it is evident that amounts of Q much lower than normal can nonetheless function to rescue the clk-1 mutant growth, fertility and lifespan phenotypes.
Lifespan extension mediated by the GD1 E. coli diet is not due to the absence of Q
We report the surprising finding that the long lifespan of C. elegans fed the GD1 Q-less E. coli diet is in fact not dependent on the absence of Q per se. Supplementation of the Q-less E. coli diet with NovaSOL® Q, a water-soluble micelle-based formulation of Q10, did not rescue the long lifespan. Two lines of evidence indicate that the Q10 in the NovaSOL Q supplement is taken up by the worms: (i) when added to the GD1 E. coli diet this Q10 formulation rescues both clk-1 mutant growth arrest and sterility (Fig. 1); and (ii) the Q10 provided is readily detected in isolated mitochondria prepared from either N2 or clk-1 mutant animals (Fig. 3). We then suspected that the shorter lifespan of N2 worms fed the standard Q8-replete E. coli diet might be due to detrimental effects of the shorter Q8 isoform, since C. elegans normally produce Q9. However, this was not the case as N2 worms fed a diet of E. coli engineered to produce Q8, Q9, or Q10 isoforms showed lifespans that were no different from the standard Q8-replete OP50 diet (Fig. 5). These results indicate that the shorter lifespan of N2 worms fed a Q-replete E. coli diet cannot be attributed to the content of exogenously supplied Q.
Therefore, other attributes of the Q-less E. coli diet must be responsible for the lifespan extension. We considered the possibility that C. elegans fed the GD1 E. coli diet may experience dietary restriction, which is known to extend C. elegans lifespan (Klass, 1977; Bishop & Guarente, 2007). However, N2 young adult worms consume Q-replete and Q-less E. coli at equivalent rates (Fig. 3). Moreover, the brood size of N2 worms fed the GD1 E. coli diet is the same as worms fed the Q-replete OP50 E. coli diet (Jonassen et al., 2001). In contrast, dietary restriction regimes are known to decrease C. elegans brood size (Klass, 1977; Bishop & Guarente, 2007). Based on these observations, it seems unlikely that the GD1 E. coli diet is imposing a dietary restriction, although the experiments presented here do not rule this out. The slow growth of the GD1 E. coli might be another potential factor enhancing lifespan, because antibiotic treatment or UV irradiation of Q-replete E. coli diets are known to extend N2 worm lifespan (Gems & Riddle, 2000; Garigan et al., 2002; Walker et al., 2005). The lifespan extension produced by these treatments is thought to result from a decreased toxicity of E. coli in the worm intestine, although the exact mechanism accounting for the phenomenon is still unclear. Intriguingly, antibiotic treatment of the GD1 E. coli diet further augmented C. elegans lifespan, whereas UV irradiation of the GD1 E. coli shortened C. elegans lifespan (Figs 7 and 8). The opposing effects of these two treatments suggested that the slow growth of GD1 E. coli was not the important parameter. The abrogation of lifespan extension by UV treatment of GD1 E. coli suggests that the UV irradiation destroyed a beneficial component and/or the metabolic activity of viable GD1 E. coli is important.
E. coli respiratory metabolism modulates C. elegans lifespan
Wild-type C. elegans fed another respiratory deficient strain of E. coli, an ATP synthase mutant, have significantly longer lifespan than worms fed the standard E. coli diet. The ATP synthase mutant fails to grow on succinate media and is respiratory defective, although it has an intact respiratory chain, with normal content of cytochromes and Q8 (Butlin et al., 1971). We propose that one or more metabolites produced by the respiratory defective E. coli enhance C. elegans lifespan. It is also possible that respiratory competent E. coli produce detrimental metabolites. It is important to note that the lifespan extension provided by axenic media is abrogated by metabolically active, but not by killed, E. coli (Lenaerts, I., Walker, G., Van Hoorebeke, L., Gems, D., and Vanfleteren, J., personal communication). Intriguingly, this abrogation required a threshold concentration of E. coli. The changes in worm lifespan in response to decreased E. coli density could be interpreted as a form of dietary restriction. However, the findings presented here suggest that E. coli metabolism itself may modulate nematode lifespan. Thus, the physiology of E. coli cultured at high density, rather than E. coli ‘Calories’ per se, may be an important parameter modulating worm lifespan. The respiratory incompetent E. coli strains used here may either lack products normally produced by respiratory competent E. coli strains, or may produce fermentation products such as acetate, ethanol, lactate, formate and succinate (Bock & Sawers, 1996), that impact C. elegans metabolism and, hence, lifespan. This may be a phenomenon specific to C. elegans since feeding respiratory incompetent strains of S. cerevisiae (both Q-less and Q-replete) to Drosophila shortened lifespan as compared to the respiratory competent parental yeast strain (Palmer & Sackton, 2003). Unlike the variety of acids produced by respiratory incompetent E. coli, ethanol is the primary fermentation product of respiratory incompetent S. cerevisiae.
Q biosynthesis vs. uptake – phenotypes and pharmacology
A growing body of evidence indicates that manipulation of Q content via endogenous synthesis as opposed to dietary intervention produce dramatically different outcomes and may be in part related to intra- and extracellular systems of Q trafficking. Intriguingly, both C. elegans and mice heterozygous for a clk-1 gene disruption show increased lifespan (Liu et al., 2005). It is tempting to attribute the enhanced longevity to the decreased levels of endogenously produced Q in these two models. In contrast, although uptake was not monitored, addition of Q and Tween 80 to the growth medium extends lifespan of both wild-type and mev-1 mutants (Ishii et al., 2004). C. elegans mev-1 mutants suffer from a defect in respiratory Complex II and are hypersensitive to oxidative stress (Ishii et al., 1998; Senoo-Matsuda et al., 2001). Dietary Q supplementation in this manner also decreases superoxide anion levels in mev-1 mutants and affects the localization of DAF-16 in both mev-1 mutants and in gas-1 mutants (Kondo et al., 2005). C. elegans gas-1 mutants have a defect in respiratory Complex I (Kayser et al., 1999). DAF-16 is normally present in the cytoplasm and moves to the nucleus in response to environmental stressors such as oxidative stress (Henderson & Johnson, 2001). These data suggest that exogenous Q counteracts the stress caused by the gas-1 and mev-1 mutations. How can these findings be reconciled with the results presented here? Since Q10 is a very hydrophobic molecule, it is likely that the mode of presentation is of critical importance in the experimental outcome. A recent review of pharmacological interventions in C. elegans suggests that the variable lifespan effects produced by treatment with superoxide dismutase (SOD) mimetics may be highly sensitive to dosage, uptake, and to conditions of culture, including the degree of imposed stress (Collins et al., 2006). Effects of Q supplementation also depend on the species; lifespan of mice fed diets supplemented with Q10 showed no alteration (Sohal et al., 2006).
The mechanism of Q uptake remains unclear. In C. elegans it seems likely that the exogenous Q is ingested, absorbed via the gut, and distributed to the gonad in order to restore fertility. Intracellular transport of exogenous Q in HL-60 cells is sensitive to inhibition by brefeldin-A and requires the endomembrane system (Fernandez-Ayala et al., 2005). It is possible that in C. elegans cellular uptake of Q relies on this same system. The yolk proteins (vitellogenins) and their receptor, RME-2, have been shown to be important for the proper uptake and transport of cholesterol in C. elegans (Matyash et al., 2001). Vitellogenins are generated in the intestine, secreted into the body cavity, and then taken up by oocytes in adult hermaphrodites (Kimble & Sharrock, 1983). Q may also bind to the vitellogenins as it is transported from the gut throughout the body of the animal. Once Q is taken into cells it must be assimilated within a variety of organelles including the mitochondrial inner membrane in order to function in respiration. The C. elegans homolog of the yeast Coq10p could be involved in its proper delivery to the mitochondria respiratory transport complexes (Barros et al., 2005). Providing exogenous Q via the NovaSOL formulation will facilitate further studies elucidating the mechanism of uptake and mode of intracellular transport of Q in C. elegans.
Experimental procedures
Culture conditions and strains
Caenorhabditis elegans strains used were the wild-type control N2 (Bristol strain) (Brenner, 1974), and the clk-1 mutant strains CB4876 clk-1(e2519) III and MQ130 clk-1(qm30) III (Wong et al., 1995). C. elegans were cultured in S medium (liquid) [0.1 M sodium chloride, 0.05 M potassium phosphate (pH 6), 5 μg mL−1 cholesterol, 0.01 M potassium citrate (pH 6), 3 mM calcium chloride, 3 mM magnesium sulfate, 50 μM EDTA, 25 μM ferrous sulfate, 10 μM manganese chloride, 10 μM zinc sulfate, 1 μM copper sulfate] or on standard NGM plates with various bacteria food sources (Sulston & Hodgkin, 1988).
Escherichia coli strains used in this study are listed in Table 1. The Q-less strain, GD1 (ubiG::Kan, zei::Tn10dTet) (Hsu et al., 1996), contains a deletion of the ubiG gene required for Q biosynthesis. Succinate defined medium (SDM) contained 0.5% casamino acids supplement and was prepared as described (Poole et al., 1989). E. coli strains harboring antibiotic resistance were streaked on NGM plates with corresponding antibiotics (e.g. 50 μg mL−1 kanamycin and 100 μg mL−1 ampicillin). The water-soluble 22% coenzyme Q10 solubilisate and vehicle control, hereafter referred to as NovaSOL Q and NovaSOL vehicle control, respectively, were kindly provided by AQUANOVA AG (Darmstadt, Germany).
Escherichia coli growth assay
GD1 and GD1:pAHG, which harbors the E. coli ubiG gene on a plasmid and produces Q8 (Hsu et al., 1996), were inoculated in liquid Luria-Bertani and grown overnight. Serial dilutions were plated onto Luria-Bertani, SDM alone, SDM supplemented with NovaSOL Q, and SDM supplemented with NovaSOL vehicle control. NovaSOL Q was added to the SDM at a concentration of 682 μg solubilisate/milliliter medium to achieve a final Q10 concentration of 150 μg mL−1. NovaSOL vehicle control was added at a concentration of 532 μg solubilisate/milliliter medium. E. coli growth was assessed after an overnight incubation at 37 °C. All media in which the GD1 and GD1:pAHG strains were cultured contained 50 μg mL−1 kanamycin.
Brood size determination
Eggs were isolated from gravid N2, clk-1(e2519), and clk-1(qm30) worms with an alkaline hypochlorite treatment as described (Sulston & Hodgkin, 1988). The eggs were placed on NGM supplemented with either NovaSOL Q or NovaSOL vehicle control. The concentrations of the NovaSOL Q and NovaSOL vehicle control were the same as those used in the supplemented SDM plates. All plates contained a lawn of GD1. Individual worms that developed to the L4 larval stage were transferred daily to fresh plates, and the number of eggs laid by each worm was recorded.
Mitochondria purification
Mitochondria were isolated from young adult worms as described (Curran et al., 2004). L1 larvae were prepared by treating gravid adult nematodes with sodium hypochlorite, rinsing the liberated eggs, and allowing the eggs to hatch overnight in S medium without E. coli. Fresh cultures were then started with starved L1 larvae from N2 or clk-1(qm30) at 2000 larvae/milliliter of S medium supplemented with either NovaSOL Q or with NovaSOL vehicle control at the concentrations stated above. These nematodes were fed GD1 and allowed to develop into young adults at 20 °C. The animals were cleaned via sucrose floatation and suspended in STEG buffer [250 mM sucrose, 5 mM Tris-HCl (pH 7.4) and 1 mM EGTA] containing 1 mM PMSF and protease inhibitor mixture (PIC, Roche Diagnostics, Indianapolis, IN, USA). The worm suspension was homogenized with a Kontes ground glass tissue grinder, and unbroken worm bodies were removed by centrifugation at 750 g for 10 min. An aliquot of the 750 g supernatant was saved as the ‘total lysate’, and the remainder was then centrifuged at 12 000 g for 10 min. An aliquot of the 12 000 g supernatant was saved as the ‘post mitochondrial supernatant’. The 12 000 g pellet was resuspended in STEG buffer without protease inhibitors, and centrifuged at 750 g for 10 min, and the supernatant centrifuged at 12 000 g for 10 min. A portion of this pellet was saved as ‘crude mitochondria’.
An enriched mitochondrial fraction, which we designated ‘pure mitochondria’, was generated by resuspending the remaining ‘crude mitochondria’ and loading onto a discontinuous sucrose gradient composed of layers of 1.2 M, 1.3 M, 1.6 M, and 1.8 M solutions of sucrose. This gradient was centrifuged at 25 000 g for 90 min, and the band migrating at the 1.3 M and 1.6 M sucrose interface was resuspended in STEG buffer and saved as ‘pure mitochondria’. All worm lysate fractions were stored at −80 °C until use. Protein concentrations were measured by the bicinchoninic acid assay (Pierce, Rockford, IL, USA). Enzyme assays were performed as described (Jonassen et al., 2003). The enzymatic rates reported were calculated by subtracting signal produced in the absence of substrate from the signal produced when substrate was present at various time points. The band migrating at the 1.3 M and 1.6 M sucrose interface was enriched in succinate-cytochrome c reductase (mitochondria-specific) activity, while the mannosidase II (Golgi-specific) and glucose 6-phosphatase (endoplasmic reticulum-specific) activities in this band were lower than those present in the ‘crude mitochondria’ pellet (Table 4).
Table 4.
Separation of crude mitochondria on a discontinuous sucrose gradient results in a fraction specifically enriched in succinate-cytochrome c reductase activity
| Subcellular fraction | Succinate-cytochrome c reductase activity (mitochondria) | Mannosidase II activity (Golgi) | Glucose 6-phosphatase activity (endoplasmic reticulum) |
|---|---|---|---|
| nmol cytochrome c | nmol para-nitrophenol | nmol phosphate | |
| reduced/min/mg protein | produced/h/mg protein | produced/min/mg protein | |
| Crude mitochondria | 296.6 ± 8.6 | 73.9 ± 16.4 | 60.7 ± 7.5 |
| Pure mitochondria | 559.4 ± 10.8 | 44.2 ± 3.3 | 36.3 ± 11.5 |
Crude mitochondria isolated from synchronous young adult N2 worms were separated on a discontinuous sucrose gradient (1.2 M, 1.3 M, 1.6 M, and 1.8 M). The band at the interface between 1.3 M and 1.6 M was isolated as the pure mitochondria fraction. Activities represent the average of three assays ± the standard deviation.
Isolation and quantification of quinones
Aliquots of the total lysate, post mitochondrial supernatant, crude mitochondria, and pure mitochondria from N2 and clk-1(qm30) worms were subjected to lipid extraction and analyzed for Q content (Jonassen et al., 2002). Q6 was used to gauge quinone recovery in the lipid extracts and was added to all standards and worm lysate samples. Standards and samples were extracted by adding 0.5 mL water, 9 mL methanol, and 6 mL petroleum ether to each. The mixtures were vortexed for 1 min, centrifuged at 910 g, and the top layer of petroleum ether was removed from each mixture and saved in a separate vial. Fresh petroleum ether (6 mL) was added to each vial containing the aqueous phase and vortexed for 1 min The vials were subjected to centrifugation as before and the second petroleum ether layer removed. The process was repeated once more, and the three pooled petroleum ether fractions were dried under nitrogen and resuspended in 150 μL methanol.
The rhodoquinone-9 standard was purified from whole Ascaris suum tissue with the lipid extraction procedure described above, and rhodoquinone-9 was isolated by normal phase thin layer chromatography (Whatman, Florham Park, NJ, USA, cat. no. 4861-830) developed with 1 : 1 benzene/chloroform. The Q6 and Q10 standards were from Sigma-Aldrich (St. Louis, MO, USA), and Q9 was a gift from Dr Youssef Hatefi.
The quinones present in extracted standards and samples were separated and quantified by HPLC connected to an electrochemical detector as described (Jonassen et al., 2002), with the following exceptions: the precolumn electrode was the only electrode used and was set at +650 mV to oxidize all quinones, and a Gilson 118 UV/Vis detector was utilized to detect quinones as they eluted from the column. The amounts of rhodoquinone-9, Q9, demethoxyubiquinone-9, and Q10 in the standards and samples was normalized to the amount of Q6 recovered in the individual lipid extracts.
The content of Q isoforms was determined for OP50 and in the KO229 E. coli strains designed to produce predominantly Q7, Q8, Q9, or Q10. In order to determine whether incubation on NGM plate media affected the predominant Q isoform, each strain was subjected to incubation conditions that would simulate the conditions of a lifespan experiment. Each E. coli strain was applied to an NGM plate, and after incubation at 37 °C overnight, the plates were stored at 4 °C for 4 weeks, followed by 1 week at 20 °C. Each plate was then rinsed with M9, and E. coli recovered from five NGM plates were collected in one tube. Preparation of E. coli lipid extracts and determination of the quinone content was performed as described (Jonassen et al., 2003).
Food consumption assay
The rate of bacterial consumption by adult N2 C. elegans was measured as previously described (Jones et al., 1996). Young adult worms were cultured in S medium containing equivalent amounts of either GD1 or OP50. Aliquots were taken from these cultures at 30-min intervals, and the OD600 of each aliquot was measured after briefly allowing the worms to settle on ice.
Preparation of UV irradiated bacteria
NGM plates with E. coli strains (OP50 or GD1) were irradiated with UV light for 5 min with a DNA Transfer Lamp (FOTODYNE, Inc., Hartland, WI, USA). Following irradiation, at least one of the treated plates was checked to verify complete loss of colony formation when replica plated to Luria-Bertani plate media.
Lifespan analysis
All measurements were performed at 20 °C, with the zero time point taken at the late fourth larval stage. During the egg-laying period hermaphrodites were transferred daily to new plates. These animals were transferred to freshly streaked bacterial plates every 4 days after the egg-laying period. Animals that did not respond when prodded by a platinum wire pick were counted as dead. If an animal was suspected dead, the worm was straightened out with the pick and checked to determine whether it remained straight (dead) or it had moved (alive) the next day. Worms showing abnormal death such as vulva explosion, hatched progeny inside the hermaphrodite or climbing up the plate wall were excluded from the lifespan analysis. Statview 5.0.1 (SAS) software was used to construct lifespan curves and to perform statistical analysis.
Supplementary Material
Table for data of Fig. 4
Table for data of Fig. 5
Table for data of Fig. 7(A)
Table for data of Fig. 7(B)
Table for data of Fig. 8
Table for data of Fig. 9
Acknowledgments
We wish to thank D. Behnam at AQUANOVA AG for providing us with NovaSOL Q and NovaSOL vehicle control. We are also indebted to N. Ishii, S. Curran, M. Jackson, and T. Kruse for their technical advice and help, to M. Kawamukai for providing the E. coli strains engineered to produce Q7–Q10, and to D. Bowman for providing us with Ascaris suum tissue as a source of rhodoquinone-9. We thank A. Frand, A. van der Bliek, D. Gems, S. Clarke, J. Vanfletern and I. Lenaerts for their input on drafts of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported in part by NIH grant GM45952 and National Institutes of Aging (NIA) grant AG19777 to C.F.C., by a grant from the Ellison Medical Foundation to P.L.L. and C.F.C., and by a grant from the Glenn Medical Foundation to P.L.L. A.L.L. received support from the Ruth L. Kirschstein National Research Service Award issued from NIH training grant GM007185.
References
- Back EI, Frindt C, Ocenaskova E, Nohr D, Stern M, Biesalski HK. Can changes in hydrophobicity increase the bioavailability of α-tocopherol? Eur J Nutr. 2006;45:1–6. doi: 10.1007/s00394-005-0556-9. [DOI] [PubMed] [Google Scholar]
- Barros MH, Johnson A, Gin P, Marbois BN, Clarke CF, Tzagoloff A. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem. 2005;280:42627–42635. doi: 10.1074/jbc.M510768200. [DOI] [PubMed] [Google Scholar]
- Bentinger M, Brismar K, Dallner G. The antioxidant role of coenzyme Q. Mitochondrion. 2007;7S:S41–S50. doi: 10.1016/j.mito.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Bhagavan HN, Chopra RK, Craft NE, Chitchumroonchokchai C, Failla ML. Assessment of coenzyme Q10 absorption using an in vitro digestion-Caco-2 cell model. Int J Pharm. 2007;333:112–117. doi: 10.1016/j.ijpharm.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Science. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bock A, Sawers G. Fermentation. In: Neidhardt FC, Curtiss R, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 262–282. [Google Scholar]
- Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr Biol. 1999;9:493–496. doi: 10.1016/s0960-9822(99)80216-4. [DOI] [PubMed] [Google Scholar]
- Branicky R, Nguyen PA, Hekimi S. Uncoupling the pleiotropic phenotypes of clk-1 with tRNA missense suppressors in Caenorhabditis elegans. Mol Cell Biol. 2006;26:3976–3985. doi: 10.1128/MCB.26.10.3976-3985.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess J, Hihi AK, Benard CY, Branicky R, Hekimi S. Molecular mechanism of maternal rescue in the clk-1 mutants of Caenorhabditis elegans. J Biol Chem. 2003;278:49555–49562. doi: 10.1074/jbc.M308507200. [DOI] [PubMed] [Google Scholar]
- Butlin JD, Cox GB, Gibson F. Oxidative phosphorylation in Escherichia coli K12. Mutations affecting magnesium ion- or calcium ion-stimulated adenosine triphosphatase. Biochem J. 1971;124:75–81. doi: 10.1042/bj1240075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Evason K, Kornfeld K. Pharmacology of delayed aging and extended lifespan of Caenorhabditis elegans. Exp Gerontol. 2006;41:1032–1039. doi: 10.1016/j.exger.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Curran SP, Leverich EP, Koehler CM, Larsen PL. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J Biol Chem. 2004;279:54655–54662. doi: 10.1074/jbc.M409618200. [DOI] [PubMed] [Google Scholar]
- Do TQ, Hsu AY, Jonassen T, Lee PT, Clarke CF. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- Dutton PL, Ohnishi T, Darrouzet E, Leonard MA, Sharp RE, Gibney BR, Daldal F, Moser CC. Coenzyme Q oxidation reduction reactions in mitochondrial electron transport. In: Kagan VE, Quinn PJ, editors. Coenzyme Q: Molecular Mechansisms in Health and Disease. Boca Raton, FL: CRC Press; 2000. pp. 65–82. [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, Brea-Calvo G, Lopez-Lluch G, Navas P. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim Biophys Acta. 2005;1713:129–137. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Galpern WR, Cudkowicz ME. Coenzyme Q treatment of neurodegnerative diseases of aging. Mitochondrion. 2007;7S:S146–S153. doi: 10.1016/j.mito.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. Respiration. In: Neidhardt FC, Curtiss R, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 217–261. [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hihi AK, Kebir H, Hekimi S. Sensitivity of Caenorhabditis elegans clk-1 mutants to ubiquinone side-chain length reveals multiple ubiquinone-dependent processes. J Biol Chem. 2003;278:41013–41018. doi: 10.1074/jbc.M305034200. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Vanfleteren JR. Public and private mechanisms of life extension in Caenorhabditis elegans. Mol Genet Genomics. 2007;277:601–617. doi: 10.1007/s00438-007-0225-1. [DOI] [PubMed] [Google Scholar]
- Hsu AY, Poon WW, Shepherd JA, Myles DC, Clarke CF. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- Humbert R, Brusilow WS, Gunsalus RP, Klionsky DJ, Simoni RD. Escherichia coli mutants defective in the uncH gene. J Bacteriol. 1983;153:416–422. doi: 10.1128/jb.153.1.416-422.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Ishii N, Senoo-Matsuda N, Miyake K, Yasuda K, Ishii T, Hartman PS, Furukawa S. Coenzyme Q10 can prolong C. elegans lifespan by lowering oxidative stress. Mech Ageing Dev. 2004;125:41–46. doi: 10.1016/j.mad.2003.10.002. [DOI] [PubMed] [Google Scholar]
- James AM, Cocheme HM, Smith RA, Murphy MP. Interactions of mitochondria-targeted and untargeted ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. Implications for the use of exogenous ubiquinones as therapies and experimental tools. J Biol Chem. 2005;280:21295–21312. doi: 10.1074/jbc.M501527200. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Proft M, Randez-Gil F, Schultz JR, Marbois BN, Entian KD, Clarke CF. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Larsen PL, Clarke CF. A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc Natl Acad Sci USA. 2001;98:421–426. doi: 10.1073/pnas.021337498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen T, Marbois BN, Faull KF, Clarke CF, Larsen PL. Development and fertility in Caenorhabditis elegans clk-1 mutants depend upon transport of dietary coenzyme Q8 to mitochondria. J Biol Chem. 2002;277:45020–45027. doi: 10.1074/jbc.M204758200. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Davis DE, Larsen PL, Clarke CF. Reproductive fitness and quinone content of Caenorhabditis elegans clk-1 mutants fed coenzyme Q isoforms of varying length. J Biol Chem. 2003;278:51735–51742. doi: 10.1074/jbc.M308760200. [DOI] [PubMed] [Google Scholar]
- Jones D, Stringham EG, Babich SL, Candido EP. Transgenic strains of the nematode C. elegans in biomonitoring and toxicology: effects of captan and related compounds on the stress response. Toxicology. 1996;109:119–127. doi: 10.1016/0300-483x(96)03316-1. [DOI] [PubMed] [Google Scholar]
- Kawamukai M. Biosynthesis, bioproduction and novel roles of ubiquinone. J Bioscience Bioengineering. 2002;94:511–517. doi: 10.1016/s1389-1723(02)80188-8. [DOI] [PubMed] [Google Scholar]
- Kayser EB, Morgan PG, Sedensky MM. GAS-1: a mitochondrial protein controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. Anesthesiology. 1999;90:545–554. doi: 10.1097/00000542-199902000-00031. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Brusilow WS, Simoni RD. Assembly of a functional F0 of the proton-translocating ATPase of Escherichia coli. J Biol Chem. 1983;258:10136–10143. [PubMed] [Google Scholar]
- Kondo M, Senoo-Matsuda N, Yanase S, Ishii T, Hartman PS, Ishii N. Effect of oxidative stress on translocation of DAF-16 in oxygen-sensitive mutants, mev-1 and gas-1 of Caenorhabditis elegans. Mech Ageing Dev. 2005;126:637–641. doi: 10.1016/j.mad.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kumbhakar M, Nath S, Mukherjee T, Pal H. Solvation dynamics in triton-X-100 and triton-X-165 micelles: effect of micellar size and hydration. J Chem Phys. 2004;121:6026–6033. doi: 10.1063/1.1784774. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Clarke CF. Extension of life span in C. elegans by a diet lacking coenzyme Q. Science. 2002;295:120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- Levavasseur F, Miyadera H, Sirois J, Tremblay ML, Kita K, Shoubridge E, Hekimi S. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J Biol Chem. 2001;276:46160–46164. doi: 10.1074/jbc.M108980200. [DOI] [PubMed] [Google Scholar]
- Linnane AW, Kios M, Vitetta L. Coenzyme Q10 – its role as a prooxidant in the formation of superoxide anion/hydrogen peroxide and the regulation of the metabolome. Mitochondrion. 2007;7S:S51–S61. doi: 10.1016/j.mito.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio MB, Pagan J, Parsonage D, Hatch L, Senior AE. The defective proton-ATPase of uncA mutants of Escherichia coli. Identification by DNA sequencing of residues in the alpha-subunit which are essential for catalysis or normal assembly. J Biol Chem. 1987;262:8981–8984. [PubMed] [Google Scholar]
- Matyash V, Geier C, Henske A, Mukherjee S, Hirsh D, Thiele C, Grant B, Maxfield FR, Kurzchalia TV. Distribution and transport of cholesterol in Caenorhabditis elegans. Mol Biol Cell. 2001;12:1725–1736. doi: 10.1091/mbc.12.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, Miyoshi H, Sakamoto K, Ishii N, Hekimi S, Kita K. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J Biol Chem. 2001;276:7713–7716. doi: 10.1074/jbc.C000889200. [DOI] [PubMed] [Google Scholar]
- Nakai D, Yuasa S, Takahashi M, Shimizu T, Asaumi S, Isono K, Takao T, Suzuki Y, Kuroyanagi H, Koseki H, Shirasawa T. Mouse homologue of coq7/clk-1, longevity gene in Caenorhabditis elegans is essential for coenzyme Q synthesis, maintenance of mitochondrial integrity and neurogenesis. Biochem Biophys Res Commun. 2001;289:463–471. doi: 10.1006/bbrc.2001.5977. [DOI] [PubMed] [Google Scholar]
- Okada K, Suzuki K, Kamiya Y, Zhu X, Fujisaki S, Nishimura Y, Nishino T, Nakagawa T, Kawamukai M, Matsuda H. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim Biophys Acta. 1996;1302:217–223. doi: 10.1016/0005-2760(96)00064-1. [DOI] [PubMed] [Google Scholar]
- Okada K, Minehira M, Zhu X, Suzuki K, Nakagawa T, Matsuda H, Kawamukai M. The ispB gene encoding octaprenyl diphosphate synthase is essential for growth of Escherichia coli. J Bacteriol. 1997;179:3058–3060. doi: 10.1128/jb.179.9.3058-3060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Kainou T, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur J Biochem. 1998;255:52–59. doi: 10.1046/j.1432-1327.1998.2550052.x. [DOI] [PubMed] [Google Scholar]
- Olson RE, Rudney H. Biosynthesis of ubiquinone. Vitam Horm. 1983;40:1–43. doi: 10.1016/s0083-6729(08)60431-8. [DOI] [PubMed] [Google Scholar]
- Palmer MR, Sackton TB. The effects of dietary coenzyme Q on Drosophila life span. Aging Cell. 2003;2:335–339. doi: 10.1046/j.1474-9728.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- Pepe S, Marasco SF, Haas SJ, Sheeran FL, Krum H, Rosenfeldt FL. Coenzyme Q10 in cardiovascular disease. Mitochondrion. 2007;7S:S154–S167. doi: 10.1016/j.mito.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Poole RK, Williams HD, Downie JA, Gibson F. Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: identification and mapping of a fourth locus, cydD. J Gen Microbiol. 1989;135:1865–1874. doi: 10.1099/00221287-135-7-1865. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, Nakazawa H, Hartman PS, Ishii N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J Biol Chem. 2001;276:41553–41558. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Kamzalov S, Sumien N, Ferguson M, Rebrin I, Heinrich KR, Forster MJ. Effect of coenzyme Q10 intake on endogenous coenzyme Q content, mitochondrial electron transport chain, antioxidative defenses, and life span of mice. Free Radic Biol Med. 2006;40:480–487. doi: 10.1016/j.freeradbiomed.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack AE, Cain BD. Mutations in the delta subunit influence the assembly of F1F0 ATP synthase in Escherichia coli. J Bacteriol. 1994;176:540–542. doi: 10.1128/jb.176.2.540-542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7S:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Shukitt-Hale B, Kalt W, Ingram DK, Joseph JA, Wolkow CA. Blueberry polyphenols increase lifespan and thermotolerance in Caenorhabditis elegans. Aging Cell. 2006;5:59–68. doi: 10.1111/j.1474-9726.2006.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table for data of Fig. 4
Table for data of Fig. 5
Table for data of Fig. 7(A)
Table for data of Fig. 7(B)
Table for data of Fig. 8
Table for data of Fig. 9