Abstract
Introduction
Ritonavir is a potential therapeutic agent in lung cancer, but its targets in lung adenocarcinoma are unknown, as are candidate biomarkers for its activity.
Methods
RNAi was used to identify genes whose expression affects ritonavir sensitivity. Synergy between ritonavir, gemcitabine and cisplatin was tested by isobologram analysis.
Results
Ritonavir inhibits growth of K-ras mutant lung adenocarcinoma lines A549, H522, H23 and K-ras wild type line H838. Ritonavir causes G0/G1 arrest and apoptosis. Associated with G0/G1 arrest, ritonavir down-regulates cyclin dependent kinases, cyclin D1 and Rb phosphorylation. Associated with induction of apoptosis, ritonavir reduces survivin mRNA and protein levels more than 2-fold. Ritonavir inhibits phosphorylation of c-Src and STAT3, which are important events for survivin gene expression and growth, and induces cleavage of PARP1. While knock down of survivin, c-Src or STAT3 inhibits cell growth, only survivin knock down enhances ritonavir inhibition of growth and survivin over-expression promotes ritonavir resistance. Ritonavir was tested in combination with gemcitabine or cisplatin exhibiting synergistic and additive effects, respectively. The combination of ritonavir/gemcitabine/cisplatin is synergistic in the A549 line and additive in the H522 line, at clinically feasible ritonavir concentrations (<10 μM).
Conclusions
Ritonavir is of interest for lung adenocarcinoma therapeutics and survivin is an important target and potential biomarker for its sensitivity. Ritonavir cooperation with gemcitabine/cisplatin might be explained by involvement of PARP1 in repair of cisplatin-mediated DNA damage and survivin in repair of gemcitabine-mediated double stranded DNA breaks (DSB).
Keywords: Non-small cell lung cancer, ritonavir, survivin, chemotherapy
Introduction
A potentially successful approach for cancer drug development consists of re-purposing existing drugs, with known tolerability, toxicity and pharmacology 1. Ritonavir, originally developed as an HIV drug, exhibits anti-cancer activity against epithelial malignancies including prostate, breast cancer and ovarian cancer 2–4 as well as Kaposi’s sarcoma 5 and glioma 6, but its potential inhibitory activity against lung adenocarcinoma is unknown as are its mechanisms of action. The objective of these studies was to determine whether ritonavir exhibits effects on lung adenocarcinoma growth and survival and, if so, which cancer-associated signaling pathways may be affected in this malignancy. Identification of signaling pathways affected by ritonavir in lung adenocarcinoma could assist the design of clinical trials for recurrent/metastatic disease, where there is an unmet need for new therapeutic options.
One target of particular importance in lung cancer is the anti-apoptotic protein survivin 7. Survivin regulates multiple cancer-relevant pathways and most notably inhibits apoptosis as an Inhibitor of Apoptosis (IAP) family member, while promoting cell cycle progression through the G1/S and G2/M checkpoints 8–11. Because of its nodal role in cancer networks, its short half-life 12, and its exclusive expression in tumor tissues 13, 14, survivin is a particularly attractive target in cancer therapy 15. Various approaches to down-regulate survivin expression, including anti-sense oligonucleotides 16, ribozymes 17, siRNA 18, 19, and dominant-negative protein 20, have been used successfully in pre-clinical setting in NSCLC and other types of cancer. Furthermore a survivin antisense approach, alone or in combination with chemotherapy, is being tested in clinical trials 21, 22. Survivin is highly expressed in NSCLC 7, 13, 23 and the level of its gene expression may be prognostic of 5 year survival 24, although correlation between survivin expression and outcomes remains to be determined conclusively 25. Survivin may also play a role in the development of chemotherapy resistance in NSCLC 26–28 and its down-regulation may sensitize lung cancer to chemotherapy 29.
Ritonavir is known to reduce the levels of survivin in T cell leukemia 30, but it is unknown whether ritonavir affects the level or function of survivin in lung adenocarcinoma. Because of the critical importance of survivin in NSCLC biology and its outcomes, it was determined whether ritonavir inhibits survivin also in NSCLC, specifically in lung adenocarcinoma cell lines. Survivin transcription is regulated by c-Src and STAT3, both known to be important for NSCLC growth 31–34. Thus, the effects of ritonavir on these potential targets were also studied. Finally, combinations of ritonavir and chemotherapy that is not expected to interact pharmacokinetically, were tested for efficacy. Specifically, ritonavir was combined with gemcitabine and cisplatin, because both drugs have already been used in combination with ritonavir and other HIV protease inhibitors in case reports and clinical trials 35–37.
Materials and Methods
Antisera
Antibodies raised against human phospho-Rb (#9308), phospho-c-Src (#2101S), STAT3 (clone 124H6; #9139), phospho-STAT3 (clone 3E2; # 9138S), and PARP1 (#9542) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies raised against p53 (clone DO1; #sc-126), CDK2 (#sc-163), CDK4 (#sc-260), CDK6 (#sc-177), cyclin D1 (sc-718), cyclin A (clone C-19; #sc-596) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody anti-c-Src (clone GD11; #05-184) was purchased from Millipore (Billerica, MA). Antibodies anti-p27 (#610242), Rb (#554136) and Phospho-Rb were purchased from BD Biosciences (San Jose, CA), antibody anti-survivin (#AF886) from R&D Systems (Minneapolis, MN), and antibody anti-GAPDH (clone 6C5; 10R-G109A) from Fitzgerald Industries International (Concord, MA).
Cell lines and drugs
The lung adenocarcinoma lines A549, H522, H23 were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The lung adenocarcinoma line H838 was a kind gift from Dr. Manish Patel (University of Minnesota). Purified ritonavir, gemcitabine and cisplatin were purchased ‘from Sequoia Research Products (Pangbourne, UK).
siRNA
siRNA duplex SMARTpool upgrades of c-Src (#M-003175-03), STAT3 (#M-003544-02), and survivin (#M-003459-02), and siControl (# D-001210-01) were purchased from Dharmacon Research, Inc (Lafayette, CO). The sequences for siRNA duplexes used in these studies are given in Table S1.
Western blotting
Cell lysates were prepared with standard methodology. Samples containing the same amount of protein were separated on a 4–20% gradient SDS gel, blotted on a nitrocellulose membrane and probed with specific antibodies. Densitometry analysis of the bands was performed using the software Alpha Imager (Alpha Innotech; San Leandro, CA) and the relative amount of protein was normalized to the corresponding signal for glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
MTT assay
The cells were cultured in 96-well plates for 48 h in the presence of increasing concentration of ritonavir (ranging from 5 to 60 μM) or DMSO. Cell growth was determined by measuring the mitochondrial reduction of the tetrazolium salt, 3, [4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (Sigma; St Luis, MO). Each concentration of ritonavir was tested in octuplicate, and each MTT assay was repeated three times.
Measurement of clonogenic efficiency
Clonogenic efficiency of lung adenocarcinoma lines was measured by exposing cell monolayers (20% confluent) to varying concentrations of ritonavir (ranging from 10 to 100 μM) or DMSO for 24 h. Each ritonavir concentration was tested in triplicate. At the end of the 24 h treatment, cell monolayers were treated with trypsin and re-plated (500 cells per 100 mm plate) in the absence of ritonavir in complete medium (CM) consisting of RPMI 1640 (ATCC; Manassas, VA) supplemented with 10% fetal calf serum (Hyclone/Thermo Fisher Scientific; Logan, UT). The medium was changed every 3 days for 21 days. At the end of the 21-day incubation period, the colonies were fixed, stained with Wright-Giemsa stain (Thermo Fisher Scientific; Waltham, MA) and counted.
Cell cycle analysis
Lung adenocarcinoma cell lines 30–50% confluent were treated with trypsin, washed and resuspended in CM. Cells were plated at a density of 5×105 cells/100 mm plate, and grown for 24 and 48 h in CM in the presence of ritonavir (IC50) or DMSO. Profiling of propidium iodide (PI) incorporation was performed by FACScan analysis (Becton Dickinson; San Jose, CA). Adherent and non-adherent cells were included in the profile. Cell cycle distribution was determined using ModFit software (Becton Dickinson; San Jose, CA).
Measurement of apoptosis
Apoptotic cells were detected using an Annexin-V-FITC/PI apoptosis detection kit (Oncogene; Boston, MA). Cells were plated at a density of 5×105 cells/100 mm plate, and grown for 48 h in CM in presence of ritonavir (IC50) or DMSO. After 48 h, cells were harvested and stained with PI and Annexin-V-FITC. Cells were then acquired by FACScan (1 × 104 cells/assay) and analyzed using CellQuest software (Becton Dickinson; San Jose, CA). Each assay was performed in duplicate.
Real time PCR
RNA was isolated from A549 and H522 cell lines treated for 48 h with ritonavir (40μM) or DMSO by RNeasy mini kit (QIAGEN; Valencia, CA), following manufacturer’s instructions. First strand cDNA was synthesized with high capacity cDNA archive kit (Applied Biosystems; Foster City, CA). The PCR reaction consisted in 40 cycles (2 min at 50°C, 10 min at 95°C, 15 sec at 95°C, 1 min at 60°C).
siRNA transfection and cell survival assay
Cells were plated in 6 well poly-D-Lysine coated tissue culture plate (Becton Dickinson; Bedford, MA) at 2.5 × 104 cells/well, grown for 24 h and then transfected with 50 nmoles/well of siRNA duplexes for c-Src, STAT3, survivin, or control siRNA using Oligofectamine (Invitrogen; Carlsbad, CA) and OPTI-MEM (Invitrogen; Carlsbad, CA). The efficacy of gene silencing was evaluated by examining the protein product with Western blotting 72 h after transfection. Each assay was performed in triplicate. 24 h after the siRNA transfection, the cells were exposed for 48 h to ritonavir (ranging from 0 to 60 μM), and cell viability was measured with an MTT assay.
Generation of survivin expression MIEG3 vectors
Human survivin cDNA was cloned upstream of the internal ribosomal entry site for enhanced green fluorescence protein. The survivin sense orientation expression plasmid (S-IRES-EGFP-MIEG3) was constructed using an EcoRI adaptor as described 10. Empty MIEG3 plasmid was used as control vector. Flow cytometry was used to sort A549 or H522 cells transiently transfected with the S-IRES-EGFP-MIEG3.
Effects of drug combination on cell growth
Cells were exposed to ritonavir and/or gemcitabine at a fixed constant ratio corresponding to the ratio of the IC50 values for each single drug. The CalcuSyn software (Biosoft; Cambridge, U.K.) – which uses the median-effect analysis method of Chou and Talalay – was used to determine the possible synergic effect of the drugs. According to this method a combination index (CI) can be calculated from dose-response curves obtained using the drugs as single agent or in combination. CI < 1.0 indicates synergy, CI =1.0 indicates that the drugs act in an additive fashion, and CI > 1.0 indicates antagonism.
Statistical analysis
Statistical analysis was conducted using two-tailed Student’s t test (Excel, Microsoft). A difference was considered significant if p < 0.05.
Results
Ritonavir inhibits growth and colony formation of lung adenocarcinoma lines
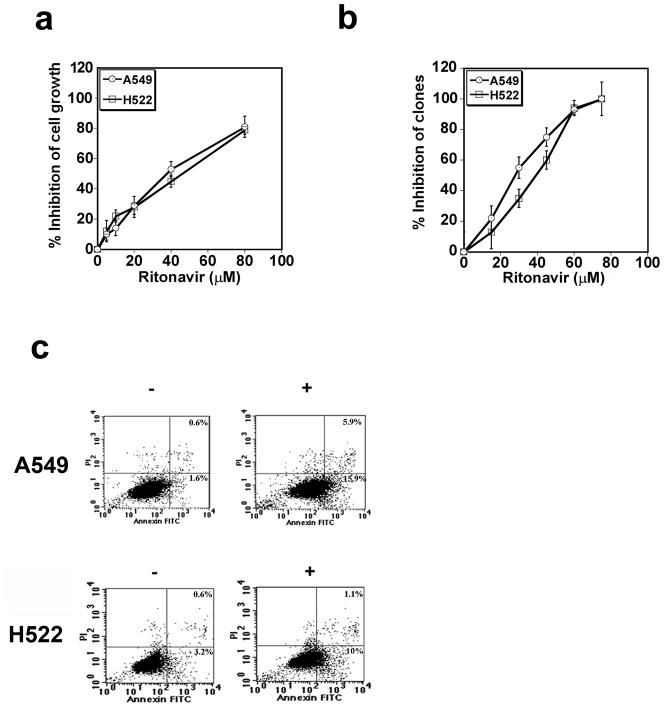
The effect of ritonavir on the growth of the A549 and H522 lines was assayed by MTT assay and compared to vehicle (DMSO). Ritonavir inhibits the growth of the lung adenocarcinoma lines A549 and H522 lines, exhibiting IC50 values of 35 and 42 μM, respectively (Fig. 1a). Ritonavir also inhibits the growth of the lung adenocarcinoma lines H23 and H838, exhibiting IC50 values of 44 and 35 μM, respectively, confirming that ritonavir is active across a range of lung adenocarcinoma lines (results for H23 not shown; results for H838 shown in Fig. S1). Ritonavir was also effective at inhibiting adhesion-dependent colony formation of the A549 and H522 lines, exhibiting IC50 values of 30 and 40 μM, respectively (Fig. 1b).
Figure 1.
Panel a. Ritonavir inhibits growth of the lung adenocarcinoma lines A549 and H522. Ritonavir inhibited the growth of both lines, exhibiting an IC50 of 35μM for A549 (open circles) and 42μM for H5222 (open squares). Results are representative of three consistent experiments.
Panel b. Ritonavir reduces the clonogenic efficiency of the lung adenocarcinoma lines A549 and H522. Ritonavir was efficient in reducing the clonogenic efficiency in both the A549 (open circles) and H522 (open squares) lines, exhibiting an IC50 of 30 and 40 μM, respectively.
Panel c. Ritonavir induces apoptosis in the lung adenocarcinoma lines A549 and H522. Ritonavir significantly increased early and late apoptotic events in both lines. The results are representative of two consistent experiments.
Ritonavir induces apoptosis of lung adenocarcinoma lines
To determine whether ritonavir induces apoptosis, the adenocarcinoma lines A549 and H522 were exposed to ritonavir at the IC50 for 48 h, stained with PI/Annexin-V-FITC and analyzed by flow cytometry. Ritonavir treatment increased the percentage of early apoptotic cells 10-fold from 1.6 to 16%, for the A549 line, and 3-fold, from 3.2 to 10%, for the H522 line (Fig. 1c). The percentage of late apoptotic cells increased 10-fold, from 0.6 to 5.9% for the A549 line, and two-fold from 0.6 to 1.1% for the H522 line.
Ritonavir induces a G0/G1 cell cycle arrest
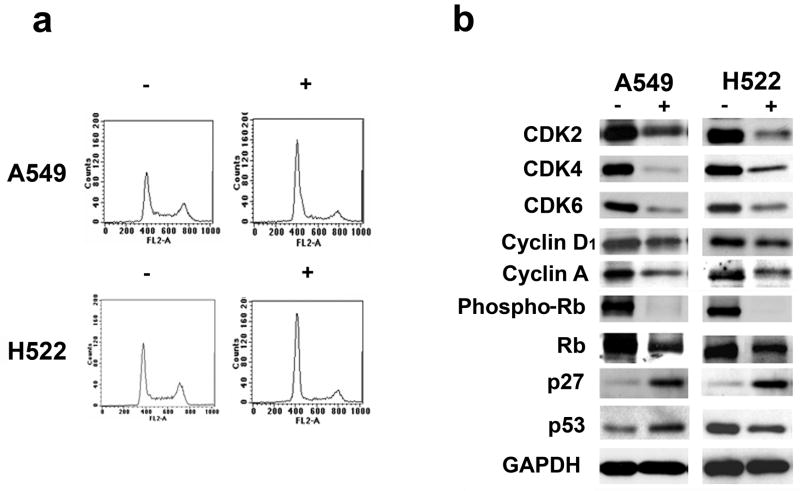
To determine whether ritonavir inhibits cell cycle progression of lung adenocarcinoma cells, the A549 and H522 lines were grown in the presence of ritonavir at either its IC50 (40 μM) or half the IC50 (20 μM) or in the presence of vehicle (DMSO) for 24 and 48 h. The cycle distribution was determined by flow cytometry. For both lines, incubation with ritonavir for 48 h resulted in a dose-dependent G0/G1 cell cycle block, as demonstrated by increase of the G0/G1 population and by a corresponding reduction of the S and G2/M populations (Fig. 2a, Table S2). Similar results were obtained after 24 h incubation with ritonavir (results not shown).
Figure 2.
Panel a. Ritonavir causes G0/G1 cell cycle arrest in the A549 and H522 lung adenocarcinoma lines. Ritonavir was able to induce a G0/G1 block as demonstrated by increase of the G0/G1 population and by a corresponding reduction of the S and G2/M populations. Cells growth with ritonavir or vehicle are indicated with (+) and (−), respectively. For quantitation of the flow cytometry analysis see Table 2S).
Panel b. Ritonavir down-regulates the expression of numerous cell cycle-associated proteins in the lung adenocarcinoma lines A549 and H522. CDK2, CDK4, CDK6, cyclin D1, cyclin A, phospho-Rb, and Rb levels were reduced in both lines treated with ritonavir. Levels of p53 were reduced in the H522 line but increased in the A549 line. The levels of p27Kip1 were increased in both lines. For quantitation of the Western blots see Table S3.
Ritonavir down-regulates CDK2, CDK4, CDK6, and cyclin D1 levels and inhibits pRb, while up-regulating p27Kip1and wild type p53
Because ritonavir induces a G0/G1 block in lung adenocarcinoma lines (Fig. 2a), it was investigated whether ritonavir affects the levels of proteins known to regulate the G1/S checkpoint. Both the A549 and H522 lines exhibit reduction of G1/S check point regulators CDK2, CDK4, CDK6, and cyclin D1, following 48 h of treatment at the ritonavir IC50 (Fig. 2b; Table S3). Correlating with reduction of CDKs and cyclin D1, phospho-Rb levels were reduced by more than 80% (Fig. 2b; Table S3). Cyclin A, involved in S phase progression, was also decreased by ritonavir (Fig. 2b; Table S3). Furthermore, p27Kip1, an important inhibitor of cyclin E/CDK2 complexes was increased in both lines. The A549 line is p53 wild type, while the H522 line is p53 mutant (codon 191) and the effects of ritonavir differed, with p53 increasing in the wild type line and decreasing in the mutant line (Fig. 2b and Table S3).
Effects of ritonavir on survivin, c-Src, STAT3, and cleaved PARP levels in lung adenocarcinoma lines
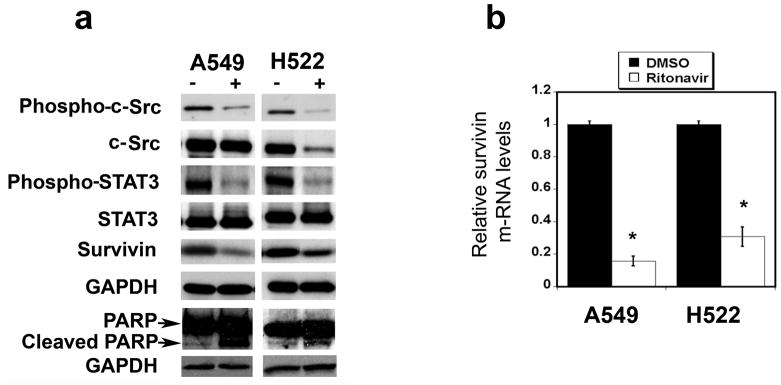
The potential roles of survivin, its up-stream regulatory proteins c-Src and STAT3 and their phosphorylated forms, and its down-stream signaling molecule PARP were studied in the A549 and H522 lines. Ritonavir treatment significantly reduced survivin expression in both cell lines (Fig. 3a; Table S4a). In both lines, ritonavir reduced fractional phosphorylation of c-Src by more than half (Fig. 3a; Table S4a). Total c-Src was also reduced in the H522 line, but not A549 (Fig. 3a; Table S4a). Ritonavir treatment reduced the fractional phosphorylation of STAT3 by 40% in both cell lines, but did not affect STAT3 levels (Fig. 3a; Table S4a). In both lines, ritonavir significantly increased the levels of cleaved PARP (Fig. 3a; Table S4a).
Figure 3.
Panel a. Ritonavir down-regulates the expression of phospho-c-Src, Src, phospho-STAT3, survivin, and increases cleaved PARP in the lung adenocarcinoma lines A549 and the H522. Both in the A549 and H522 line, ritonavir reduced phospho-c-Src, phospho-STAT3, and survivin levels while increased the cleaved PARP levels; it did not affect total STAT3 levels and it down-regulated total c-Src only in the H522 line. Cells growth with ritonavir or vehicle are indicated with (+) and (−), respectively. For quantitation of the Western blots see Table S4a.
Panel b. Ritonavir down-regulates survivin mRNA in the lung adenocarcinoma lines A549 and H522. After 48 h treatment, ritonavir drastically reduced the levels of survivin mRNA in both lines.
Effects of ritonavir on survivin mRNA levels in lung adenocarcinoma lines
To determine whether reduction in survivin protein levels by ritonavir corresponds to a down-regulation of its message, survivin mRNA levels were measured by real-time PCR. Survivin mRNA was reduced by 70 to 80% in ritonavir-treated lung adenocarcinoma lines (Fig. 3b). These results indicate that ritonavir-mediated reduction of survivin is mediated largely through reduction of survivin mRNA.
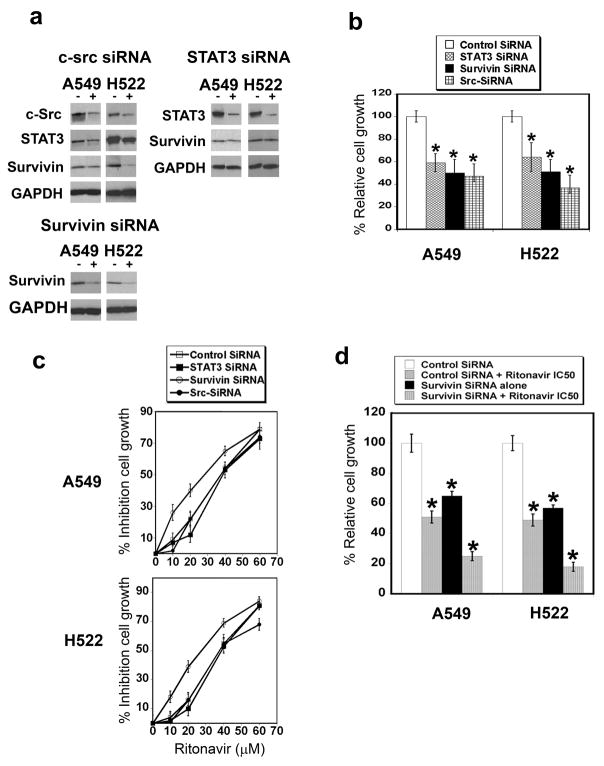
RNAi profiling of survivin or its regulators c-Src and STAT3 reveals that survivin is the most important ritonavir target
Recently, RNAi silencing has been used to identify genes whose expression affects drug responses in cancer 38. In the present study, the regulatory pathway for survivin expression, which is mediated by c-Src and STAT3, was profiled by RNAi with the purpose of testing the relative importance of the members of this pathway as ritonavir targets. To verify the effectiveness of the RNAi approaches, the protein levels of survivin, c-Src and STAT3 were determined. Knock down of c-Src, STAT3 and survivin reduced their corresponding proteins by 50, 70 and 50%, respectively (Fig. 4a, Table S4b). Knock down of c-Src significantly reduced survivin and STAT3 levels in both lines, indicating that there is a direct relationship between c-Src levels and its downstream signaling proteins (Fig. 4a, Table S4b). Knock down of STAT3 reduced survivin in the H522 line only (Fig. 4a, Table S4b). These results indicate that survivin levels are determined, in part, by c-Src and STAT3 levels, confirming a model of a signaling hierarchy between these regulatory proteins in NSCLC. Inhibitory effects of survivin, c-Src and STAT3 knock down on the ritonavir-sensitive lung adenocarcinoma lines were verified by MTT assay. siRNA targeting c-Src, STAT3, or survivin significantly inhibits the growth of both the A549 and H522 lines at 48 h (Fig. 4b). After establishing the effectiveness of the RNAi silencing of c-Src, STAT3 and survivin, this approach was used to determine the relative importance of the survivin pathway members as ritonavir targets. Only survivin knock down shifted the MTT dose response curve significantly to the left for the A549 and H522 lines, indicating the importance of survivin levels for ritonavir sensitivity (Fig. 4c). In contrast, despite the hierarchy placing c-Src and STAT3 up-stream of survuvin, c-Src, STAT3 and non-target RNAi failed to affect the ritonavir IC50 for either line. After survivin RNAi treatment, the ritonavir IC50 decreased to 25 μM for both lines (Fig. 4c). Testing ritonavir at its IC50 in combination with survivin siRNA confirmed that reduction of survivin sensitizes lung cancer lines to ritonavir (Fig. 4d).
Figure 4.
Panel a. c-Src, STAT3, and survivin siRNA reduce the levels of their target proteins. The (−) column indicates non-target siRNA, while the (+) column indicates specific siRNA as labeled. The siRNA to c-Src, STAT3 and survivin reduced the level of the corresponding protein in all three lines. For quantitation of the Western blots see Table S4b.
Panel b. siRNA targeting c-Src, STAT3, and survivin reduces growth of the lung adenocarcinoma lines A549 and H522. Cells treated with siRNA targeting c-Src, STAT3, and survivin exhibited reduced growth when compared to cells treated with non-targeting siRNA. P value of <0.05 is indicated with an asterisk.
Panel c. siRNA targeting c-Src or STAT3 do not reduce ritonavir IC50 for the lung adenocarcinoma lines A549 and H522. A549 and H522 cells transfected with siRNA targeting c-Src, STAT3, survivin, or non-targeting siRNA were grown in the presence of increasing concentrations of ritonavir. Silencing of survivin mRNA expression increased the cell sensitivity to ritonavir, reducing its IC50 to 25μM for the A459 line and 27μM for the H522 line.
Panel d. siRNA targeting survivin increases the sensitivity to ritonavir of the lung adenocarcinoma lines A549 and H522. A549 and H522 cells transfected with siRNA targeting survivin were treated with ritonavir at the appropriate IC50. Survivin siRNA and ritonavir acted together to reduce cell growth in both lines. P value of <0.05 is indicated with an asterisk.
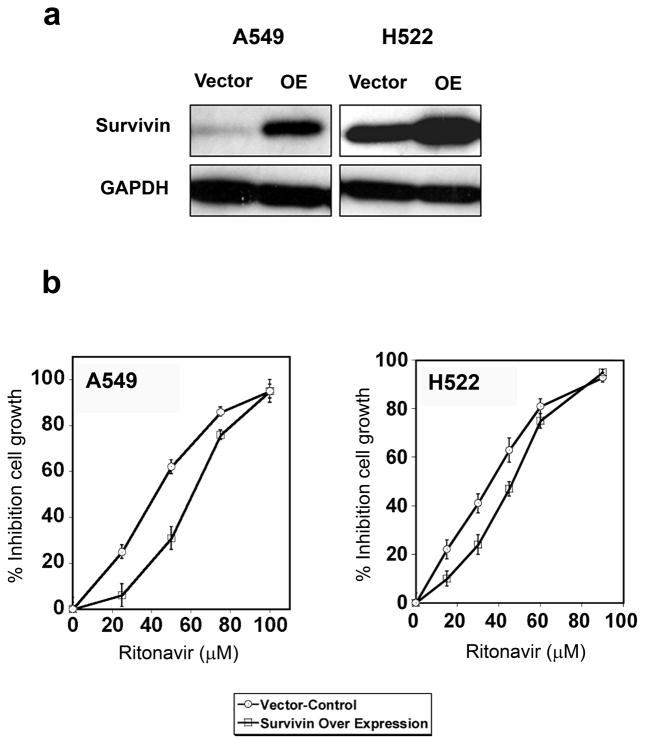
Over-expression of survivin induces resistance to ritonavir in lung adenocarcinoma lines
Because survivin knock down sensitizes lung adenocarcinoma lines to ritonavir, it was determined whether survivin over-expression leads to ritonavir resistance. Survivin expression was increased three-fold in both lines by the S-IRES-EGFP-MIEG3 transfection (Fig. 5a). Transfection with the survivin expression plasmid resulted in significant increases of the ritonavir IC50 for both lines (10 to 20 μM) (Fig. 5b). Therefore survivin over-expression causes resistance to ritonavir while survivin knock down causes sensitivity, identifying survivin as a critical target for this drug.
Figure 5.
Panel a. Over-expression of survivin in the lung adenocarcinoma lines A549 and H522. Cells transiently transfected with human survivin cDNA had significantly higher levels of survivin when compared to cells transfected with the empty vector.
Panel b. Over-expression of survivin increases resistance the lung adenocarcinoma lines A549 and H522 to ritonavir. Over-expression of survivin reduced the cell sensitivity to ritonavir, increasing its IC50 to 45μM for the A549 line and 55μM for the H522 line.
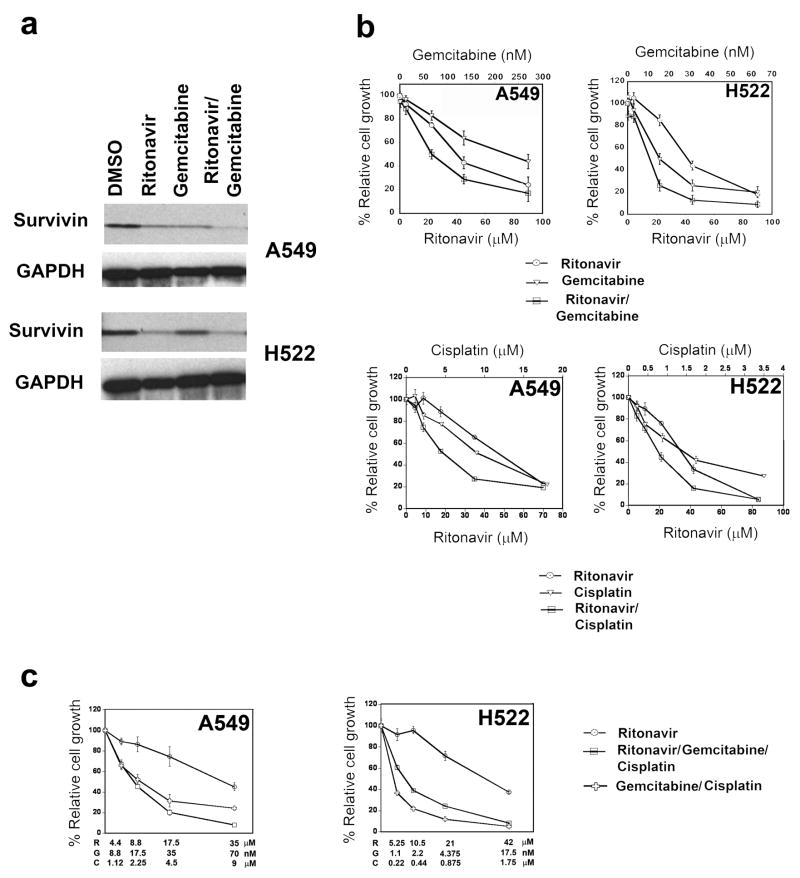
Ritonavir exhibits enhanced anti-cancer activity when combined with gemcitabine and or cisplatin
After identifying ritonavir inhibitory activity in lung adenocarcinoma lines and survivin as its major target, it was important to learn effective ways of combining ritonavir with other chemotherapy agents. This goal can be accomplished by identifying chemotherapy drugs that do not interact pharmacokinetically with ritonavir, thereby avoiding drug interactions that could cause unpredictable toxicity. Ritonavir is known to interact with drugs metabolized by CYP3A4, by either decreasing or increasing their metabolism 39. For instance, as a CYP3A4 inhibitor, ritonavir is known to boost the levels of chemotherapy drugs, such as taxanes, vinca alkaloids, and etoposide, making toxicity difficult to predict 40, 41. In contrast, the widely used lung cancer drug gemcitabine is not known to interact with ritonavir and as such it represents a promising candidate for combined therapy. When gemcitabine was combined with ritonavir at the respective IC50 for each drug, reduction of survivin was still observed, indicating that gemcitabine does not interfere with ritonavir-mediated reduction of survivin (Fig. 6a, Table S5). Furthermore, in the A549 gemcitabine alone significantly reduced survivin levels and such reduction was enhanced by the combination ritonavir/gemcitabine. When tested for synergy by Chou Talalay isobologram analysis 42, the combination ritonavir/gemcitabine exhibited strong synergy (Fig. 6b) with combinatorial index (CI) values for the A549 and H522 lines of 0.57 ± 0.33 and 0.16 ± 0.09, respectively. Ritonavir exhibits an IC50 of 15–20 μM in combination with gemcitabine concentrations of 15 to 60 nM, which are well below Cmax concentrations attained clinically with gemcitabine monotherapy 43, 44. Also cisplatin, which is commonly used in lung cancer chemotherapy, is not known to interact with ritonavir. The ritonavir/cisplatin combination was less active in inhibiting the growth of lung adenocarcinoma lines than the ritonavir/gemcitabine combination, exhibiting CI values of 1 ± 0.55 and 1.1 ± 1.16 for the A549 and H522 lines, respectively (Fig. 6b). These CI values indicate that cisplatin and ritonavir are additive rather than synergistic for the A549 and H522 lines 42. Similarly to what we observed for the combination ritonavir/gemcitabine, ritonavir IC50 was reduced to ~20 μM in combination with cisplatin at 4.5 μM.
Figure 6.
Panel a. Gemcitabine and ritonavir down-regulate the survivin levels of the lung adenocarcinoma lines A549, H522. Ritonavir was able to reduce the levels of survivin either when used alone or together with gemcitabine. For quantitation of the Western blots see Table S5.
Panel b. Ritonavir and gemcitabine synergize in inhibiting the growth of the lung adenocarcinoma lines A549, H522. The combination gemcitabine and ritonavir had a synergic effect and was significantly more effective in reducing growth compared to each drug alone. On the other hand, the combined effect of ritonavir and cisplatin was additive for the A549 and H522 lines.
Panel c. The combination ritonavir/gemcitabine/cisplatin is synergistic in the A549 line and additive in the H522 line. The concentrations of ritonavir (R), gemcitabine (G) and cisplatin (C) are indicated on the X axis. The combination ritonavir/gemcitabine/cisplatin acted synergistically in the A549 line and additively in the H522 line. The combination gemcitabine/cisplatin was superior to ritonavir/gemcitabine/cisplatin in the H522 line but not in the A549 line.
Since the combination gemcitabine/cisplatin is a standard regimen for the treatment of lung adenocarcinoma 45, we also tested the effect of ritonavir in combination with gemcitabine/cisplatin in the A459 and H522 lines. The ritonavir/gemcitabine/cisplatin combination acted synergistically in the A549 line (CI=0.6 ± 0.2) and was slightly superior to the combination gemcitabine/cisplatin (Fig. 6c). On the other hand, in the H522 line, the combination ritonavir/gemcitabine/cisplatin acted additively (CI=0.9 ± 0.2) but it was inferior to the gemcitabine/cisplatin combination (Fig. 6c). Importantly, in both cell lines, ritonavir IC50 was < 10 μM in combination with concentrations of gemcitabine and cisplatin well below their IC50.
Discussion
Survival of recurrent/metastatic NSCLC with palliative chemotherapy fails to exceed 1 y and there is an unmet need for new drugs and drug combinations that work through novel mechanisms 46. In the present study, we propose ritonavir as a candidate drug for metastatic lung adenocarcinoma clinical trials, based on its inhibition of adenocarcinoma lines at concentrations in the 35–45 μM range. Such concentrations are clinically attainable, albeit with significant gastrointestinal toxicity 47, with ritonavir monotherapy at 600 mg twice daily 48.
Study of signaling pathways affected by ritonavir by siRNA profiling revealed that survivin is an important target, whereas c-Src and STAT3 appear to be of lesser importance. Furthermore, forced over-expression of survivin confers relative resistance to ritonavir, confirming importance of survivin as a ritonavir target. Ritonavir reduces survivin, in part, by reducing its mRNA levels. Because survivin is regulated in cancer primarily through its mRNA expression 49 these results suggest that ritonavir is likely attacking a basic mechanism of survivin transcriptional regulation.
Ritonavir inhibits lung adenocarcinoma growth and anchorage-dependent clonogenicity, in part, by inducing G0/G1 cell cycle arrest and, in part, by inducing apoptosis. Survivin is implicated in regulation of the G1/S 8, 9, 11 as well as G2/M checkpoint and therefore its reduction by ritonavir is expected to cause inhibition of the cell cycle. Ritonavir may also inhibit the G1/S checkpoint through down-regulation of CDKs, cyclin D1 and associated Rb phosphorylation, as well as by induction of p27Kip and wild type p53. While reduction of survivin by ritonavir is expected to promote apoptosis in lung cancer presumably related to loss of the antiapoptotic effect of survivin 50, the remaining mechanisms by which ritonavir induces apoptosis remain to be determined. These mechanisms could include induction of DNA damage, in part, through increased cleavage of PARP1.
Importantly, drug combination studies revealed that ritonavir is active at lower concentrations when combined with gemcitabine, cisplatin and the gemcitabine/cisplatin combination. In combination with gemcitabine or cisplatin, ritonavir exhibits IC50 values in the range of 15–20 μM. With the gemcitabine/cisplatin combination, the ritonavir IC50 is in the 8 μM range, which should be attainable with 100 mg twice daily dosing 48. While the gemcitabine/cisplatin combination in advanced NSCLC resulted in the longest median time to progression compared to three other chemotherapy doublets, this was only 4.2 months 45. We hypothesize, based on in vitro synergy, that addition of low dose ritonavir to the gemcitabine/cisplatin combination may improve time to progression, with acceptable toxicity. Furthermore, because ritonavir is not myelosuppressive and potentially could be continued through the period of gemcitabine/cisplatin treatment, ritonavir could potentially inhibit re-growth of lung adenocarcinoma between cycles of chemotherapy. Therefore, a phase I study of daily ritonavir in combination with the established gemcitabine and cisplatin schedule is an important next step. While K-ras mutation status did not affect sensitivity to ritonavir, for the H838 K-ras wild type line there was lack of synergy with gemcitabine and antagonism with cisplatin. These results suggest that K-ras mutant lung adenocarcinoma is the best candidate histology for future clinical trials.
Although the mechanisms behind cooperation between ritonavir and gemcitabine and/or cisplatin are not known, it is likely these mechanisms involve survivin effects on DNA repair pathways. Gemcitabine is a DNA strand-terminator that stalls replication forks, causes S phase arrest 51 and double strand breaks (DSB) while inhibiting homologous recombination repair (HRR), which is required for repairing DSB 52, 53. Survivin has been reported to enhance DSB repair and we hypothesize that reduction of survivin by ritonavir may increase sensitivity to gemcitabine through this mechanism 54. Survivin reduction may also explain sensitivity of lung adenocarcinoma to ritonavir in combination with cisplatin due to increased PARP1 cleavage.
PARP1 may be involved in repair of cisplatin-induced DNA damage. PARP1 is known to recruit XRCC1 to sites of DNA damage 55. XRCC1 is a scaffolding factor required for base excision repair (BER) 56 and recently, nucleotide excision repair (NER) 57. Of interest, interference with NER interferes with repair of cisplatin-induced DNA damage 53. Although PARP1 has not been implicated as a key regulator of NER, it has been recently been located at sites of cisplatin-induced DNA damage, by two photoaffinity labeling studies 58, 59. This finding potentially implicates PARP1 in repair of cisplatin-mediated DNA interstrand crosslinks by NER. In addition, PARP1 reduction has also recently been demonstrated to play a critical role in chemosensitivity to the gemcitabine/cisplatin combination in triple negative breast cancer 60. Future studies will determine the mechanisms by which ritonavir may enhance DNA damage by cisplatin and gemcitabine.
Based on the importance of survivin as a ritonavir target in lung adenocarcinoma, we propose that survivin may be a useful biomarker for ritonavir sensitivity. We hypothesize that among tumors expressing survivin, those exhibiting lower survivin levels will be more likely to respond to ritonavir. Our results from forced survivin over-expression are artificial and may not reflect survivin levels in tumors occurring in patients and therefore we would not recommend excluding patients with high survivin levels from clinical trials of ritonavir. Only the analysis of data from such trials would reveal whether there is a relationship between survivin levels and ritonavir sensitivity.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health [grants P20-GM66403 and R01 CA113570 to DAP; grant HL-079654 to LMP]. DAP acknowledges a Walther Cancer Research Prize, the Flight Attendant’s Medical Research Institute Clinical Innovator Award 042257, and support from the Walther Oncology Center at Indiana University, the Thoracic Oncology Program at Indiana University, a Clarian Values Foundation grant and equipment grant from the Indiana Elks.
We thank Drs. Lawrence Einhorn, Hal Broxmeyer and Patrick Loehrer for support and encouragement of this work. We thank Drs. Anja Bielinsky, Robert Kratzke, Manish Patel, David Donner, Janice Blum, Ann Roman, Maureen Harrington, Christine Clouser, and Reuben Harris for helpful discussions. We thank Dr. Manish Patel for the H838 line. We thank Susan Rice for help with flow cytometry experiments.
Nonstandard abbreviations used
- CDK
cyclin dependent kinase
- CI
combination index
- CM
complete medium
- DMSO
dimethyl sulfoxide
- EGFP
enhanced green fluorescence protein
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IC50
half maximal inhibitory concentration
- MTT
3, [4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- NSCLC
non small cell lung cancer
- PCR
polymerase chain reaction
- PI
propidium iodide
- STAT
signal transducer and activator of transcription protein
- Rb
retinoblastoma protein
References
- 1.Chong CR, Xu J, Lu J, et al. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem Biol. 2007;2:263–70. doi: 10.1021/cb600362d. [DOI] [PubMed] [Google Scholar]
- 2.Ikezoe T, Hisatake Y, Takeuchi T, et al. HIV-1 protease inhibitor, ritonavir: a potent inhibitor of CYP3A4, enhanced the anticancer effects of docetaxel in androgen-independent prostate cancer cells in vitro and in vivo. Cancer Res. 2004;64:7426–31. doi: 10.1158/0008-5472.CAN-03-2677. [DOI] [PubMed] [Google Scholar]
- 3.Srirangam A, Mitra R, Wang M, et al. Effects of HIV protease inhibitor ritonavir on Akt-regulated cell proliferation in breast cancer. Clin Cancer Res. 2006;12:1883–96. doi: 10.1158/1078-0432.CCR-05-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Bryant CS, Chamala S, et al. Ritonavir blocks AKT signaling, activates apoptosis and inhibits migration and invasion in ovarian cancer cells. Mol Cancer. 2009;8:26. doi: 10.1186/1476-4598-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pati S, Pelser CB, Dufraine J, et al. Antitumorigenic effects of HIV protease inhibitor ritonavir: inhibition of Kaposi sarcoma. Blood. 2002;99:3771–9. doi: 10.1182/blood.v99.10.3771. [DOI] [PubMed] [Google Scholar]
- 6.Laurent N, de Bouard S, Guillamo JS, et al. Effects of the proteasome inhibitor ritonavir on glioma growth in vitro and in vivo. Mol Cancer Ther. 2004;3:129–36. [PubMed] [Google Scholar]
- 7.Kren L, Brazdil J, Hermanova M, et al. Prognostic significance of anti-apoptosis proteins survivin and bcl-2 in non-small cell lung carcinomas: a clinicopathologic study of 102 cases. Appl Immunohistochem Mol Morphol. 2004;12:44–9. doi: 10.1097/00129039-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Ai Z, Yin L, Zhou X, et al. Inhibition of survivin reduces cell proliferation and induces apoptosis in human endometrial cancer. Cancer. 2006;107:746–56. doi: 10.1002/cncr.22044. [DOI] [PubMed] [Google Scholar]
- 9.Song J, So T, Cheng M, et al. Sustained survivin expression from OX40 costimulatory signals drives T cell clonal expansion. Immunity. 2005;22:621–31. doi: 10.1016/j.immuni.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda S, Foster RG, Porter SB, et al. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 2002;100:2463–71. doi: 10.1182/blood.V100.7.2463. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki A, Hayashida M, Ito T, et al. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–34. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Tenev T, Martins LM, et al. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;113(Pt 23):4363–71. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]
- 13.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka K, Iwamoto S, Gon G, et al. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127–34. [PubMed] [Google Scholar]
- 15.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 16.Shinohara ET, Hallahan DE, Lu B. The Use of Antisense Oligonucleotides in Evaluating Survivin as a Therapeutic Target for Radiation Sensitization in Lung Cancer. Biol Proced Online. 2004;6:250–256. doi: 10.1251/bpo95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennati M, Binda M, Colella G, et al. Radiosensitization of human melanoma cells by ribozyme-mediated inhibition of survivin expression. J Invest Dermatol. 2003;120:648–54. doi: 10.1046/j.1523-1747.2003.12082.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhu H, Quan L, et al. Downregulation of survivin by RNAi inhibits the growth of esophageal carcinoma cells. Cancer Biol Ther. 2005;4:974–8. doi: 10.4161/cbt.4.9.1914. [DOI] [PubMed] [Google Scholar]
- 19.Song H, Xin XY, Xiao F, et al. Survivin gene RNA interference inhibits proliferation, induces apoptosis, and enhances radiosensitivity in HeLa cells. Eur J Obstet Gynecol Reprod Biol. 2008;136:83–9. doi: 10.1016/j.ejogrb.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 20.Asanuma K, Kobayashi D, Furuya D, et al. A role for survivin in radioresistance of pancreatic cancer cells. Jpn J Cancer Res. 2002;93:1057–62. doi: 10.1111/j.1349-7006.2002.tb02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimmer AD, Dalili S. Targeting the IAP family of caspase inhibitors as an emerging therapeutic strategy. Hematology Am Soc Hematol Educ Program. 2005:215–9. doi: 10.1182/asheducation-2005.1.215. [DOI] [PubMed] [Google Scholar]
- 22.Ryan BM, O’Donovan N, Duffy MJ. Survivin: a new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–62. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Falleni M, Pellegrini C, Marchetti A, et al. Survivin gene expression in early-stage non- small cell lung cancer. J Pathol. 2003;200:620–6. doi: 10.1002/path.1388. [DOI] [PubMed] [Google Scholar]
- 24.Karczmarek-Borowska B, Filip A, Wojcierowski J, et al. Survivin antiapoptotic gene expression as a prognostic factor in non-small cell lung cancer: in situ hybridization study. Folia Histochem Cytobiol. 2005;43:237–42. [PubMed] [Google Scholar]
- 25.Fan J, Wang L, Jiang GN, et al. The role of survivin on overall survival of non-small cell lung cancer, a meta-analysis of published literatures. Lung Cancer. 2008;61:91–6. doi: 10.1016/j.lungcan.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Dasgupta P, Kinkade R, Joshi B, et al. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc Natl Acad Sci U S A. 2006;103:6332–7. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgillo F, Woo JK, Kim ES, et al. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66:10100–11. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- 28.Morgillo F, Kim WY, Kim ES, et al. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 29.Olie RA, Simoes-Wust AP, Baumann B, et al. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60:2805–9. [PubMed] [Google Scholar]
- 30.Dewan MZ, Uchihara JN, Terashima K, et al. Efficient intervention of growth and infiltration of primary adult T-cell leukemia cells by an HIV protease inhibitor, ritonavir. Blood. 2006;107:716–24. doi: 10.1182/blood-2005-02-0735. [DOI] [PubMed] [Google Scholar]
- 31.Mazurenko NN, Kogan EA, Zborovskaya IB, et al. Expression of pp60c-src in human small cell and non-small cell lung carcinomas. Eur J Cancer. 1992;28:372–7. doi: 10.1016/s0959-8049(05)80056-5. [DOI] [PubMed] [Google Scholar]
- 32.Song L, Turkson J, Karras JG, et al. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–65. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 33.Dalwadi H, Krysan K, Heuze-Vourc’h N, et al. Cyclooxygenase-2-dependent activation of signal transducer and activator of transcription 3 by interleukin-6 in non-small cell lung cancer. Clin Cancer Res. 2005;11:7674–82. doi: 10.1158/1078-0432.CCR-05-1205. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Kalyankrishna S, Wislez M, et al. SRC-family kinases are activated in non-small cell lung cancer and promote the survival of epidermal growth factor receptor-dependent cell lines. Am J Pathol. 2007;170:366–76. doi: 10.2353/ajpath.2007.060706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato T, Ieki R, Saito E, et al. A long-term survival case of small cell lung cancer in an HIV-infected patient. Jpn J Clin Oncol. 2005;35:349–52. doi: 10.1093/jjco/hyi093. [DOI] [PubMed] [Google Scholar]
- 36.Brunner TB, Geiger M, Grabenbauer GG, et al. Phase I trial of the human immunodeficiency virus protease inhibitor nelfinavir and chemoradiation for locally advanced pancreatic cancer. J Clin Oncol. 2008;26:2699–706. doi: 10.1200/JCO.2007.15.2355. [DOI] [PubMed] [Google Scholar]
- 37.Okuma Y, Hosomi Y, Takagi Y, et al. Long-term survival following metachronous intratumoral hemorrhage in an HIV-infected patient with lung cancer. Int J Clin Oncol. 2010 doi: 10.1007/s10147-010-0072-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YW, Jones TL, Martin SE, et al. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem. 2009;284:18085–95. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barry M, Gibbons S, Back D, et al. Protease inhibitors in patients with HIV disease. Clinically important pharmacokinetic considerations. Clin Pharmacokinet. 1997;32:194–209. doi: 10.2165/00003088-199732030-00003. [DOI] [PubMed] [Google Scholar]
- 40.Bardelmeijer HA, Ouwehand M, Buckle T, et al. Low systemic exposure of oral docetaxel in mice resulting from extensive first-pass metabolism is boosted by ritonavir. Cancer Res. 2002;62:6158–64. [PubMed] [Google Scholar]
- 41.Makinson A, Pujol JL, Le Moing V, et al. Interactions between cytotoxic chemotherapy and antiretroviral treatment in human immunodeficiency virus-infected patients with lung cancer. J Thorac Oncol. 5:562–71. doi: 10.1097/JTO.0b013e3181d3ccf2. [DOI] [PubMed] [Google Scholar]
- 42.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 43.Kroep JR, Giaccone G, Voorn DA, et al. Gemcitabine and paclitaxel: pharmacokinetic and pharmacodynamic interactions in patients with non-small-cell lung cancer. J Clin Oncol. 1999;17:2190–7. doi: 10.1200/JCO.1999.17.7.2190. [DOI] [PubMed] [Google Scholar]
- 44.Fogli S, Danesi R, De Braud F, et al. Drug distribution and pharmacokinetic/pharmacodynamic relationship of paclitaxel and gemcitabine in patients with non-small-cell lung cancer. Ann Oncol. 2001;12:1553–9. doi: 10.1023/a:1013133415945. [DOI] [PubMed] [Google Scholar]
- 45.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 46.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–14. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 47.Gatti G, Di Biagio A, Casazza R, et al. The relationship between ritonavir plasma levels and side-effects: implications for therapeutic drug monitoring. Aids. 1999;13:2083–9. doi: 10.1097/00002030-199910220-00011. [DOI] [PubMed] [Google Scholar]
- 48.Hsu A, Granneman GR, Witt G, et al. Multiple-dose pharmacokinetics of ritonavir in human immunodeficiency virus-infected subjects. Antimicrob Agents Chemother. 1997;41:898–905. doi: 10.1128/aac.41.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao R, Connolly DC, Murphy M, et al. Activation of cancer-specific gene expression by the survivin promoter. J Natl Cancer Inst. 2002;94:522–8. doi: 10.1093/jnci/94.7.522. [DOI] [PubMed] [Google Scholar]
- 50.Ulukus EC, Kargi HA, Sis B, et al. Survivin expression in non-small-cell lung carcinomas: correlation with apoptosis and other apoptosis-related proteins, clinicopathologic prognostic factors and prognosis. Appl Immunohistochem Mol Morphol. 2007;15:31–7. doi: 10.1097/01.pai.0000201808.35931.78. [DOI] [PubMed] [Google Scholar]
- 51.Ewald B, Sampath D, Plunkett W. ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance. Cancer Res. 2008;68:7947–55. doi: 10.1158/0008-5472.CAN-08-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wachters FM, van Putten JW, Maring JG, et al. Selective targeting of homologous DNA recombination repair by gemcitabine. Int J Radiat Oncol Biol Phys. 2003;57:553–62. doi: 10.1016/s0360-3016(03)00503-0. [DOI] [PubMed] [Google Scholar]
- 53.Crul M, van Waardenburg RC, Bocxe S, et al. DNA repair mechanisms involved in gemcitabine cytotoxicity and in the interaction between gemcitabine and cisplatin. Biochem Pharmacol. 2003;65:275–82. doi: 10.1016/s0006-2952(02)01508-3. [DOI] [PubMed] [Google Scholar]
- 54.Jiang G, Ren B, Xu L, et al. Survivin may enhance DNA double-strand break repair capability by up-regulating Ku70 in human KB cells. Anticancer Res. 2009;29:223–8. [PubMed] [Google Scholar]
- 55.Dantzer F, Ame JC, Schreiber V, et al. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 56.Dianova II, Sleeth KM, Allinson SL, et al. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 2004;32:2550–5. doi: 10.1093/nar/gkh567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moser J, Kool H, Giakzidis I, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–23. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 58.Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–25. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guggenheim ER, Xu D, Zhang CX, et al. Photoaffinity isolation and identification of proteins in cancer cell extracts that bind to platinum-modified DNA. Chembiochem. 2009;10:141–57. doi: 10.1002/cbic.200800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res. 2010;70:7970–80. doi: 10.1158/0008-5472.CAN-09-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.