Abstract
Trypanosoma brucei undergoes two clearly distinct develomental stages: in the insect vector (procyclic stage) the cells generate the bulk of their energy through respiration, while in the bloodstream of the mammalian host (bloodstream stage) they grow mostly glycolytically. Several mitochondrial respiratory proteins require iron-sulfur clusters for activity, and their activation coincides with developmental changes. Likewise some tRNA modification enzymes either require iron-sulfur clusters or use components of the iron-sulfur cluster assembly pathway for activity. These enzymes affect the anticodon loop of various tRNAs and may impact protein synthesis. Herein, the possibility of these pathways being integrated and exploited by T. brucei to carefully coordinate energy demands to translational rates in response to enviromental changes is examined.
Assembling iron-sulfur clusters in proteins
Every extant organism synthesizes iron-sulfur (Fe/S) clusters de novo as cofactors for the function of dozens of proteins. Not surprisingly, the assembly of Fe/S clusters is invariably essential for viability. In Eukarya, a substantial fraction of Fe/S clusters is incorporated into mitochondrial proteins involved in electron transport, such as subunits of respiratory complexes I, II and III, and ferredoxin. Moreover, nuclear and cytosolic proteins, including the ribosomal protein Rli1, primase Pri2 and xanthine oxidoreductase, to name a few, depend functionally on these co-factors [1, 2].
A key component of the Fe/S cluster assembly machinery is the cysteine desulfurase (Nfs), which removes sulfur from cysteine, converting it into alanine, and together with three other proteins, IscU, Isd11, and frataxin, plays a crucial role in Fe/S cluster assembly within the mitochondrion [3, 4]. Therefore, in Trypanosoma brucei as in other eukaryotes, depletion of any of these proteins leads to impairment of the Fe/S cluster assembly pathway and lethality [5–7].
In the course of its life cycle, the single mitochondrion of T. brucei, the causative agent of African sleeping sickness, alternates between a cristae-rich reticulated organelle of the procyclic stage, which parasitizes the tsetse fly vector, and a much reduced vesicle-like mitochondrion of the bloodstream stage, which occurs in the vertebrate host. The mitochondrion of the former stage contains cytochrome c-carrying respiratory complexes, an alternative terminal oxidase (TAO), acetate:succinate CoA oxidase and an incomplete Krebs cycle. Due to its reliance on glycolysis and substrate-level phosphorylation, the bloodstream stage represses many functions of its mitochondrion, including oxidative phosphorylation [8–10]. Yet even in this organelle DNA replication, transcription, RNA editing and processing, as well as tRNA and protein import remain functional and essential [11–14]. However, since neither TAO [15] nor the ATPase complex, which are still functional in the bloodstream stage, contain any Fe/S clusters, the demand for these co-factors dramatically drops in the mitochondrion as compared to its procyclic counterpart. These interstagial differences make the assembly of Fe/S clusters in the mitochondrion of T. brucei of special interest.
Moreover, this dual mitochondrial metabolism, in two life cycle stages containing a single organelle per cell, makes T. brucei a suitable model for exploring mechanisms that govern the switch from a fully active to a repressed mitochondrion. Transformation from the bloodstream to the procyclic stage requires almost instant mitochondrial activation and concomitantly the up-regulation of the mitochondrial Fe/S assembly pathway, which is essential for many subunits of respiratory complexes and enzymes such as aconitase and fumarase. The clearly marked metabolic switches that occur during development must therefore be somehow coordinated with environmental changes to match metabolic demands of the parasite. These developmental transformations are known to involve similarly drastic changes in gene expression, mediated primarily at the post-transcriptional level [16].
tRNA editing at an important position and thiolation at an unusual position
Recent findings have shown that the Nfs complex is not only important for the assembly of Fe/S clusters and their incorporation into a growing list of enzymes involved in various metabolic pathways, but also for pathways that could potentially affect gene expression, including tRNA maturation [13,17,18]. In all organisms tRNAs undergo numerous post-transcriptional modifications. Of these, some are required for tRNA function and cell viability, while others are not by themselves indispensable, but in conjunction with other modifications serve important roles in ensuring proper tRNA folding [19,20]. In general, modifications that occur away from the anticodon loop play more of a structural role, and their absence may lead to destabilization of the tRNA. Modifications that affect the anticodon nucleotides usually have a direct bearing on decoding. A special type of modification known as RNA editing may replace one nucleotide for another, directly changing the amino acid decoding capacity of the tRNA and effectively reassigning codons without the need for changes at the DNA level.
A now classic example of tRNA editing was first discovered in Leishmania tarentolae [21] and also later found in T. brucei [22], where it occurs at the first position of the anticodon in tRNATrp. Since trypanosomatids (Leishmania and Trypanosoma) do not contain any tRNA genes in their mitochondrial genomes [21], the complete set of tRNAs used in both cytoplasmic and mitochondrial protein synthesis is encoded solely by the nuclear genome. In these organisms tRNAs are transcribed in the nucleus, exported to the cytoplasm, and later a subset of cytoplasmic tRNAs is actively imported into the mitochondrion [23]. However, translation of tryptophan codons represents a potential problem because, like in many other eukaryotes, in the trypanosomatid mitochondrial genome the canonical UGG tryptophan codon is often replaced by UGA, a stop codon in cytoplasmic translation. This led to the question of how organisms with a single tRNATrp with the anticodon CCA could decode UGG as tryptophan and UGA as a stop codon in the cytoplasm and UGA as tryptophan upon import into the mitochondrion [21]. Trypanosomatids solved this decoding conundrum in a simple yet elegant way. Approximately 50% of the tRNATrp found in mitochondria undergoes cytidine to uridine (C to U) editing at the first position of the anticodon, effectively creating two versions of the tRNA: one with a UCA anticodon that can now decode UGA and a second with the standard CCA anticodon for UGG decoding [21] (Figure 1). This 50/50 split has raised questions as to how such a balance is maintained. An earlier observation demonstrated that tRNATrp in L. tarentolae had an additional unusual feature, beyond C to U editing; this tRNA was found to be thiolated at position 33 (to form 2-thiouridine, s2U33) of the anticodon loop, a position that was presumably never modified in tRNAs of any organism. Even more unusual was the fact that this modification only occurred in the edited tRNA, prompting a model by which thiolation at U33 was required for editing [24]. Although it was later shown in T. brucei that both the edited and unedited tRNAs were thiolated [25], the question as to how editing levels were regulated and also what was the role of the thiolation event remained open. Recently, the down-regulation of Nfs was shown to cause the expected reduction of thiolation, but also, surprisingly, an almost 100% increase in editing. This led to the conclusion that thiolation was a negative determinant for editing, thus implying editing and thiolation were competing reactions [16,18] (Figure 1).
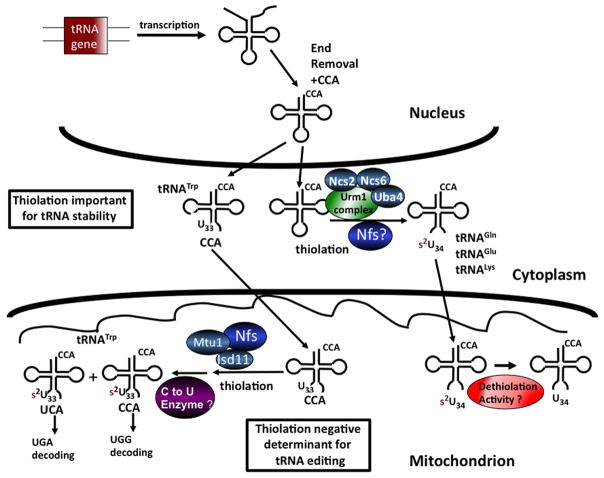
Figure 1.
Maturation pathway of thiolated tRNAs in T. brucei. A schematic of the maturation pathway of thiolated tRNAs and the role that thiolation plays in the different cellular compartments. In T. brucei all tRNAs are transcribed in the nucleus where they undergo end-trimming and CCA addition prior to export to the cytoplasm. Once in the cytoplasm, a portion of the tRNAs are modified and kept for cytoplasmic translation of nucleus-encoded mRNAs. Another portion are imported into the mitochondrion already bearing cytoplasmic modifications and are further modified in the organelle. Among these modifications, C to U editing is essential for decoding of UGA codons as tryptophan. Thiolation plays two different roles in T. brucei; in mitochondria it serves as a negative determinant for editing. In the cytoplasm, thiolation is important for the stability of tRNAGln, tRNAGlu and tRNALys. Question marks denote activities which are inferred to exist but for which the enzyme involved is not yet known. It is still not clear if all of the components of the Urm1 cytoplasmic thiolation pathway exist in T. brucei.
What then is the connection between editing and Fe/S cluster assembly? Interestingly, in yeast and probably other organisms including trypanosomes, Nfs plays crucial roles in both tRNA thiolation and Fe/S cluster assembly [26]. The key differentiating feature between T. brucei and other organisms, however, is the unique s2U33 thiolation confined to tRNATrp [21,25]. Nfs also plays a role in the cytoplasmic thiolation of tRNAGln, tRNAGlu and tRNALys in T. brucei [18] (Figure 1). Therefore, the key desulfurase in the Fe/S assembly pathway plays an essential function in uniquely controlling two different thiolation pathways in T. brucei: one mitochondrial and the other cytoplasmic [17,18,27]. Nfs achieves this despite the fact that the enzymes involved in mitochondrial thiolation are not known to require Fe/S, but rather are the recipients of the sulfur-donor activity of Nfs (Table 1). This then leads to the question of whether Fe/S clusters are found as cofactors in other modification enzymes. The surprising answer is that indeed at least four different tRNA modification pathways involve a key enzymatic reaction harboring an Fe/S cluster [28–30] (Table 1).
Table 1.
tRNA modification enzymes that either require an Fe/S cluster or depend on the Nfs desulfurase for activity
| Enzymea | Fe/S cluster | Localization | Function |
|---|---|---|---|
| MiaB | Yes | ?b | Methyl thiolation at position 37 of tRNAs |
| Tyw1 | Yes | Cytoplasmic | First step of wybutosine biosynthesis at position 37 of tRNAPhe |
| Urm1, Uba4, Ncs2, Ncs6 | ? | Cytoplasmic | Thiolation of tRNAGln, tRNAGlu, and tRNALys |
| Elp3 | Yes | Cytoplasmic | Alkylation of tRNAs at position 34 |
| Mtu1 | No, but requires Nfs | Mitochondrial | Thiolation of various tRNAs at position 34 and tRNATrp at position 33 in T. brucei |
Various potential homologs of these enzymes are found in the T. brucei sequence database
Intracellular localization not known
The common denominator of these unrelated pathways is their capacity to modify target nucleotides which, as stated above, play key roles either directly affecting decoding, influencing translational efficiency and/or fidelity by impacting the tRNA anticodon-loop structure [31]. For example, the hypermodified nucleotide wybutosine found at position 37 of tRNAPhe in most, if not all, eukaryotes has been shown to prevent translational frameshifting [30,32]. Wybutosine biosynthesis involves a series of enzymatic activities, which include addition of an extra ring to the purine ring of m1G (normally found at position 37 of other tRNAs). This reaction is catalyzed by Tyw1, an enzyme that contains and presumably requires an Fe/S cluster for its activity [33]. Similar arguments can be made for the role of other Fe/S-containing enzymes listed in Table 1.
Could seemingly unrelated pathways help couple metabolic and translational rates in trypanosomes?
A curious corollary of the story of tRNA thiolation and Fe/S cluster assembly pathways in T. brucei is the remarkable finding that the same desulfurase is required for the Fe/S assembly and thiolation of both cytoplasmic and mitochondrial tRNAs [18]. The fact that respiratory complexes depend on numerous Fe/S clusters in their subunits means that down-regulation of Nfs could lead to a drop of respiratory rates. Here a model is suggested by which the divergence of the two pathways (Fe/S assembly and tRNA editing/thiolation) from a common key enzyme may be exploited by these organisms to carefully match respiration rates to mitochondrial translation, perhaps by offsetting the 50/50 ratio for edited and unedited tRNATrp. In this model, decreasing activity of the Fe/S pathway is paralleled with decreased tRNA thiolation, which will lead to an increase in tRNATrp editing, eventually affecting translational efficiency, assuming that the thiolated versions of the unedited and edited tRNAs are required for proper protein synthesis (Figure 1). Perhaps, this could be used by the parasites for the differential expression of certain genes that may contain different numbers of UGG and UGA codons. Currently, however, in the absence of an in vitro mitochondrial translation and/or transformation system, these will remain open questions. These types of regulatory circuits may be particularly relevant in trypanosomatids; these organisms lack most transcriptional control with the bulk of the regulation of gene expression occuring post-transcriptionally.
Surprisingly, down-regulation of Mtu1 in T. brucei mitochondria produced no obvious phenotype [6,17]. This is despite the fact that the lack of Mtu1 caused the expected impaired thiolation and also increased editing to nearly 100%. Currently, we do not have an explanation for this observation, especially when in other systems lack of mitochondrial thiolation leads to serious physiological defects. Perhaps as previously suggested, the lack of phenotype may be more a reflection of laboratory cultivation conditions, as opposed to growth in the insect gut or mammalian bloodstream, depending on the developmental stage of the parasite.
tRNA utilization in the cytoplasm may also be affected by the lack of certain modifications (Table 1), which directly depend on the presence of Fe/S clusters. Thus, this hypothetical coupling of translational rates to metabolic rates may even include cytoplasmic tRNA modification systems in connection with the assembly of Fe/S clusters and global metabolic regulation [34]. Interestingly, the effect of thiolation itself is not limited to mitochondrial function. A recent report showed that ablation of Nfs in T. brucei also leads to the specific destabilization of the cytoplasmic thiolated tRNA species [18], potentially providing another means of down-regulating protein synthesis in response to changes in nutrient levels and environmental signals. In addition, a connection between folate-dependent pathways and Fe/S assembly has been established [35]. Folate is also a co-factor required for some modification enzymes. What is not clear is if tRNA modifications indirectly form part of the response to changes in the levels of nutrients, for example cysteine pools and/or iron availability, or in fact are part of the signal for such changes. Nevertheless, the possible impact on overall translational rates should not be at all surprising.
The question then becomes how to test this simple and appealing hypothesis. The study of tRNA modifications and their changing levels is by itself challenging, especially when in most organisms the complete set of modifications is not even known, let alone the enzymes responsible for them. Luckily, efforts from various laboratories have already produced a set of modification enzymes that require Fe/S for their activity and whose homologs exist in T. brucei. These could be genetically manipulated and their overall effect on the T. brucei proteome analyzed, potentially revealing trends in gene expression patterns that depend on changes in modification patterns. In the end, given how little we know about the Fe/S assembly and tRNA modifications in trypanosomes, these proposals are of course largely speculative but their exploration may reveal important aspects of a higher order in the coordination of these aspects of cellular metabolism.
Acknowledgments
We would like to thank all members of the Alfonzo and Lukeš laboratories for helpful discussions. This work was supported by GACR 204/09/1667, the Ministry of Education of the Czech Republic (LC07032 and 2B06129 and 6007665801) and the Praemium Academiae award to J.L., and a GM084065 grant from NIH to J.D.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 2.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 3.Gerber J, et al. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramazzotti A, et al. Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett. 2004;557:215–220. doi: 10.1016/s0014-5793(03)01498-4. [DOI] [PubMed] [Google Scholar]
- 5.Smid O, et al. Knock-downs of iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- 6.Paris Z, et al. The Fe/S cluster assembly protein Isd11 is essential for tRNA thiolation in Trypanosoma brucei. J Biol Chem. 2010;285:22394–22402. doi: 10.1074/jbc.M109.083774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long S, et al. Ancestral roles of eukaryotic frataxin: mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologs in trypanosomes. Mol Microbiol. 2008;69:94–109. doi: 10.1111/j.1365-2958.2008.06260.x. [DOI] [PubMed] [Google Scholar]
- 8.Hannaert V, et al. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis. 2003;2:11. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coustou V, et al. ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J Biol Chem. 2003;278:49625–49635. doi: 10.1074/jbc.M307872200. [DOI] [PubMed] [Google Scholar]
- 10.Tielens AG, van Hellemond JJ. Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 2009;25:482–490. doi: 10.1016/j.pt.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Schnaufer A, et al. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 12.Hashimi H, et al. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristodero M, et al. Mitochondrial translation is essential in bloodstream forms of Trypanosoma brucei. Mol Microbiol. 2010;78:757–769. doi: 10.1111/j.1365-2958.2010.07368.x. [DOI] [PubMed] [Google Scholar]
- 14.Paris Z, et al. Futile import of tRNAs and proteins into the mitochondrion of Trypanosoma brucei evansi. Mol Biochem Parasitol. 2010;176:116–120. doi: 10.1016/j.molbiopara.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhuri M, et al. Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 2006;22:484–491. doi: 10.1016/j.pt.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruske EI, et al. Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J Biol Chem. 2009;284:36491–36499. doi: 10.1074/jbc.M109.064527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wohlgamuth-Benedum JM, et al. Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J Biol Chem. 2009;284:23947–23953. doi: 10.1074/jbc.M109.029421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfonzo JD, et al. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charriere F, et al. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2006;103:6847–6852. doi: 10.1073/pnas.0602362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfonzo JD, Soll D. Mitochondrial tRNA import--the challenge to understand has just begun. Biol Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crain PF, et al. Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA. 2002;8:752–761. doi: 10.1017/s1355838202022045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charriere F, et al. Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J Biol Chem. 2009;284:16210–16217. doi: 10.1074/jbc.M109.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai Y, et al. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- 27.Paris Z, et al. Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA. 2009;15:1398–1406. doi: 10.1261/rna.1589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierrel F, et al. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J Biol Chem. 2002;277:13367–13370. doi: 10.1074/jbc.C100609200. [DOI] [PubMed] [Google Scholar]
- 29.Paraskevopoulou C, et al. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 30.Noma A, et al. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto-Ito S, et al. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr D Biol Crystallogr. 2007;63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki Y, et al. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Lillig CH, Lill R. Lights on iron-sulfur clusters. Chem Biol. 2009;16:1213–1214. doi: 10.1016/j.chembiol.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Waller JC, et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc Natl Acad Sci U S A. 2010;107:10412–10417. doi: 10.1073/pnas.0911586107. [DOI] [PMC free article] [PubMed] [Google Scholar]