Abstract
This study combines metabolic oligosaccharide engineering (MOE), a technology where the glycocalyx of living cells is endowed with chemical features not normally found in sugars, with custom-designed three dimensional biomaterial substrates to enhance the adhesion of cancer cells and control their morphology and gene expression. Specifically, Ac5ManNTGc, a thiol-bearing analogue of N-acetyl-d-mannosamine (ManNAc) was used to introduce thiolated sialic acids into the glycocalyx of human Jurkat T-lymphoma derived cells. In parallel 2D films and 3D electrospun nanofibrous scaffolds were prepared from polyethersulfone (PES) and (as controls) left unmodified or aminated. Alternately, the materials were malemided or gold-coated to provide bioorthogonal binding partners for the thiol groups newly expressed on the cell surface. Cell attachment was modulated by both the topography of the substrate surface and by the chemical compatibility of the binding interface between the cell and the substrate; a substantial increase in binding for normally non-adhesive Jurkat line for 3D scaffold compared to 2D surfaces with an added degree of adhesion resulting from chemoselective binding to malemidede-derivatived or gold-coated surfaces. In addition, the morphology of the cells attached to the 3D scaffolds via MOE-mediated adhesion was dramatically altered and the expression of genes involved in cell adhesion changed in a time-dependent manner. This study showed that cell adhesion could be enhanced, gene expression modulated, and cell fate controlled by introducing the 3D topograhical cues into the growth substrate and by creating a glycoengineered binding interface where the chemistry of both the cell surface and biomaterials scaffold was controlled to facilitate a new mode of carbohydrate-mediated adhesion.
1. Introduction
This study combines two emerging research foci – three dimensional (3D) growth substrates and metabolic oligosaccharide engineering (MOE) – to gain enhanced control over cancer cells. This work builds on seminal experiments that established that adhesion has a dramatic impact on cell fate and behavior [1, 2] by including contributions made through the manipulation of glycosylation and the exploitation of a metabolically engineered carbohydrate-mediated cell attachment to the growth substrate. This study exemplifies today’s great interest in the design of cell niches where high throughput screening methods are being used to tune the biochemistry at the binding interface between a cell and its growth substrate by probing scaffolds displaying a broad diversity of chemical and biophysical properties [3-5].
Despite remarkable progress, precise control of cell attachment to matrix materials remains challenging [4]. By contrast to most extant efforts to control adhesion, which are concentrated on the growth substrate, we are pursuing a fundamentally different approach by modulating the chemistry of the cell surface by using MOE to incorporate chemical functional groups not naturally found in carbohydrates into the glycocalyx of living cells (Scheme 1) [6-9]. By virtue of its location on the outer periphery of the cell surface, the glycocalyx is ideally situated to interact with complementary chemical functional groups located on the growth substrate allowing us to co-engineer the cell and scaffold to achieve carbohydrate-based modes of cell adhesion [10, 11].
Scheme 1.
(a) Two-step scheme for metabolically engineering a cell surface with complementary chemical properties to bind to a custom-designed tissue engineering scaffold, as described in more detail in panels (b) and (c). (b) In the first step, chemical functional groups – thiol groups situated at the N-acyl position of the ManNAc analog Ac5ManNTGc – are metabolically incorporated into the sialic acid pathway and replace Neu5Ac (the natural form of sialic acid most commonly found on human cells) with the thiolated Sia5TGc (ThioGlycolyl) form of this sugar. (c) In the second step, the thiolated cells are incubated with a chemically compatible biomaterial – in this case PES. As negative controls, neither unmodified PES (i) or animated surfaces (ii) were expected to bind the metabolically engineered cells. By contrast, bio-orthogonal bonds form between the thiolated sialic acids found on the cell surface and (iii) gold coated or (iv) malemide-derivatized surfaces promote new modes of carbohydrate-based cell adhesion.
Previously, our laboratory demonstrated the concept that a “glycoengineered” binding interface can be constructed by employing peracetylated N-thiolglycolyl-d-mannosamine (Ac5ManNTGc, Scheme 1b), to biosynthetically endow cell surface sialic acids with thiol groups [12]. Unlike natural thiols found in cell surface proteins, the sialic acid-carried thiols are located at the outer periphery of the glycocalyx, which allows them to be highly accessible and spontaneously form high affinity interactions with a complementary growth substrate. In one example of this strategy, normally non-adhesive Jurkat cells (a human T-lymphoma line) were attached to malemide-derivatized glass [10]. Extending this methodology to human embryonic cells, incubation with Ac5ManNTGc combined with growth on a flat (2D) gold surface resulted in the up-regulation of the Wnt pathway and provoked differentiation of the cells into neural lineages [10]. Interestingly, Wnt proteins (e.g., Wnt 3 or Wnt 10) normally required for canonical Wnt pathway activation were not present in this system. Moreover, neither the ManNAc analogue nor the gold substrate by themselves were sufficient to trigger the full repertoire of observed responses. These results provided compelling evidence that a glycoengineered binding interface where both the cell surface and growth substrate are simultaneously designed to support carbohydrate-mediated adhesion can not only be used to attach cells to a growth substrate, but also can control signaling, downstream behaviors, and cellular outcomes.
In this study, we extend MOE-based technology designed to create custom-designed cell niches in two directions. First, we move from 2D surfaces to 3D systems that employ electrospinning to fabricate synthetic nanofiber scaffolds designed to mimic certain topographical features (e.g., collagen fibrils) found in the extracellular matrix (ECM); these scaffolds were maleimide-functionalized or gold-coated to enhance the adhesion of cells displaying thiolated sialic acids (Scheme 1c). The second direction is the application of this technology for the creation of 3D tumor niches [13] that are useful in understanding cancer biology [14] and testing drugs in a realistic context [15]. By using Jurkat cells – a T-lymphoma derived line that is normally non-adhesive when grown in tissue culture conditions (even in the presence of ManNAc analogues used in MOE, as documented in our previous publications [16-19]), we demonstrated a hierarchical response where scaffold topography alone affected cell adhesive properties while MOE further influenced cellular responses to the microenvironment.
2. Materials and Methods
2.1. Electrospinning of polyethersulfone (PES) fiber
All chemicals were purchased from Sigma-Aldrich (St. Louis, USA) unless otherwise stated. PES granules (Mw=55,000, Goodfellow Cambridge Limited, UK) were dissolved in dimethyl sulfoxide (DMSO) at 15-25 wt% concentration and placed in a plastic syringe fitted with a 27G needle. A syringe pump (KD Scientific, USA) was used to feed the polymer solution into the needle tip. The feed rate of the syringe pump was fixed at 0.6 ml/h. The PES nanofiber meshes were fabricated by electrospinning at 13 kV using a high voltage power supply (Gamma High Voltage Research, USA). Nanofibers were collected directly onto grounded 15 mm diameter glass coverslips located at a fixed distance of 160 mm from the needle tip. PES films as a 2D control were fabricated on glass coverslips by spin-coating 8.0 wt% PES in dimethylformamide (DMF). The deposited nanofiber and film samples were washed thoroughly in distilled water and then in ethanol to remove any residue DMSO, and subsequently dried and stored in a desiccator. To quantify the fiber size and scaffolds thickness, the electrospun samples were measured by NIH Image J software analysis of SEM images obtained from a JEOL JSM-5200 scanning electron microscope.
2.2. Incorporation of thiol functionalities into cell surface with sugar analogue
Peracetylated N-thiolglycolyl-d-mannosamine (Ac5ManNTGc) was synthesized and characterized using published methods [20] and a 50 mM stock solution was prepared in ethanol (which could be stored at 4 °C for 4-6 weeks before use). Analogue, or an equal volume of ethanol as the “negative” control, was added to culture plated prior to cell seeding (generally at the optimized concentration of 25 μM) and cells were allowed to incubate in its presence for two to three days to maximize thiol expression [10]. The resulting cell-surface thiols (CSTs) were detected and quantified by labeling the cells with (+)-biotinyl-3-maleimidopropionamidyl-3,6-dioxaoctanediamine (MB) followed by staining with fluorescein-conjugated avidin and quantification by flow cytometry [12].
2.3. Electrospinning of complementary functionalized PES substrates
Ac5ManNTGc-treated, thiol-expressing cells were expected to have enhanced affinity to either a maleimide-modified or a gold-coated surface (Scheme 1). To prepare the maleimided substrate, the PES matrixes were carboxylated first by UV-initiated poly(acrylic acid) (PAAc) grafting, following amination of PAAc-grafted PES [21], and finally maleimide ligand was functionalized via covalent conjugation to the aminated materials. Briefly, samples were immersed in aqueous solution containing 5.0% AAc solution and 0.5 mM NaIO4, and exposed to a 400 W mercury UV lamp (5000-EC, Dymax, Germany) for 4.0 min at a distance of 25 cm under 5-8 °C. The PAAc-grafted meshes were then thoroughly washed with deionised water for over 36 h. Following, the scaffolds were immersed in acetonitrile containing 50 mM N-hydroxysuccinimide (NHS) and dicyclohexylcarbodiimide (DCC) for 6.0 h, and then further conjugated with 0.03 mM ethylene diamine (EtDA) for 12 h to obtain the aminated PES fiber. Furthermore, the maleimidation was performed by functionalized aminated fibers with 0.2 mM sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC, Pierce Biotechnology Inc.). The various chemical surfaces were characterized by FT-infrared spectra (data not shown). The density of conjugated primary amino and maleimide groups on PES nanofiber were ~50 and ~13 nmol/cm2, as quantified by the colorimetric titration [22] and photometric determination [23], respectively. The gold-coated substrates were obtained by sputter-coating PES fibers with approximately 20 nm gold using a JEOL JFC-1100E sputtering device (the coating thickness had been optimized by varying the gold layer upwards from 0.6 nm; no increase in cell adhesion was observed above 6 nm but 20 nm was used to ensure full coverage of surfaces). The obtained substrates were subsequently thoroughly washed in phosphate buffered saline (PBS) and sterilized in 70% ethanol, then loaded into 24-well tissue culture plates and stored in sterile PBS until use.
2.4. Cell Culture
Throughout these experiments, Jurkat human T-lymphoma wild-type (JWT, Clone E-6, ATCC, Manassas, VA) were typically incubated in 500 μl of RPMI 1640 medium supplemented with 5.0% fetal bovine serum (FBS) and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin) in 24-well plates (37 °C, 5.0% CO2). To optimize the adhesion to the electrospun fiber scaffolds, we quantified cell attachment to 2D PES film and 3D fibrous scaffolds with thickness of 10, 50, and 80 μm three days after the prepared scaffolds had been seeded with 500,000 cells by measuring DAPI nuclei staining or by DNA quantification (as described in more detail below). In subsequent experiments testing the effects of MOE, unless otherwise stated, the cells were pre-incubated with the optimized concentration of 25 μM Ac5ManNTGc for two days in suspension cultures (in normal tissue culture flasks) and then transferred to well containing either the 2D or 3D chemically-modified surfaces (the initial concentration of Ac5ManNTGc was maintained after the transfer of the cells for the duration of the experiment).
2.5. DNA quantification
The quantity of DNA in the cells attached to each scaffold was determined using a Hoescht dye assay based on the methods of Yang and coworkers [24]. After removal of the supernatant, the scaffolds were gently washed with PBS twice to remove the unbound cells. To disassociate the DNA from the scaffolds, each sample was digested by 500 μl of papain in digestion buffer (125 μg/ml papain in 0.1 M sodium phosphate, 5.0 mM Na2EDTA, and 5.0 mM cystein-HCl, pH 6.0) for 16 h at 60 °C. Following digestion, each sample was centrifuged at 10,000 × g for 30 s to remove insoluble or undigested material. The DNA content was measured fluorometrically using Hoechst Dye 33258 solution (Molecular Probes, Eugene, OR), and determined by Hoefer® DyNA Quant® 200 fluorometer (Amersham Pharmacia Biotech) at 656 nm.
2.6. Cell viability on scaffolds
Cell viability assays were based on the integrity of cellular membranes using the Live/Dead Viability/Cytotoxicity Kit (Molecular Probes, Eugene, OR) that contains calcein-AM (‘Live’ dye) and ethidium homodimer-1 (‘Dead’ dye). The dye solution was prepared with 1.0 μl of calcein-AM dye and 2.0 μl of ethidium homodimer-1 dye in 1.0 ml RPMI 1640 medium. After cells were cultured in the presence of 2D or 3D constructs as described above, the medium was aspirated and the scaffolds were incubated in 500 μl of the ‘Live/Dead’ dye solution for 30 min at 37 °C. Fluorescence microscopy then was performed using a fluorescein optical filter (485±10 nm) for calcein-AM and a rhodamine optical filter (530±12.5 nm) for ethidium homodimer-1. The percentage of viable cells was determined by counting the live cells, which were stained with green dye, relative to the total cells (i.e., green plus red-stained cells) present in three different fields.
2.7. SEM characterization of 3D scaffolds and adhered cells
After removal of the culture medium, the specimens were rinsed with PBS twice and the cells were fixed with 3.0% glutaraldehyde solution for 30 min, and rinsed again with PBS. After cell fixation, the specimens were dehydrated in an ethanol/PBS and then hexamethyldisilazane (HMDS)/ethanol, at varying concentration (25%, 50%, 75%, 90%, and 100%) for 5.0 min, respectively. After being completely dried under vacuum, the specimens were mounted on copper stubs, coated with gold using a JEOL JFC-1100E sputtering device, and observed by a JEOL JSM-5200 scanning electron microscope. To ensure a representative count, each sample was divided into quarters and two fields per each quarter were photographed.
2.8. Hematoxylin and Eosin staining
After three days of incubation with the scaffolds (and Ac5ManNTGc when appropriate), cell cultures were stained with hematoxylin by using the following procedures. The samples were rinsed with tap water for 3.0 min, incubated with Harris hematoxylin (Polyscientific) for 3.0 min, washed 10 times with ddH2O for 10 s each, and rinsed with tap water for 5.0 min. The samples were then exposed to Eosin Y for 30 s, rinsed 10 times in a 1.0% acid EtOH dip, washed twice with tap water for 1.0 min and then two more times with ddH2O for 2.0 min. Finally the samples were immersed in 95% EtOH for 1.0 min followed by 100% EtOH for 1.0 min and xylene for 1.0 min. The samples were dried by blotting on paper wipes, placed in permount mounting solution, and covered with a glass slide taking care to exclude any visible air bubbles.
2.9. Sulfated glycosaminoglycan (sGAG) analysis
Sulfated glycosaminoglycan content was determined using the Blyscan sGAG Kit (Biocolor Ltd., UK) that utilizes the quantitative dye-binding method to analyze the glycosaminoglycan component of sulfated proteoglycans. Samples were digested by papain and collected as described in Section 2.5 taking care to conduct the PBS washes gently so as to not dislodge ECM-like deposits associated with the scaffolds. Blyscan protocols were followed for sample analysis and O.D. readings were obtained using an ultraviolet spectrophotometer (Biotech Powerwave 340). The Hoechst DNA assay was used to determine cell number to normalize protein and sGAG content.
2.10. Quantitative real-time polymerase chain reaction (qRT-PCR) evaluation of gene expression
RNA was extracted by the TRIzol (Invitrogen, Carlsbad, CA) method following the manufacturer’s instruction from cells obtained from both 2D and 3D cultures. For the 2D cultures only non-adherent suspension cells were analyzed because too few cells attached to the substrate to obtain reliable results. For the 3D cultures, the cells in the supernatant were collected and analyzed separately from cells that were attached to the 3D fibrous scaffolds, which were harvested as described above in Section 2.5. Total RNA (2.0 μg) per 20 μl of reaction volume was reverse transcribed into cDNA using the SuperScript First-Strand Synthesis System (Invitrogen). Amplifications for the cDNA samples were carried out at 65°C for 5.0 min; 50 °C for 50 min, and 85 °C for 5.0 min. Real-time polymerase chain reactions were performed and monitored using the SYBR Green PCR Mastermix (Roche Diagnostics, Mannheim, Germany) and Bio-Rad iQ™ 5 Detection System with the thermal cycle conditions of 50 °C for 2.0 min; 95 °C for 10 min; 40 cycles of 95°C for 15 s and 60°C for 1.0 min. cDNA samples (1.0 μl for total volume of 23 μl per reaction) were analyzed for gene of interest and for the reference gene glyceraldehydes-3-phosphate-dehydrogenase (GAPDH). The level of expression of each target gene was normalized to GAPDH with the fold difference calculated using the ΔΔCt method (as described in detail elsewhere [25, 26]). Analysis of each sample was repeated at least three times (with 3 to 4 replicates each time) for each gene of interest.
2.11. Statistical analysis
Unless otherwise specified, all values were presented as the mean ± SD. All experiments were independently repeated three to five times with different cell preparations and all measurements were done in triplicate. Comparisons between groups were performed using the unpaired Student’s t-test. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. Fabrication and optimization of PES substrates
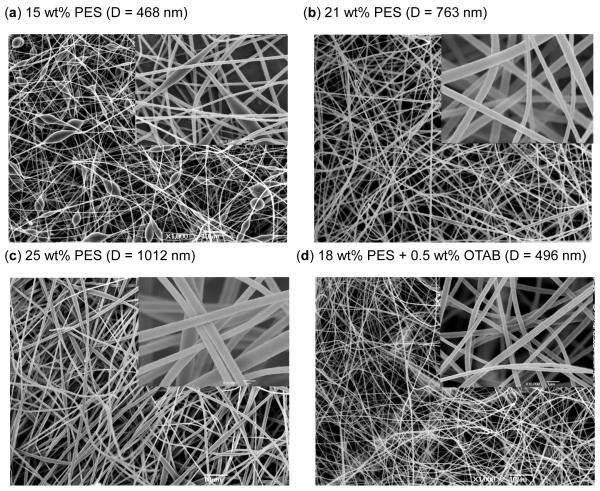
To create growth substrates with 3D topographical features, non-woven fibrous substrates were prepared by electrospinning PES at concentrations ranging from 15 to 25 wt% based on published information [21]. In our experiments, PES solutions with concentration between 15 and 20 wt% yielded beaded fibers with the minimum concentration required to achieve non-beaded fibers was 21 wt% (Fig. 1a). This concentration produced fibers with an average diameter of 763 nm (Fig. 1b) while 25 wt% produced fibers with an average diameter of 1012 nm (Fig. 1c). The fiber diameter could be reduced to 496nm (Fig. 1d) without beading by adding 0.5 wt% OTAB (octadecyltrimethylammonium bromide) [27] into 18wt% PES solution.
Figure 1.
Electrospun fibers produced in dimethyl sulfoxide (DMSO) at a flow rate of 0.6 ml/h and the indicated concentrations of PES (panels (a) – (c)) with the addition of OTAB in (d). The fibers were analyzed by scanning electron microscopy and representative images are shown here. Scale bars represent 10 μm and 1 μm in each panel and inset, respectively.
Once fiber diameter could be controlled reproducibly, the thickness of the scaffold (i.e., the depth in the “z” direction) was varied. Electrospinning for longer time periods increased the amount of fiber deposition and, thereby, the thickness of the fiber layer; for example at 5.0 min the thickness of a 1012 nm scaffold was ~10 μm. The thickness of the scaffold increased to ~50 μm after 15 min of electrospinning and to ~80 μm at 30 min.
3.2 Evaluation of cell infiltration into the 3D scaffolds
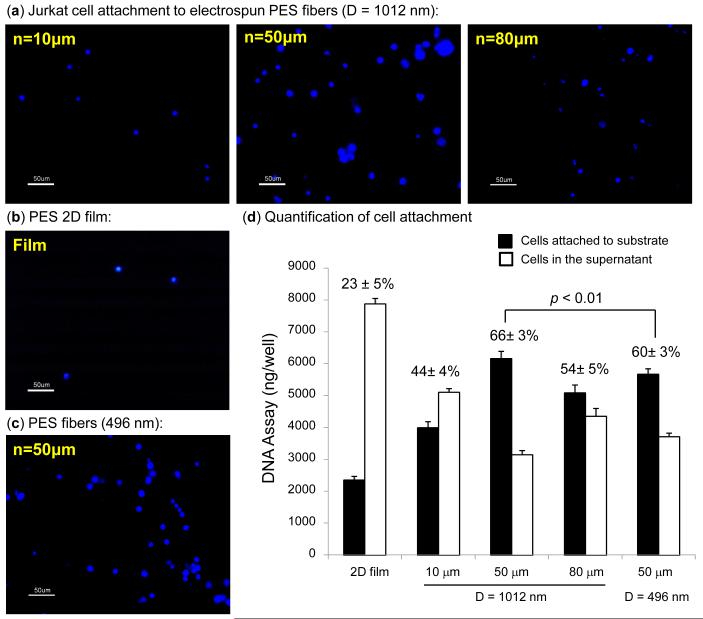
In theory, cells could interact with the nanofibers in two ways. First, they could sit on top of the mesh; in this case similar biological responses would be expected from any of the 10, 50, or 80 μm thick layers. Alternatively, the cells could migrate into, and subsequently infiltrate, the fiber layer; this option was only possible for the ~50 and ~80 μm thick layers because the 10 μm scaffolds were too thin to provide a truly three-dimensional microenvironment for Jurkat cells, which have an average diameter of ~12 μm. To test these possibilities, Jurkat cell attachment to 2D film and 3D fiber scaffolds was qualitatively visualized three days post-seeding by fluorescent staining of cell nuclei and quantified with a DNA-based assay. By using PES fibers with a diameter of 1012 nm, DAPI staining demonstrated that the cells preferentially adhered to the thicker 50 and 80 μm 3D layers compared to the 10 μm layer (Fig. 2a) or to the 2D film (Fig. 2b). Similar trends were seen for fibers of other diameters where the 50 μm layers supported the highest level of cell attachment (representative adhesion to 496 nm fibers is shown in Fig. 2c).
Figure 2.
Binding of Jurkat cells, which were stained with DAPI for visualization, to PES 3D electrospun fibers and 2D film. (a) Comparison of cells attached to three different layer thicknesses of 1012 nm PES fibers, (b) as a control; 2D film is shown. (c) In general, cells attached best to 50 μm thick layers; representative data is shown for 496 nm-diameter fibers. For (a)-(c), D and n indicate the diameter and thickness of the fibers, respectively. (d) Quantification of the effect of fiber diameter and layer thickness effect on cell attachment (the data shown represents the mean standard deviation of 5 independent experiments, each conducted in triplicate). The percentage values shown represent the fraction of attached cells in each condition and the p value shows that a statistical difference exists between the two indicated layer thicknesses.
Next, DNA quantification was performed to quantify the binding trends shown by Jurkat cells to the PES fibers of various diameters and thicknesses (Fig. 2d). These results showed a significant difference between the 2D film and any of the 3D scaffolds with substantially more cells attached to 3D matrices (44-66%) compared with the 2D film (23%). In the case of 3D scaffolds with a fiber diameter of 1012 nm, the results indicated more cells attached to the 50 μm thick nanofiber matrix (66%) compared to the other two thicknesses (i.e., 10 μm (44%) and 80μm (54%)). A similar – but slightly lower – level of cell attachment was observed (60%) for scaffolds having a diameter of 496 nm with the optimal thickness of 50 μm. The enhanced attachment to 3D scaffolds was attributed to the highly porous structure with interconnected spaces of electrospun fibers, which provide structural space for cell accommodation and migration. More specifically, for cell migration or infiltration to occur, a pore size of 10 μm [28] or the size a cell [29] has been suggested as necessary for cellular infiltration; these considerations are met by the porosity of our fibers.
3.3. Cell viability and cell adhesion of Ac5ManNTGc-treated cells
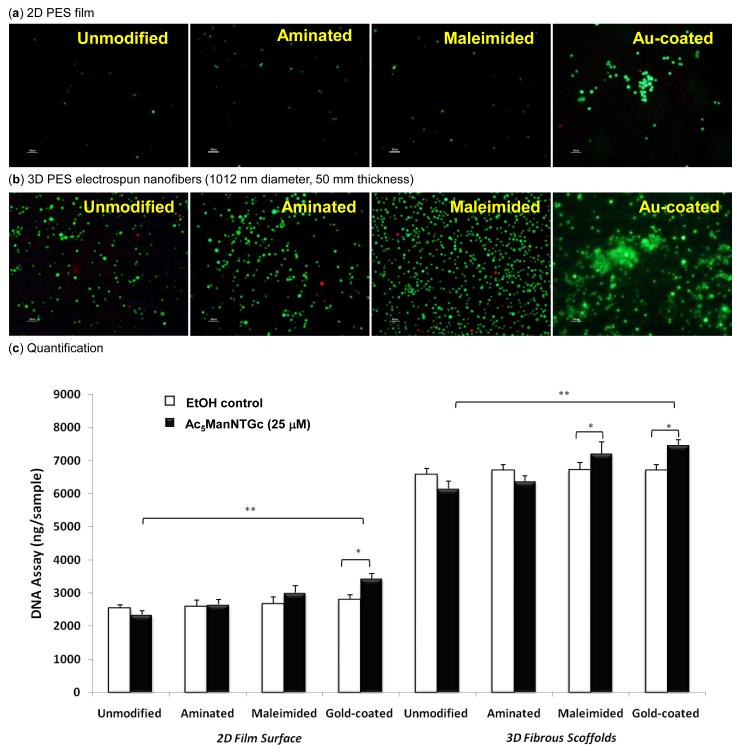
Having evaluated both the nanofiber diameter and the thickness of deposition, we found that the fibrous mesh with 1012 nm average diameter and a thickness of 50 μm provided optimal cell adhesion in the absence of thiolated sugar analog; these conditions were therefore selected for subsequent studies that added in the contributions of MOE. Turning to cell viability, a key prerequisite for Ac5ManNTGc-based oligosaccharide engineering to be useful for tissue engineering, we investigated cytotoxicity in cells undergoing carbohydrate-based adhesion. By using a maximum of 25 μM of Ac5ManNTGc, analogue-mediated cytotoxicity that can occur at higher concentrations [10] was avoided under routine 2D culture conditions as well as in the presence of chemically-modified flat surfaces (Fig. 3a). Extending the evaluation of cytotoxicity to the 3D scaffolds used in this study, the intensive green fluorescence observed in a live/dead assay indicated that > 95% of the cells remained viable after three days of culture in the presence of the electrospun nanofibers (Fig. 3b). Similar to the untreated control cells discussed above (and shown in Fig. 2), the number of analogue-treated cells attached to the 3D scaffold appeared to be significantly higher than to the 2D planar surfaces and more cells attached to the modified substrates than to the unmodified PES surfaces, especially to the gold-coated substrates where large cell clusters were observed.
Figure 3.
Viability of Ac5ManNTGc treated Jurkat cells after being cultured for three days on (a) 2D PES film and (b) 3D PES electrospun fibers with surfaces of various chemical composition (i.e., unmodified, aminated, maleimided and Au-coated as indicated). For panels (a) and (b), the scale bar is 50 μm and green represents live cells and red represents dead cells. (c) Cell attachment to the 2D film and 3D fibers was measured using a DNA quantification assay for glycoengineered (with Ac5ManTGc treated) and untreated control cells (the data shown represents the mean standard deviation of 5 independent experiments, each conducted in triplicate; *p < 0.05; **p < 0.01).
After verifying that cytotoxicity could be avoided over extended incubation of cells with Ac5ManNTGc and ensuring that an increase in accessible cell surface thiols levels occurred (i.e. upon incubation with 25 μM Ac5ManNTGc for 48 h the signal increased ~17-fold) we quantitatively evaluated cell attachment to the substrates with different chemical and topographical surfaces (e.g., unmodified and aminated, maleimided and gold-coated 2D PES film vs. 3D PES fibers) with and without treatment with Ac5ManNTGc. DNA quantification verified that significantly more cells attached to the 3D fibrous scaffolds (66-81%) compared to the 2D film (23-35%) and there was no statistical difference for cell adhesion to either 2D or 3D surfaces of different chemical composition in the absence of Ac5ManNTGc. By contrast, the incorporation of thiol groups into cell surface glycans by treatment with Ac5ManNTGc altered the adhesive properties of the glycocalyx by installing thiol groups that were expected to have enhanced affinity for the malemide-derivatized or gold-coated surfaces but not for the unmodified or animated substrates. Consistent with these expectations, ~10% more cells were detected on either maleimide-derivatized or gold-coated 2D surfaces compared to unmodified or aminated on 2D surfaces; a similar trend with ~ 15% more attached cells occurred for the 3D scaffolds. Interestingly, samples treated with Ac5ManNTGc contained ~5-9 % fewer cells regardless whether they were incubated in the presence of complementary (gold or malemided) or non-complementary (unmodified and aminated) scaffolds; this slight reduction is likely due to the growth inhibitory character of Ac5ManNTGc at sub-toxic concentrations [10]. Considering for this factor, the increase in attachment to the complementary scaffolds is even more impressive, increasing by ~25% when accounting for cells that were “missing” due to analogue-mediated growth inhibition; this enhanced binding can be attributed to chemoselective ligation between cells and the substrates that promote cell adhesion.
3.4. “Glycoengineered” binding dramatically alters cell morphology
The results presented above demonstrated that the absolute level of adhesion was modestly enhanced by treatment of the cells with thiol-displaying sugar analogs followed by incubation of the cells with a chemically complementary growth substrate. More importantly, changes in cell morphology indicated that this non-natural mode of carbohydrate-mediated adhesion modulated larger-scale cell processes. The changes on 3D scaffold were particularly interesting (on 2D film, the sparsely scattered cells that attached adopted a flattened and polygonal shape reminiscent of attachment to any lectin-coated surface – an example is shown in Fig. 4f for gold). Compared to the 2D film, not only were more cells attached to the 3D scaffolds, but the morphology of these cells was more or less “normal” (i.e., they showed spherical morphology similar to their shape when grown in suspension) in the absence of Ac5ManNTGc regardless of the surface chemistry of the growth substrate.
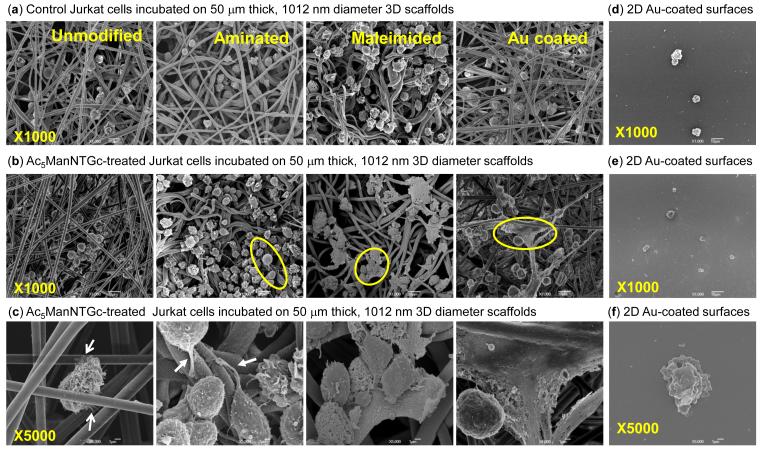
Figure 4.
Jurkat morphology when the cells are grown on different substrates with and without thiolated sialic acid display on their surfaces. Cells incubated with 3D scaffolds with the indicated surface chemistries in (a) the absence or (b) presence of Ac5ManNTGc at a magnification of ×1000 (x5000 for Ac5ManNTGc-treated cells is shown in (c)). Representative data for each of these conditions for 2D gold surfaces is shown in (d)-(f), respectively.
When incubated in the presence of Ac5ManNTGc, the response of the cells to the 3D scaffolds was even more dramatic. Overall, the cells appeared spherical when attached to a 3D scaffold but in each case had distinct morphological features that were tuned by the surface chemistry of the growth substrate. For example, the cells had small but noticeable filopodia that extended along the fiber surface when grown on unmodified scaffolds (Fig. 4c, 1st panel). On aminated fibers, the analog-treated cells assumed somewhat elliptical shapes and displayed tail-like structures (Fig. 4c, 2nd panel). When the cells were grown on complementary substrates (gold-coated or maleimide-derivatized) a highly unusual “spreading” morphology was observed wherein a sizeable proportion of the cells remained as a more or less intact spheroids but were surrounded by material spread or draped over the growth substrates. Furthermore, this effect was exacerbated on gold compared to malemide (Fig. 4c, 3rd and 4th panels) consistent with the higher density of binding partners for glycan-displayed thiol groups on the cell surface.
3.5 Analysis of the basement membrane like material deposited at the co-engineered interface
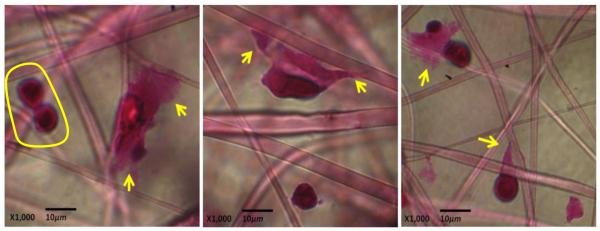
Hematoxylin and eosin (H & E) staining (hematoxylin solutions stain cellular nuclei dark blue while eosinophilic structures, which are generally composed of intracellular or extracellular protein, stain pink) was performed to analyze the composition of the material deposited on the scaffolds by cells located at binding interfaces co-engineered for high affinity adhesion. H & E staining of Jurkat cells pretreated with 25 μM Ac5ManNTGc for two days followed by seeding of the resultant thiol displaying cells on the gold coated PES fibrous scaffold revealed cells in approximately spherical morphology with relatively large nuclei compared to their cytoplasm (Fig. 5). The extracellular material associated with the cells that was spread across the scaffolds was stained bright pink, indicating that it was largely proteinaceous in nature.
Figure 5.
H & E staining of Jurkat cells that exhibit distinctive basement membrane-like morphology. Jurkat cells were pretreated with 25 μM Ac5ManNTGc for two days and cultured on gold coated PES fiber scaffolds for an additional three days. Arrows indicate the unusual spreading observed in a portion of the cells (cells in the yellow circle represent the normal spherical morphology of other cells present in the same culture).
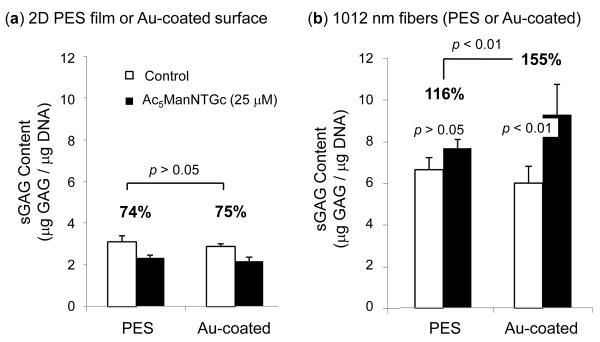
The protein deposition by Ac5ManNTGc-treated Jurkat cells observed when they were grown on complementary (e.g., gold-coated) 3D scaffolds was consistent with extracellular matrix (ECM) production; to verify this possibility we tested glycosaminoglycan levels. There was no statistically significant difference when untreated Jurkat cells were cultured on gold-coated or unmodified PES surfaces when comparing between 2D surfaces (Fig. 6a). However, an increase in sGAG content was found when comparing any of the 2D conditions with the 3D cultures (Fig. 6b; note that the two panels in Fig. 6 retain the same y-axis scale for easy comparison between 2D and 3D conditions). When the cells were grown on the 3D fibrous scaffolds, there was no statistical difference observed between the PES and gold surfaces in the absence of Ac5ManNTGc but in the presence of this sugar analogue the sGAG content increased significantly. One way to look at this endpoint is to compare the increase of sGAG in Ac5ManNTGc treated compared to untreated cells; for PES fibers this value is 116% (Fig. 6b). By comparison, this value is 155% on the gold coated fibers, which is a statistically significant increase (p < 0.01). Another way to evaluate sGAG production is to compare levels on gold coated fibers with and without Ac5ManNTGc; the level with the sugar analogue is ~9.2 μg/μg DNA, which is statistically (p < 0.01) higher than without analogue (~5.9 μg/μg DNA).
Figure 6.
Sulfated glycosaminoglycan (sGAG) content of Jurkat cells incubated (a) on 2D or (b) with 3D substrates (the data shown represents the mean standard deviation of 3 samples; in addition to the statistical comparisons indicated on the graph, comparisons of 2D v. 3D samples are statistically significant with p < 0.01). The percentage values provided above each set of data indicates the relative sGAG production in the Ac5ManNTGc-treated compared to untreated cells.
3.6. Gene expression
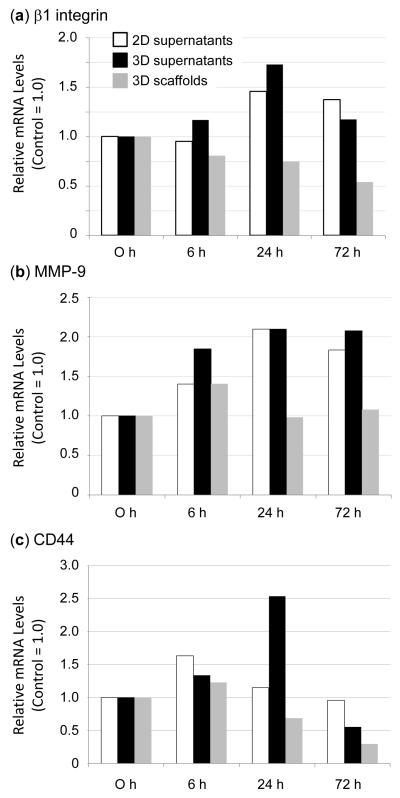
The expression of β1 integrin, MMP-9, and CD44, three genes implicated in cancer progression and the attachment of cancer cells to the ECM during metastasis, was measured by qRT-PCR and the results are summarized in Figure 7. In suspension culture (i.e., when the cells were cultured in the presence of a 2D surface to which very few Jurkat cells attached or when they were obtained from the supernatant of 3D culture systems), cells treated with Ac5ManNTGc displayed an increasing trend in gene expression for both β1 integrin (Fig. 7a) and MMP9 (Fig. 7b) that reached the highest level at 24 h and then slightly decreased (if anything) over the next 48 h. In contrast to β1 integrin and MMP9, a slight increase was observed for CD44 expression at 6 h followed by a time dependent decrease up to 72 h (Fig. 7c). An important finding from these experiments was that gene expression was markedly decreased for all three genes for Ac5ManNTGc treated cells that were attached to the 3D scaffolds compared to the cells that were harvested from the supernatants in either the 2D or 3D culture systems. This result showed that the glycoengineered binding interface engages signaling pathways and modulates different genes (e.g., β1 integrin, MMP-9, and CD44) in unique and long-lived ways.
Figure 7.
Time course of mRNA levels for (a) β1integrin, (b) MMP-9, and (c) CD44 in Jurkat cells pretreated with 25 μM Ac5ManNTGc cultured in the presence of 2D film or gold coated PES fibrous scaffold (D = 1012 nm with 50 μm layer thickness). Representative data from at least three experiments (with 3 to 4 replicates for each experiment) are shown for each gene of interest; all of the values are corrected to arbitrarily set time = 0 as 1.0.
4. Discussion
Adhesion affects multiple biological processes and plays many vital roles in cell proliferation, viability, and differentiation. Over the past two decades, numerous experiments have shown that by controlling the nature and density of the adhesive ligand molecules and their spatial molecular patterning on the growth substrate of a cell, downstream signaling can be modulated and cell fate altered [1, 2, 30]. We recently demonstrated that the properties of the cell surface could be engineered in tandem with the scaffold material to create a binding interface with chemically compatible functional groups on the respective surfaces. In particular we exploited metabolic oligosaccharide engineering (see Scheme 1) to install thiol groups into the glycocalyx of human cells and demonstrated that the sialic acid displayed thiols could support adhesion with a gold substrate or undergo chemoselectively ligation reactions with maleimide groups displayed on a flat surface. The former situation, where human embryonic cells were treated with Ac5ManNTGc and grown on gold demonstrated that neural differentiation could be influenced by this method while the latter experiment demonstrated the rapid attachment of normally non-adhesive Jurkat cells to a maleimided glass surface [10].
Our previous work with Jurkat cells – while demonstrating that metabolically engineered carbohydrate-based adhesion could be exploited to attach these normally non-adhesive cancer cells to a surface – shed no light on whether this method could modulate cancer cell biology (because the experiment was discontinued immediately after binding). In the current work we address the long term impact on cell fate by first considering that, although traditional cell culture almost always employs 2D surfaces for reasons of convenience and accessibility, three-dimensional (3D) culture of cancer cells has long been advocated as a better model of tumorigenicity in vivo [31]. Culturing cells in 3D radically alters the mechanical transduction compared to 2D systems, thus affecting cell receptor ligation, intercellular signaling and cellular migration [5]. A 3D environment also influences the diffusion and adhesion of proteins, growth factors, and enzymes, which ensures cell viability and can influence function [32].
For these reasons, in this study we moved cell culture conditions used in MOE experiments from 2D surfaces to 3D electrospun fibrous scaffolds that were designed to have a high surface to volume ratio, an interconnected pore structure, and to resemble the extra-cellular matrix present in our body. In the current experiments relatively few cells (e.g., always ≤ 25%, usually many fewer) attached to 2D planar surfaces under any condition; nevertheless, the numbers were substantially higher than for tissue culture plastic or unmodified glass where virtually no (e.g., ≤ 1%) cells attached. Interestingly, even in the absence of Ac5ManNTGc and a complementary growth substrate (e.g., maleimide or gold) the 3D scaffolds resulted in dramatically increased cell adhesion with as many as 66% of the cells attached. One explanation for the enhanced adhesion is the increased surface area available for the cells on the 3D scaffold. However, because the cells are sparsely located on the 2D films, there is no lack of “space” available for attachment and this argument is not convincing.
Another explanation for the enhanced cell adhesion in a 3D environment is that the cells have an ability to sense the nano- to micron-scale 3D topographical features of their microenvironment and become adhesive in the presence of ECM-like structures. Although the mechanism remains unknown, speculatively the enhanced responses observed in the 3D environment could result from functional motifs on the nanofiber scaffold that surround the cell in multiple dimensions thus allowing cells to receive external stimuli in novel ways compared to when they are in contact with a 2D surface. To elaborate briefly, highly sialylated species such as CD43 have been described to cluster to one pole of a Jurkat cell [33], therefore this dominant species could (or could not) be primarily responsible for adhesion to a 2D surface. However, in a 3D construct, both a dominant species (e.g., CD43) – as well as some other minor sialylated species situated on a different face of the cell – could be simultaneously engaged in cell adhesion, thereby triggering complementary signaling pathways that lead to cell outcomes not available if only one type of adhesion occurred.
Bringing the contributions of MOE into the picture where cells displayed thiolated sialic acids on their surfaces, the number of cells (~66%) attached to 3D fibrous matrices in the absence of Ac5ManNTGc or complementary surface chemistry increased (e.g., up to 81%) when the cells were incubated on scaffolds with maleimide groups or gold coating. The modest – but measurable – increase in adhesion of Ac5ManNTGc-treated cells to maleimided or gold-coated 3D nanofibers provided evidence that the normal adhesion mechanism of these cells was augmented by “glycoengineered” carbohydrate-based adhesion interactions. This type of adhesion exemplifies chemoselective bio-orthogonal interactions that can occur amid many competing chemical interactions (as reviewed elsewhere [34]) and overcome potential problems such as “fouling” of the material surface by non-specific deposition of serum proteins. The ability of the cells to detect and react to surface features of the 3D scaffolds by virtue of the chemical feature of their surfaces – as evidenced by the different responses to Ac5ManNTGc-treated cells to each of the unmodified PES, amidated, malemided, and gold coated scaffolds (e.g., see Fig. 4) – dramatically illustrates how cell fate can be controlled by not only engineering the growth substrate of cells but also the cell surface itself.
An important outcome of these experiments was that there were no significant morphological differences observed for cells that attached to any chemically-modified 2D surface with or without Ac5ManNTGc. Similarly, only negligible changes were observed in the endpoints evaluated in this study for cells that attached to any 3D fiber when incubated in the absence of sugar analog. By contrast, distinctive changes in cellular morphology and spreading were only observed by seeding Ac5ManNTGc treated cells onto the 3D fibrous scaffolds with complementary chemical groups (i.e., or maleimide or gold) capable of forming high affinity interactions with the cell surface thiols. These observations illustrate how the full repertoire of cell responses requires not only 3D topography, but also the chemical engineering of both sides of the binding interface (instead of just modification of the growth substrate, as has been done traditionally). The complementary surfaces facilitated thiol-expression cells adhesion, which mediate outside-in signaling and lead to assembly of focal adhesion contacts, exertion of contractile forces, and activation of signaling pathways that regulate cell phenotype.
By comparing the gold and maleimide functionalized surfaces, it showed that the metal enhanced the morphological changes. One possibility is that the coordinate-covalent bonds made between thiols and gold are fundamentally different than the covalent adducts made between thiols and maleimide. A second (and non-exclusive) possibility is that the relatively sparse display of thiol groups on the cells surface (if the ~2.0 × 106 accessible thiol groups were evenly distributed on the cell surface they would be ~15 nm apart) means that they are not in register with the similarly-spaced maleimide groups positioned on the scaffolds. As a consequence, only a relatively small proportion of thiol-derivatized sialic acids can form covalent bonds to the malemided surface. By contrast, steric and spatial factors allow each and every cell surface thiol that contacts a gold surface to bind due to the high adhesive capability of this surface compared to the relatively sparsely placed malemide groups.
5. Conclusion
This study showed that cell adhesion could be enhanced, gene expression modulated, and cell fate controlled by introducing the 3D topographical cues into the growth substrate while simultaneously altering the chemistry of the cell surface via MOE to form non-natural, high affinity adhesion interactions with the scaffold. First, we found that the attachment of normally non-adhesive Jurkat cells to 3D fibrous scaffolds designed and optimized to have a highly porous structure with interconnected spaces to encourage cell migration and infiltration was significantly better than to a 2D planar surface. Next, by modulating the chemistry of the cell surface by using MOE to incorporate thiol functional groups into the glycocalyx of living cells via the sugar analog Ac5ManNTGc, cell attachment was further enhanced ~15% on the 3D fibrous scaffolds. Significantly, the carbohydrate-mediated adhesion that led to this increased cell attachment was accompanied by unique cellular morphology and time-dependent changes in genes expression that indicated that signaling pathways were engaged by the interaction of the thiolated sialosides with the gold substrate. The complex interplay between altered cell-surface glycosylation and the composition of custom-designed growth substrates adds another dimension to control cell fates via interactions with their microenvironment and promises to be valuable for tissue engineering, where cell attachment to matrix materials remains a challenge.
Acknowledgements
Funding for this study was primarily provided by the National Institute for Biomedical Imaging and Bioengineering (EB 005692-04) except for ManNAc analogue synthesis, which was supported by the National Cancer Institute (CA112314-05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DIC, Whitesides GM, et al. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 2.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 3.Whitesides GM, Kriebel JK, Love JC. Molecular engineering of surfaces using self-assembled monolayers. Sci Prog. 2005;88:17–48. doi: 10.3184/003685005783238462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 5.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 6.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. Biosynthesis of a nonphysiological sialic acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J Biol Chem. 1992;267:16934–16938. [PubMed] [Google Scholar]
- 7.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 8.Campbell CT, Sampathkumar S-G, Weier C, Yarema KJ. Metabolic oligosaccharide engineering: perspectives, applications, and future directions. Mol Biosys. 2007;3:187–194. doi: 10.1039/b614939c. [DOI] [PubMed] [Google Scholar]
- 9.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VD, Yarema KJ. Metabolic glycoengineering: Sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampathkumar S-G, Li AV, Jones MB, Sun Z, Yarema KJ. Metabolic installation of thiols into sialic acid modulates adhesion and stem cell biology. Nat Chem Biol. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 11.Du J, Yarema KJ. Carbohydrate engineered cells for regenerative medicine. Adv Drug Deliv Rev. 2010;62:271–282. doi: 10.1016/j.addr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sampathkumar S-G, Jones MB, Yarema KJ. Metabolic expression of thiol-derivatized sialic acids on the cell surface and their quantitative estimation by flow cytometry. Nat Protoc. 2006;1:1840–1851. doi: 10.1038/nprot.2006.252. [DOI] [PubMed] [Google Scholar]
- 13.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, et al. Engineering tumors with 3D scaffolds. Nat Meth. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 14.Hutmacher DW, Horch RE, Loessner D, Rizzi S, Sieh S, Reichert JC, et al. Translating tissue engineering technology platforms into cancer research. J Cell Mol Med. 2009;13:1417–1427. doi: 10.1111/j.1582-4934.2009.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhimana HK, Raya AR, Panda AK. Three-dimensional chitosan scaffold-based MCF-7 cell culture for the determination of the cytotoxicity of tamoxifen. Biomaterials. 2005;26:979–986. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Jones MB, Teng H, Rhee JK, Baskaran G, Lahar N, Yarema KJ. Characterization of the cellular uptake and metabolic conversion of acetylated N-acetylmannosamine (ManNAc) analogues to sialic acids. Biotechnol Bioeng. 2004;85:394–405. doi: 10.1002/bit.10901. [DOI] [PubMed] [Google Scholar]
- 17.Kim EJ, Jones MB, Rhee JK, Sampathkumar S-G, Yarema KJ. Establishment of N-acetylmannosamine (ManNAc) analogue-resistant cell lines as improved hosts for sialic acid engineering applications. Biotechnol Prog. 2004;20:1674–1682. doi: 10.1021/bp049841q. [DOI] [PubMed] [Google Scholar]
- 18.Kim EJ, Sampathkumar S-G, Jones MB, Rhee JK, Baskaran G, Yarema KJ. Characterization of the metabolic flux and apoptotic effects of O-hydroxyl- and N-acetylmannosamine (ManNAc) analogs in Jurkat (human T-lymphoma-derived) cells. J Biol Chem. 2004;279:18342–18352. doi: 10.1074/jbc.M400205200. [DOI] [PubMed] [Google Scholar]
- 19.Sampathkumar S-G, Jones MB, Meledeo MA, Campbell CT, Choi SS, Hida K, et al. Targeting glycosylation pathways and the cell cycle: sugar-dependent activity of butyrate-carbohydrate cancer prodrugs. Chem Biol. 2006;13:1265–1275. doi: 10.1016/j.chembiol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Sampathkumar S-G, Li AV, Yarema KJ. Synthesis of non-natural ManNAc analogs for the expression of thiols on cell surface sialic acids. Nat Protoc. 2006;1:2377–2385. doi: 10.1038/nprot.2006.319. [DOI] [PubMed] [Google Scholar]
- 21.Chua KN, Chai C, Lee PC, Tang YN, Ramakrishna S, Leong KW, et al. Surface-aminated electrospun nanofibers enhance adhesion and expansion of human umbilical cord blood hematopoietic stem/progenitor cells. Biomaterials. 2006;27:6043–6051. doi: 10.1016/j.biomaterials.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Kakabakos SE, Tyllianakis PE, Evangelatos GP, Ithakissios DS. Colorimetric determination of reactive solid-supported primary and secondary amino-groups. Biomaterials. 1994;15:289–297. doi: 10.1016/0142-9612(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 23.Isaev RN, Ishkov AV. Photometric determination of dimaleimides. J Anal Chem. 2003;58:435–438. [Google Scholar]
- 24.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Sun Z, Li AV, Yarema KJ. Roles for GNE outside of sialic acid biosynthesis: modulation of sialyltransferase and BiP expression, GM3 and GD3 biosynthesis, proliferation and apoptosis, and ERK1/2 phosphorylation. J Biol Chem. 2006;281:27016–27028. doi: 10.1074/jbc.M604903200. [DOI] [PubMed] [Google Scholar]
- 27.Lin K, Chua KN, Christopherson GT, Lim S, Mao HQ. Reducing electrospun nanofiber diameter and variability using cationic amphiphiles. Polymer. 2007;48:6384–6394. [Google Scholar]
- 28.Zhang YZ, Ouyang HW, Lim CT, Ramakrishna S, Huang ZM. Electrospinning of gelatin fibers and gelatin/PCL composite fibrous scaffolds. J Biomed Mat Res B-Appl Biomat. 2005;72B:156–165. doi: 10.1002/jbm.b.30128. [DOI] [PubMed] [Google Scholar]
- 29.Pham QP, Sharma U, Mikos AG. Electrospun poly(epsilon-caprolactone) microfiber and multilayer nanofiber/microfiber scaffolds: Characterization of scaffolds and measurement of cellular infiltration. Biomacromolecules. 2006;7:2796–2805. doi: 10.1021/bm060680j. [DOI] [PubMed] [Google Scholar]
- 30.Lim SH, Liu XY, Song HJ, Yarema KJ, Mao HQ. The effect of nanofiber-guided cell alignment on the preferential differentiation of neural stem cells. Biomaterials. 2010;31:9031–9039. doi: 10.1016/j.biomaterials.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YJ, Bae HI, Kwon OK, Choi MS. Three-dimensional gastric cancer cell culture using nanofiber scaffold for chemosensitivity test. Int J Biol Macromol. 2009;45:65–71. doi: 10.1016/j.ijbiomac.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Stegemann JP, Nerem RM. Altered response of vascular smooth muscle cells to exogenous biochemical stimulation in two- and three-dimensional culture. Exp Cell Res. 2008;283:146–155. doi: 10.1016/s0014-4827(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 33.Eda S, Yamanaka M, Beppu M. Carbohydrate-mediated phagocytic recognition of early apoptotic cells undergoing transient capping of CD43 glycoprotein. J Biol Chem. 2004;279:5967–5974. doi: 10.1074/jbc.M310805200. [DOI] [PubMed] [Google Scholar]
- 34.Lemieux GA, Bertozzi CR. Chemoselective ligation reactions with proteins, oligosaccharides and cells. Trends Biotechnol. 1998;16:506–513. doi: 10.1016/s0167-7799(98)01230-x. [DOI] [PubMed] [Google Scholar]