Abstract
We describe a rapid method to probe for mutations in cell-surface ligand-binding proteins that affect the environment of bound ligand. The method uses fluorescence activated cell sorting to screen randomly-mutated receptors for substitutions that alter the fluorescence emission spectrum of environmentally-sensitive fluorescent ligands. When applied to the yeast α-factor receptor Ste2p, a G protein coupled receptor, the procedure identified 22 substitutions that red-shift the emission of a fluorescent agonist, including substitutions at residues previously implicated in ligand binding and at additional sites. A separate set of substitutions, identified in a screen for mutations that alter the emission of a fluorescent α-factor antagonist, occurs at sites that are unlikely to directly contact the ligand. Instead, they alter receptor conformation to increase ligand-binding affinity and provide signaling in response to antagonists of normal receptors. These results suggest that receptor-agonist interactions involve at least two sites, of which only one is specific for the receptor’s activated conformation.
Keywords: receptor activation, ligand binding, flow cytometry, G protein signaling, fluorescent ligands
Introduction
G-protein coupled receptors (GPCRs) are transmembrane proteins that transduce a wide variety of extracellular stimuli into intracellular responses via their interactions with the heterotrimeric guanine nucleotide-binding regulatory G proteins 1. For most GPCRs, signal transduction is initiated by binding of a specific ligand. However, additional classes of ligands are known to inhibit receptor activation. Despite the recent solution of several crystal structures of GPCRs 2; 3; 4; 5; 6; 7; 8}, the molecular mechanisms controlling activation of GPCRs remain unclear. Two prerequisites for achieving a mechanistic understanding of the basis for ligand-receptor interactions are: 1) identification of the amino acid residues of a receptor that interact with ligands; and 2) understanding differences in interactions between different ligands with different effects on receptor activation, such as agonists and antagonists. For some receptors, such as the well-studied β-adrenergic receptors, extensive analyses of the interactions of diverse ligands with normal and mutated receptors 9 and recent X-ray structures 2; 5; 6 have allowed identification of specific groups on the receptor that contact bound ligands. However, the identities of ligand-interacting elements for other classes of receptors, such as those for peptide hormones, remain less well-characterized. Furthermore, the molecular basis for the differential interactions of different classes of ligands, such as agonists and antagonists, on GPCRs remain poorly understood.
The α-mating pheromone receptor (Ste2p) of the yeast Saccharomyces cerevisiae is a GPCR that is activated by binding the 13-residue peptide α-factor (sequence: WHWLQLKPGQPMY). While Ste2p exhibits little sequence similarity to mammalian GPCRs, it is functionally interchangeable with some mammalian receptors 10; 11. Several residues in the receptor that interact with ligand have been identified by chemical crosslinking and by characterizing amino acid substitutions in the ligand and receptor 12; 13; 14; 15; 16; 17; 18. However, given the size of α-factor, it is likely that additional residues in Ste2p interact with bound peptide.
The identification of amino acid residues involved in ligand binding can be laborious. Chemical crosslinking approaches require the use of functionalized ligand analogs that retain specific binding and biological activity. Crosslinked products must be fragmented and characterized, either at low resolution, by gel electrophoresis, or at higher resolution by mass spectrometry. Genetic approaches require verification to confirm that mutations that lead to loss of binding or activation result from specific alterations in the binding interface and not from defects in synthesis, folding, intracellular trafficking, or stability of the mutant receptors. The difficulty of these approaches has generally impeded understanding the differential interactions of receptors with specific classes of ligands with different efficacies.
We describe here a new method for identifying amino acid residues of a receptor that, when mutated, result in a change in the chemical environment of a probe attached to bound ligand. This procedure recovers alleles that change the environment of the 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) fluorophore while maintaining high affinity binding to a membrane-impermeant ligand at the cell surface. Therefore, it automatically eliminates major classes of undesired loss-of-function alleles such as those affecting overall receptor folding or subcellular trafficking. The approach was developed based on the previously-characterized binding to Ste2p of the (NBD)-tagged agonist analog ([Lys7(NBD),Nle12]α-factor (see Table 1) 19; 20; 21 but is also applied to receptor-binding of fluorescent antagonist, providing a comparison of the interactions of receptors with the two types of ligands.
Table 1.
α-factor analogs
| Ligand type |
Ligand name | Sequence | Relative Bmax Ratioa |
|---|---|---|---|
| Agonist | α-factor | TrpHisTrpLeuGlnLeuLysProGlyGlnProNleTyr | 1 |
| [Dap3,Arg7,Nle12]α-factor | TrpHisDapLeuGlnLeuArgProGlyGlnProNleTyr | 0.48 ± 0.03 | |
| Antagonist | [des-Trp1,Ala3,Nle12]α-factor | HisAlaLeuGlnLeuLysProGlyGlnProNleTyr | 0.49 ± 0.03 |
| [dTyr3,Nle12]α-factor | TrpHis(dTyr)LeuGlnLeuLysProGlyGlnProNleTyr | 0.33 ± 0.02 | |
| [desTrp1,desHis2,Nle12]α-factor | TrpLeuGlnLeuLysProGlyGlnProNleTyr | 0.42 ± 0.05 |
Relative Bmax ratio is the ratio of FL1 emission to FL2 emission of the indicated ligand labeled with NBD at Lys7 when bound to normal Ste2p. The ratio is normalized to the Bmax ratio of [Lys7(NBD),Nle12]α-factor bound to normal Ste2p
Results
Mutations that affect the environment of bound agonist
[Lys7(NBD),Nle12]α-factor binds to Ste2p with an affinity similar to that for native unlabeled α-factor, and elicits similar activation of the pheromone response pathway 19; 20. Upon binding to receptor, the environmentally-sensitive NBD fluorophore of the ligand exhibits a large increase in quantum yield and a blue-shift in emission spectrum compared to its fluorescence in aqueous solution, indicative of transfer to a hydrophobic environment (Supplementary Fig. 1) 20; 21. This provides a rapid and unbiased way of detecting alterations in the receptor or ligand that affect the chemical environment of the fluorophore of bound ligand. For example substantial changes in emission spectrum have been detected upon mutation of Ste2p at residue D275 22 or alteration of the length of the side chain used to attach NBD to α-factor by as little as one CH2 group 21. This raised the possibility of using fluorescence activated cell sorting to screen a library of cells expressing randomly mutated receptors in order to identify mutations that affect the environment of bound ligand. Conducting the screen in the presence of sub-saturating concentrations of fluorescent ligand and selecting cells exhibiting high fluorescence intensities allows selective recovery of mutant receptors that retain high levels of expression and maintain the conformation required for high affinity ligand binding.
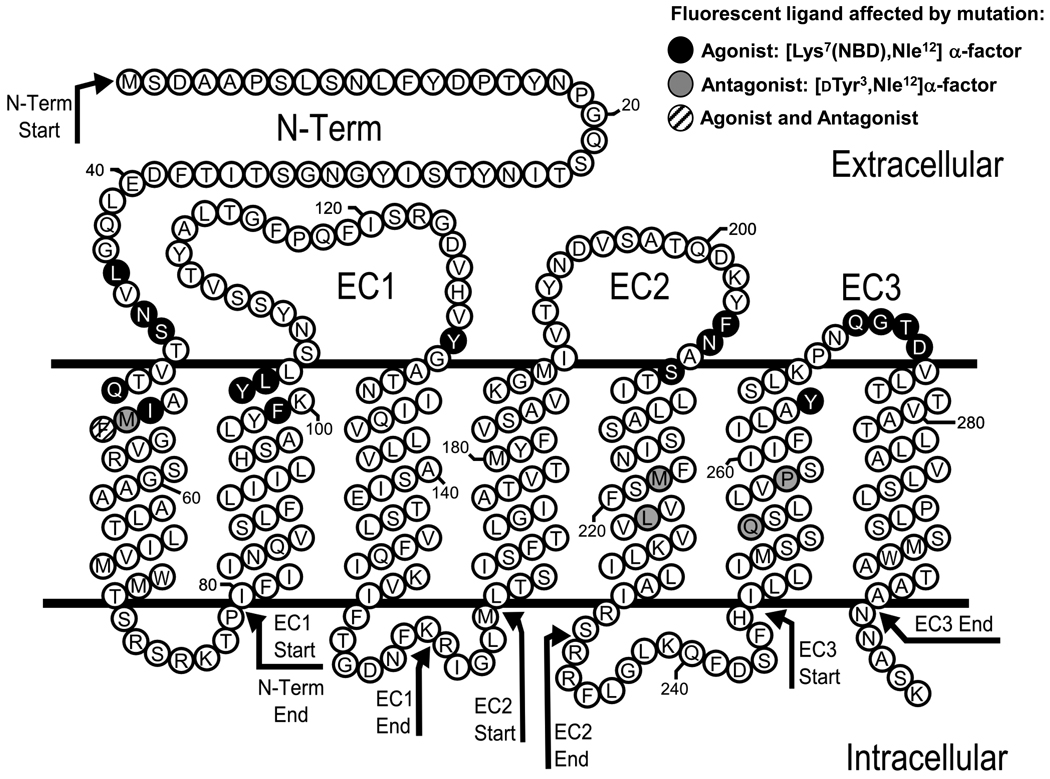
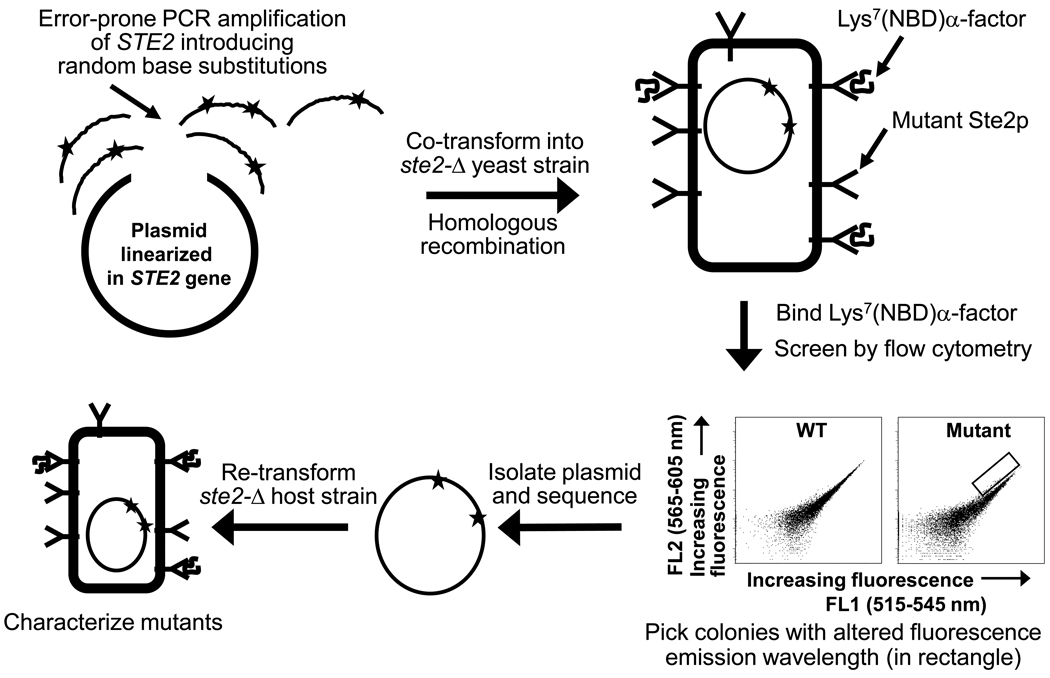
A library of genes encoding randomly mutated α-factor receptors was produced by error-prone PCR amplification of targeted regions of the STE2 gene followed by co-transformation of the PCR products and linearized vector into a ste2-Δ yeast host (see Fig. 1 and Fig. 2)23. The four regions targeted for mutagenesis each encompassed a single predicted extracellular region of the receptor as well as adjacent predicted transmembrane regions (see Supplementary Table 1; Fig. 1). Cells exhibiting altered emission spectra of bound [Lys7(NBD),Nle12]α-factor were identified based on comparisons of emission in two fluorescent channels of a flow cytometer, FL1 (515–545 nm) and FL2 (565–605 nm). Mutant populations of cells with decreased FL1/FL2 ratios could be detected by visual inspection of dot plots of FL1 vs. FL2 emission for the entire mutagenized population (Supplementary Fig. 2). No significant populations exhibiting increased FL1/FL2 ratios were detected. Following cell sorting to isolate clones exhibiting such altered ratios, receptor-encoding plasmids were sequenced. Individual substitutions were selected for further characterization based on meeting one of the following three criteria: 1) Recovery in multiple independent clones. Multiple alleles containing the same amino substitutions were recovered for mutations at 13 positions (Supplementary Table 2). Since these isolates generally also contained different additional mutations, it was clear that they originally arose from different PCR products. However, the range of possible mutations with this phenotype did not appear to be completely sampled, based on the failure to recover multiple isolates of many of the substitutions.; 2) Recovery as the only mutation in a particular clone; and 3) Recovery as a component of a double amino acid substitution in which neither mutation was recovered in any additional selected alleles. The screen led to identification of 22 amino acid substitutions at 17 different positions (Table 2) in Ste2p that, following re-transformation into a fresh yeast host, exhibited significant alterations in emission spectra compared to normal receptors. This represents about 50% of the individual substitutions selected for characterization.
Figure 1.
Predicted transmembrane topology of C-terminally truncated Ste2p. The black circles indicate residues that, when mutated, alter the FL1/FL2 ratio of fluorescence emission of [Lys7(NBD),Nle12]α-factor bound to C-terminally truncated Ste2p receptor. The gray circles indicate residues that, when mutated, alter the FL1/FL2 ratio of fluorescence emission of [dTyr3,Lys7(NBD),Nle12]α-factor bound to Ste2p receptor. The cross-hatched residue (F55) alters fluorescence ratio of both types of ligand. The arrows indicate the start- and end- points of the regions targeted for mutagenesis in the four screened libraries.
Figure 2.
Schematic view of the procedures for mutagenesis and screening. The abscissas and ordinates of the dot plots represent the fluorescence detected in the FL1 and FL2 channels of a flow cytometer. Each dot represents a cell.
Table 2.
Properties of receptors containing amino acid substitutions that affect the polarity of the environment of Ste2p-bound agonists [Lys7(NBD),Nle12]α-factor and [Dap3(NBD),Arg7,Nle12]α-factor.
| [Lys7(NBD),Nle12]α-factor | Unlabeled α-factor | [Dap3(NBD),Arg7,Nle12]α-factor | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fluorescent Ligand Binding | Receptor Function | Fluorescent Ligand Binding | ||||||||
| Library | Mutationa | Bmax Ratio (FL1/FL2) |
Relative Kd (FL1)b |
Relative Bmax (FL1)c |
Relative EC50d |
Relative FUS1-lacZ inductione |
Bmax Ratio (FL1/FL2) |
Relative Kd (FL1)b |
Relative Bmax (FL1)c |
Number of Isolatesf |
| STE2+ | 1 | 1 | 100 | 1 | 100 | 1 | 1 | 100 | ||
| N-Term | L44F | 0.6 ± 0.1 | 0.8 ± 0.1 | 71 ± 6 | 0.5 ± 0.4 | 140 ± 9 | 1.0 ± 0.04 | 0.9 ± 0.1 | 92 ± 2 | 1 |
| N46I | 0.7 ± 0.1 | 0.7 ± 0.1 | 33 ± 2 | 2.1 ± 1.4 | 96 ± 9 | 1.0 ± 0.06 | 0.6 ± 0.1 | 35 ± 1 | 1 | |
| S47R14 | 0.5 ± 0.03 | 0.9 ± 0.1 | 39 ± 2 | 1.1 ± 0.6 | 181 ± 18 | 1.0 ± 0.04 | 1.0 ± 0.1 | 79 ± 2 | 1 | |
| Q51H | 0.4 ± 0.04 | 1.9 ± 0.4 | 30 ± 2 | 1.4 ± 0.2 | 130 ± 10 | 0.9 ± 0.1 | 2.4 ± 0.4 | 60 ± 4 | 5 | |
| I53F | 0.6 ± 0.04 | 0.8 ± 0.1 | 50 ± 2 | 0.2 ± 0.1 | 152 ± 19 | 1.1 ± 0.04 | 0.6 ± 0.04 | 67 ± 2 | 2 | |
| F55S47;48 | 0.7 ± 0.1 | 1.4 ± 0.2 | 47 ± 3 | 1.1 ± 0.4 | 172 ± 11 | NDg | NDg | NDg | 1 | |
| EC1 | F99I48 | 0.7 ± 0.1 | 0.9 ± 0.3 | 69 ± 9 | 1.1 ± 0.6 | 192 ± 16 | 1.0 ± 0.1 | 0.9 ± 0.2 | 95 ± 6 | 1 |
| F99L48 | 0.6 ± 0.1 | 0.8 ± 0.1 | 51 ± 3 | 1.3 ± 0.5 | 116 ± 16 | 1.0 ± 0.1 | 0.6 ± 0.1 | 73 ± 6 | 6 | |
| F99S48 | 0.7 ± 0.1 | 0.8 ± 0.2 | 63 ± 6 | 0.4 ± 0.6 | 188 ± 22 | 1.1 ± 0.1 | 0.6 ± 0.1 | 111 ± 8 | 1 | |
| Y101C | 0.6 ± 0.1 | 0.9 ± 0.1 | 64 ± 7 | 1.0 ± 0.3 | 147 ± 17 | 1.0 ± 0.2 | 1.0 ± 0.4 | 42 ± 5 | 4 | |
| L102S48 | 0.8 ± 0.1 | 0.6 ± 0.04 | 28 ± 2 | 1.6 ± 1.2 | 115 ± 15 | 1.0 ± 0.2 | 0.7 ± 0.2 | 33 ± 4 | 1 | |
| Y128H | 0.8 ± 0.03 | 1.5 ± 0.1 | 62 ± 2 | 0.6 ± 0.3 | 121 ± 19 | NDg | NDg | 3 ± 0.3 | 1 | |
| EC2 | F204I49 | 0.7 ± 0.1 | 2.4 ± 0.6 | 19 ± 2 | 9.7 ± 2.7 | 157 ± 15 | 0.9 ± 0.1 | 1.8 ± 0.5 | 63 ± 6 | 1 |
| F204L49 | 0.7 ± 0.1 | 1.0 ± 0.2 | 11 ± 1 | 8.0 ± 1.7 | 159 ± 12 | 0.9 ± 0.1 | 1.1 ± 0.3 | 68 ± 6 | 2 | |
| N205H15;42 | 0.3 ± 0.03 | 1.1 ± 0.2 | 15 ± 1 | 10.1 ± 1.1 | 5 ± 0.4 | 1.0 ± 0.1 | 0.8 ± 0.2 | 162 ± 11 | 1 | |
| N205Y15;42 | 0.3 ± 0.04 | 1.2 ± 0.3 | 19 ± 2 | 10.9 ± 1.4 | 13 ± 2 | 1.0 ± 0.1 | 0.8 ± 0.1 | 22 ± 1 | 1 | |
| S207G | 0.8 ± 0.1 | 0.8 ± 0.1 | 67 ± 3 | 0.7 ± 0.6 | 92 ± 13 | 1.1 ± 0.2 | 0.8 ± 0.4 | 37 ± 6 | 3 | |
| EC3 | Y266C15;42 | 0.3 ± 0.04 | 0.9 ± 0.2 | 8 ± 1 | 14.6 ± 4.9 | 107 ± 7 | 1.0 ± 0.1 | 0.8 ± 0.1 | 54 ± 3 | NFSh |
| Q272P | 0.3 ± 0.1 | 1.0 ± 0.2 | 22 ± 2 | 0.6 ± 0.3 | 145 ± 11 | 1.0 ± 0.1 | 0.8 ± 0.2 | 133 ± 9 | 1 | |
| G273S | 0.5 ± 0.1 | 1.0 ± 0.2 | 42 ± 4 | 1.2 ± 0.3 | 160 ± 6 | 1.0 ± 0.1 | 0.4 ± 0.1 | 135 ± 6 | 4 | |
| T274I | 0.7 ± 0.1 | 1.0 ± 0.3 | 71 ± 5 | 0.5 ± 0.4 | 113 ± 8 | 1.0 ± 0.1 | 0.5 ± 0.1 | 71 ± 4 | 1 | |
| D275A22 | 0.3 ± 0.1 | 0.7 ± 0.2 | 5 ± 1 | 1.6 ± 0.4 | 156 ± 19 | 0.9 ± −0.1 | 0.4 ± 0.1 | 51 ± 3 | NFSh | |
| D275E22 | 0.3 ± 0.02 | 0.7 ± 0.1 | 12 ± 0 | 2.1 ± 1.0 | 173 ± 9 | 1.0 ± 01 | 0.8 ± 0.1 | 198 ± 7 | 10 | |
| D275G22 | 0.3 ± 0.03 | 1.3 ± 0.2 | 15 ± 1 | 1.3 ± 0.4 | 129 ± 19 | 1.0 ± 0.04 | 0.5 ± 0.04 | 79 ± 2 | 2 | |
superscripts are citations to previous studies that identified the same or different substitutions at the same position
computed as the ratio Kd(mutant)/Kd(wild type) (for [Lys7(NBD),Nle12]α-factor), where the absolute Kd of wild type receptor binding to [Lys7(NBD),Nle12]α-factor was 18 ± 1 nM (n = 10 triplicate assays) and the absolute Kd of wild type receptors binding to [Dap3(NBD),Arg7,Nle12]α-factor was 11 ± 1 nM (n = 11 triplicate assays).
computed as 100× Bmax(mutant)/Bmax(wild type) (for [Lys7(NBD),Nle12]α-factor)
computed as the ratio EC50(mutant)/EC50(wild type) (for [Lys7(NBD),Nle12]α-factor), where the absolute EC50 for activation of wild type receptors by α-factor was 10 ± 2 nM (n = 11 triplicate assays).
computed as 100× [maximal FUS1-lacZ induction(mutant)]/[maximal FUS1-lacZ induction(wild type)] (for [Lys7(NBD),Nle12]α-factor)
number of times independently isolated in the screen
not determined because of low levels of fluorescence
not from the screen
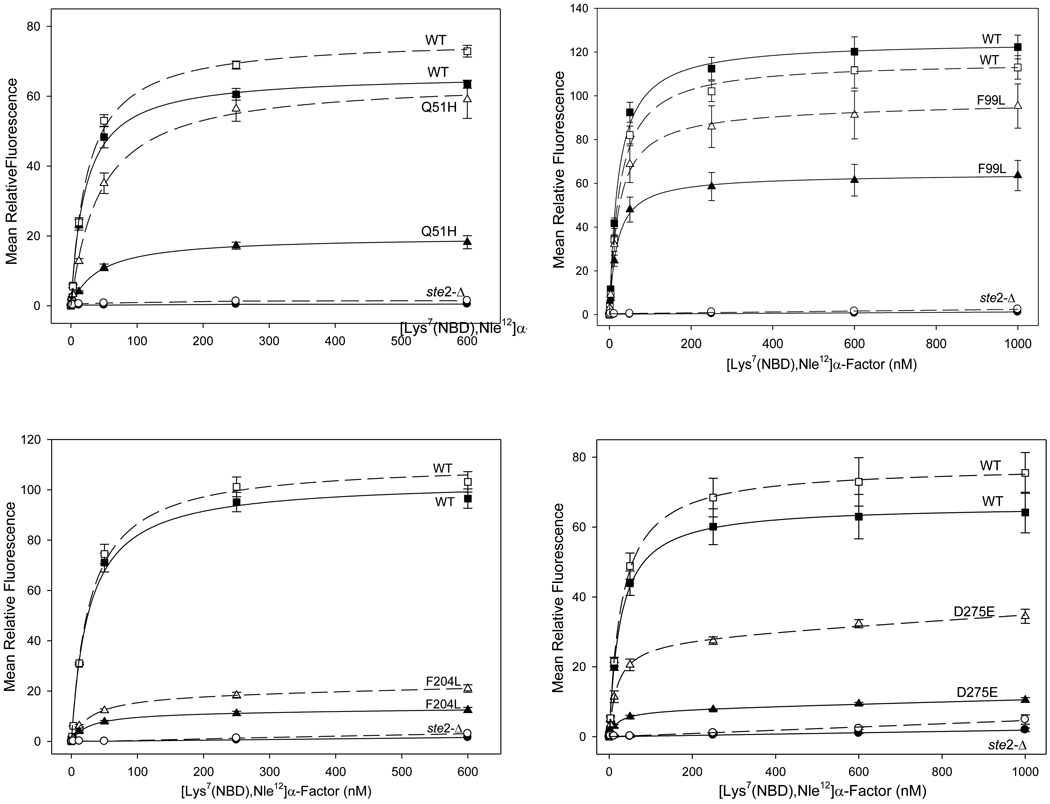
Consistent with the conditions of the screen, all of the recovered confirmed singly-substituted alleles maintained high affinity for ligand, exhibiting dissociation constants within 2.5-fold of the wild-type receptor as determined by flow cytometry-based fluorescence assays of ligand binding (Fig. 3, Table 2). In addition, most the recovered mutant receptors retained the capability for effective signal transduction based on their abilities to activate transcription of a FUS1-lacZ reporter gene in response to binding of α-factor (as indicated by an EC50 no more than ~2-fold higher than that for the wild-type receptor and a maximal response at least as great as that of the wild-type receptor (Supplementary Fig. 3; Table 2). No correction of these functional measurements for differences in receptor expression level was applied, as pheromone responsive signaling has been reported to be unaffected by up to 40-fold variations in receptor expression 24.
Figure 3.
[Lys7(NBD),Nle12]α-factor binding curves for selected Ste2p mutants from each of the four libraries. The closed symbols and the unbroken lines represent information obtained from the flow cytometer channel FL1. The open symbols and broken lines represent FL2.
To aid in evaluating the alleles derived from the screen, three classes of additional substitutions in Ste2p were created by site directed mutagenesis:
Y266C and D275A. These substitutions had previously been reported to interact with bound ligand 15; 16; 22; 25 but were not recovered from our screen. However, each of these substitutions resulted in large decreases in the FL1/FL2 ratios of bound [Lys7(NBD),Nle12]α-factor. Their low emission intensities, as indicated by low relative Bmax values (Table 2), would be expected to render these particular substitutions unlikely to be recovered from the fluorescence-based screen.
V49S, T50F, A52S, M54S, L268S, K269A, and P270S. These site-directed alterations of side chain hydrophobicity and size were introduced at sites adjacent to the positions of mutations from the screen that exhibited large shifts in fluorescence emission of bound agonist. The effects of these nearby alterations were examined in order to determine whether the fluorescence changes of the alleles derived from the screen reflect specific effects of the particular recovered substitutions at particular amino acids in the protein, or whether the fluorescence changes can arise from other substitutions in the same general region of the peptide chain. Despite their proximities to sites of mutations with strong effects on agonist fluorescence, the site-directed changes resulted in little or no shifting of fluorescence emission (Table 3). Several additional site-directed amino acid substitutions, Y98C, L277S, and T278A, that were created in the process of re-testing initial mutations recovered from the screen (Supplementary Table 2), also failed to cause any shifting of the emission of bound [Lys7(NBD),Nle12]α-factor (Table 3), despite the fact that they happen to reside at positions close to sites of mutations with strong effects on fluorescence.
F99, Y101, and Y128. Consistent with the conditions of the screen, substitutions at these sites that were recovered from the random library did not result in alteration of the affinity of Ste2p for ligand. Since these substitutions are located at the extracellular ends of the second and third transmembrane segments of Ste2p, regions that had not previously been implicated in ligand binding, we wished to determine whether altering the side chain hydrophobicity, size, and/or charge could alter the receptor’s interactions with α-factor. This was tested by creation of site directed substitutions at these sites. Several such substitutions, Y101R, Y128R, and Y128W, all significantly reduced the binding affinity of the receptor for [Lys7(NBD),Nle12]α-factor (Table 3), indicating that residues at these positions may be directly involved in contacts with α-factor. The site-directed substitution F99R resulted in a complete loss of detectable ligand binding, which suggests either that this residue directly interacts with the agonist or that the mutant receptor is expressed poorly on the cell surface.
Table 3.
Effects of site-directed substitutions in Ste2p on binding of agonist [Lys7(NBD),Nle12]α-factor.
| Mutation | Bmax Ratio (FL1/FL2) |
Rel Kda (FL1) |
Rel Bmaxb (FL1) |
|---|---|---|---|
| WT | 1.0 | 1.0 | 100 |
| Substitutions adjacent to mutations recovered in screen | |||
| V49S | 0.9 ± 0.1 | 0.8 ± 0.1 | 73 ± 5 |
| T50F | 0.8 ± 0.2 | 0.6 ± 0.2 | 74 ± 11 |
| A52S | 1.1 ± 0.3 | 1.1 ± 0.4 | 130 ± 21 |
| M54S | 0.9 ± 0.2 | 0.8 ± 0.2 | 76 ± 10 |
| Y98C | 1.1 ± 0.1 | 1.6 ± 0.3 | 60 ± 5 |
| L268S | 1.0 ± 0.2 | 0.2 ± 0.1 | 3.3 ± 0.4 |
| K269A | 1.0 ± 0.2 | 1.1 ± 0.4 | 138 ± 24 |
| P270S | 0.9 ± 0.2 | 1.0 ± 0.3 | 95 ± 14 |
| L277S | 0.9 ± 0.1 | 0.8 ± 0.1 | 67 ± 5 |
| T278A | 1.0 ± 0.2 | 1.0 ± 0.3 | 148 ± 18 |
| Alternative substitutions at F99, Y101, and Y128 | |||
| F99R | No detectable binding | ||
| F99W | 0.8 ± 0.1 | 0.7 ± 0.2 | 67 ± 5 |
| F99A | 0.7 ± 0.1 | 0.7 ± 0.1 | 71 ± 56 |
| Y101R | 0.9 ± 0.2 | 4.3 ± 0.8 | 46 ± 4 |
| Y101W | 0.9 ± 0.1 | 0.7 ± 0.1 | 47 ± 3 |
| Y101A | 0.6 ± 0.1 | 0.7 ± 0.1 | 57 ± 5 |
| Y128R | 0.8 ± 0.1 | 8.8 ± 1.5 | 56 ± 4 |
| Y128W | 0.6 ± 0.3 | 17.9 ± 4.4 | 51 ± 10 |
| Y128A | 1.0 ± 0.1 | 1.4 ± 0.3 | 79 ± 8 |
computed as the ratio Kd(mutant)/Kd(wild type) (for [Lys7(NBD),Nle12]α-factor)
computed as 100× Bmax(mutant)/Bmax(wild type) (for [Lys7(NBD),Nle12]α-factor)
To further examine the specificity of the effects of the amino acid substitutions recovered from the screen, we tested their effects on fluorescence of an alternative fluorescent ligand, [Dap3(NBD),Arg7,Nle12]α-factor, containing an NBD group attached to a 2,3 diaminopropionic acid residue replacing Trp3 of the peptide. This peptide has the highest Ste2p binding affinity of any NBD-labeled analog other than ones derivatized at Lys7 and exhibits approximately 30% the activity of native α-factor in growth arrest assays 21. Compared with [Lys7(NBD),Nle12]α-factor, the peak of fluorescence emission of free [Dap3(NBD),Arg7,Nle12]α-factor in buffer is blue-shifted (compare dashed lines in panels a) and c) of Supplementary Fig. 1). When analyzed by flow cytometry, the Dap3 analog bound to normal receptors with a approximately the same Kd as that of the Lys7-labeled agonist and an FL1/FL2 ratio that is 50% less than that of the bound Lys7-labeled agonist (Supplementary Fig. 4).
None of the mutations that alter the emission spectrum of [Lys7(NBD),Nle12]α-factor caused significant shifts in FL1/FL2 ratio of receptor bound [Dap3(NBD),Arg7,Nle12]α-factor, most likely because the wavelength of emission of NBD attached at position 3 is less environmentally sensitive than when it is at position 7 (Supplementary Fig. 1). The Dap3 derivative does, however, display a large increase in the emission intensity in a non-polar, compared to a polar, environment. In fact, several mutations that alter the emission wavelengths of [Lys7(NBD),Nle12]α-factor exhibited very different effects on the emission intensities of fluorescent agonists labeled at the two different positions. Two substitutions, F55S and Y128H, reduced the apparent Bmax values for binding of [Dap3(NBD),Arg7,Nle12]α-factor by more than 30-fold compared with the Bmax for normal receptors, whereas they result in only approximately 2-fold reductions in the apparent Bmax values for binding of [Lys7(NBD),Nle12]α-factor. Thus, these substitutions appear to specifically block binding of the Dap3-substituted ligand. In contrast, four alleles, N205H, Q272P, G273S, and D275E result in at least 30% increases in emission of bound [Dap3(NBD),Arg7,Nle12] compared with wild-type receptors, indicative of a decrease in the polarity of the bound Dap3-labeled fluorophore, despite the fact that these same mutations result in decreased Bmax values for binding of the [Lys7(NBD),Nle12]α-factor. The other mutations in Table 2 exhibit differences in emission intensities for the two different agonists that are more subtle, apparently reflecting differences in the changes in polarity at positions 3 and 7 of the ligand, as well as varying numbers of mutant receptors at the cell surface.
Mutations that affect the environment of bound antagonist
Several analogs of α-factor with truncations or alterations of residues in the N-terminal end of the peptide behave as antagonists toward normal α-factor receptors 26; 27; 28. In addition, we report here the use of a new antagonist, [dTyr3,Lys7(NBD),Nle12]α-factor, that binds to normal Ste2p with an affinity similar to that of normal α-factor, while exhibiting minimal activation of signaling (Supplementary Fig. 5). We refer to these compounds as “antagonists”, despite the fact that they exhibit very weak partial agonist activity toward normal receptors and substantial agonist activity toward some mutant receptor alleles, as discussed below.
When bound to wild-type receptors, analogs of three antagonists, all modified with NBD at Lys7, [des-Trp1,des-His2, Nle12]α-factor, [des-Trp1Ala3, Nle12]α-factor, and [dTyr3,Nle12]α-factor, all exhibit significant red-shifts compared to the Lys7-labeled agonist, indicating that their NBD groups reside in more polar environments than in the similarly labeled agonist (see Table 1). These red-shifts must reflect the fluorophores’ environments when the antagonists are bound to the receptor, since the unbound antagonists all exhibit essentially identical emission spectra to similarly labeled agonist in buffer or 2-propanol (Supplementary Fig. 1). Further evidence for differences in the binding modes of the agonists and antagonists was provided by examining binding of dTyr3[Lys7(NBD),Nle12]α-factor to a number of the mutations that exhibited strong effects on agonist emission (Table 2). None of the tested mutations resulted in any shifts in the emission of bound [dTyr3,Lys7(NBD),Nle12]α-factor (Supplementary Table 3).
To identify mutations that affect the emission of receptor-bound labeled antagonist, we conducted flow cytometry-based screens of randomly mutagenized receptors incubated in the presence of the antagonist [dTyr3,Lys7(NBD),Nle12]α-factor. These screens failed to detect any significant population of cells expressing mutant receptors with emission spectra that were red-shifted compared with the already-red-shifted emission of the antagonist bound to normal receptors. However, we recovered seven different mutant alleles that exhibited blue-shifted spectra (see Table 4). Three of these involved residues Q253 and P258, sites of the two strongest known constitutively activating mutations in Ste2p 29; 30 and re-testing of the recovered alleles confirmed ligand-independent activation of the FUS1-lacZ reporter. P258L, the strongest previously-described constitutively activating mutation of STE2, was not recovered from the current screen, most likely because of low levels of expression at the cell surface 30. However, site-directed introduction of the P258L substitution did result in slight blue-shifting of the emission of receptor-bound antagonist (Table 4). M218K, an additional mutation that shifted antagonist emission, also resulted in constitutive activation of the FUS1-lacZ reporter, despite the fact that it had not been recovered by previous screens for activating mutations.
Table 4.
Binding, emission, and pheromone response of the antagonist [dTyr3,Lys7(NBD),Nle12]α-factor in cells expressing mutant α-factor receptors.
| Fluorescent Ligand Binding | Receptor Function | |||||||
|---|---|---|---|---|---|---|---|---|
| Library | Mutation | Relative Bmax Ratioa (FL1/FL2) [dTyr3,Lys7 (NBD),Nle12] |
FL1 | Relative EC50d [dTyr3,Lys7 (NBD),Nle12] |
Relative Max FUS1-lacZ Inductione [dTyr3,Lys7 (NBD),Nle12] |
Relative Constitutive Activity |
Number of Isolatesf |
|
| Relative Kdb [dTyr3,Lys7 (NBD),Nle12]] |
Relative Bmaxc [dTyr3,Lys7 (NBD),Nle12] |
|||||||
| WT | 0.3a | 1 | 100 | 1 | 1 | 1 | − | |
| N-Term | M54I | 0.4 ± 0.06 | 0.6 ± 0.1 | 129 ± 14 | 0.3 ± 0.003 | 13 ± 0.7 | 1.4 ± 0.3 | 2 |
| F55L | 0.5 ± 0.1 | 2.2 ± 0.6 | 24 ± 3 | 1.6 ± 0.01 | 3 ± 0.5 | 0.9 ± 0.1 | 2 | |
| EC2 | M218K | 0.7 ± 0.04 | 0.4 ± 0.04 | 161 ± 6 | 0.03 ± 0.003 | 19 ± 4 | 3.7 ± 1.1 | 3* |
| L222R | 0.6 ± 0.03 | 0.5 ± 0.04 | 87 ± 3 | 0.2 ± 0.01 | 14 ± 2 | 2.1 ± 0.9 | 1 | |
| EC3 | Q253L | 0.5 ± 0.06 | 0.4 ± 0.1 | 141 ± 11 | 0.2 ± 0.004 | 10 ± 2 | 14 ± 3 | 4 |
| P258L | 0.4 ± 0.07 | 0.04 ± 0.11 | 4 ± 0.5 | N.D.f | N.D.f | 34 ± 10 | NFSg | |
| P258S | 0.9 ± 0.1 | 0.4 ± 0.2 | 56 ± 1 | 0.3 ± 0.003 | 20 ± 4 | 18 ± 2 | 1 | |
| P258T | 0.7 ± 0.1 | 0.3 ± 0.1 | 74 ± 7 | N.D.h | N.D.f | 15 ± 2 | 1 | |
| Double Mutants | P258S D275E | 0.3 ± 0.04 | 0.4 ± 0.1 | 41 ± 3 | 0.05 ± 0.002 | 8 ± 2 | 26 ± 6 | NFSg |
| Q51H P258S | 0.3 ± 0.03 | 0.4 ± 0.1 | 21 ± 2 | 0.1 ± 0.001 | 6 ± 1 | 15 ± 3 | NFSg | |
the reported Bmax ratio is normalized to that of the agonist [Lys7(NBD),Nle12]α-factor binding to normal receptors. (Note that all other results in this table are normalized to the values for normal receptor interacting with antagonist [DTyr3,Lys7(NBD),Nle12]α-factor.
computed as the ratio Kd(mutant)/Kd(wild type) or EC50(mutant)/EC50(wild type) for [dTyr3,Lys7(NBD),Nle12]α-factor where the absolute Kd for binding of wild type receptors to [DTyr3,Lys7(NBD),Nle12]α-factor) was 23 ± 1 nM (n = 13 triplicate assays).
computed as 100× Bmax(mutant)/Bmax(wild type) for the ligand [dTyr3,Lys7(NBD),Nle12]α-factor.
computed as 100× [maximal FUS1-lacZ induction(mutant)]/[maximal FUS1-lacZ induction(wild type)]for the ligand [dTyr3,Lys7(NBD),Nle12]α-factor.
computed as the ratio EC50(mutant)/EC50(wild type) for [dTyr3,Lys7(NBD),Nle12]α-factor where the absolute EC50 for activation of wild type receptors by [DTyr3,Lys7(NBD),Nle12]α-factor) was 420 ± 140 nM (n = 6 triplicate assays).
number of times independently isolated in screen
not from screen
not determined
Mutations that constitutively activate Ste2p were previously shown to simultaneously confer the ability to activate receptor signaling responses upon binding the ligand [des-Trp1Ala3]α-factor, which behaves as an antagonist toward normal receptors 30. Consistent with this, the constitutive mutations recovered from the current screen also can be activated by antagonists such as [dTyr3,Lys7(NBD),Nle12]α-factor (Table 4). Three additional mutations, recovered from the screen, M54I, F55L, and L222R, also confer the ability to be activated by antagonists without exhibiting any detectable constitutive signaling. None of the mutations recovered from the antagonist screen had any effect on the emission spectrum of bound agonist. Most of the mutations that result in altered fluorescence of bound antagonist also increase the binding affinity for both antagonist and agonist (Table 4 and Supplementary Table 4), despite their lack of effects on the emission spectrum of bound agonist. The only mutation from the antagonist screen that failed to enhance affinity for antagonist was F55L, which did, however, exhibit higher affinity for agonist. To further analyze the role of F55, we examined the binding of antagonist to receptors containing the substitution, F55S, recovered from the agonist screen. Substitution of serine at this position reduces the binding of the antagonist [dTyr3,Lys7(NBD),Nle12]α-factor to undetectable levels (Supplementary table 3), even though binding of agonist to this allele was nearly normal (Table 2).
Discussion
We have developed and tested a rapid protocol for identifying amino acid residues that, when altered, affect the chemical environment of a ligand bound to a receptor. The method uses fluorescence activated cell sorting to screen libraries of randomly mutated receptors for mutations that alter the emission wavelengths of bound fluorescent ligand analogs. It is selective for mutations that preserve high affinity ligand binding at the cell surface, thereby minimizing or eliminating confounding classes of mutations such as those that alter yields of receptor synthesis, folding, or subcellular targeting. The usefulness of the approach was evaluated in two separate screens focusing on the different interactions of the yeast α-pheromone receptor with the bound agonist and antagonist.
Mutations affecting the fluorescence emission of receptor-bound agonist all led to red-shifting of the spectrum, indicative of increased polarity of the fluorophore environment. They were all located at sites predicted to reside at the interface between the predicted transmembrane region of Ste2p and its extracellular surface. Several of the substitutions were at amino acid residues that had previously been identified as likely sites of interaction with α-factor, including: 1) S47R and F55S at the extracellular end of TM1. A previous genetic analysis implicated residues S47 and T48 of Ste2p in direct interactions with the Gln10 of α-factor 14. Furthermore, crosslinking has been observed between α-factor derivatized at Tyr13 and regions of Ste2p encompassing residues F55-R58 13 or F55-M6918 and between the unnatural amino acid p-benzoyl-L-phenylalanine incorporated at position 55 in Ste2p and α-factor 31. 2) F204I, F204L, N205H, and N205Y at the extracellular ends of TM5 and TM6. Mutational approaches and crosslinking have previously implicated residues F204, N205, and Y266 in interactions with the N-terminal region of the ligand 15; 16; 25; 32. Residue Y266 has been implicated in interactions with ligand 15; 16. Although mutations at Y266 were not recovered from the screen, presumably because of low levels of cell surface expression, the site-directed substitution at Y266C resulted in a significant emission shift of bound [Lys7(NBD),Nle12]α-factor. 3) D275E and D275G in the short extracellular loop. Mutations at D275 have previously been reported to affect ligand binding and receptor activation 22.
The screen also led to the recovery of mutations at particular residues that have not previously been implicated directly as sites of ligand interaction, even though they reside in known interacting regions. These sites include amino acids L44, N46, Q51, and I53, located in a region of TM1 in which other ligand-interacting residues have been identified. Similarly, mutations that we recovered at residues S207, Q272, G273, and T274 are located in the third extracellular loop, which mutagenic and crosslinking studies have previously implicated in interactions with ligand 15; 16; 17; 25; 32.
Additional mutations affecting the fluorescence of bound agonist were recovered at the extracellular ends of TM2 (at F99, Y101, and L102) and TM3 (at Y128) in the predicted first extracellular loop of Ste2p, which has not previously been identified as a site of contact with ligand. A previous cysteine-scanning study of residues 100–135 in the first extracellular loop of Ste2p uncovered no substitutions that affected ligand binding affinity 33, consistent with the binding data presented in Table 2 33; 34. Evidence the recovered mutations in the first extracellular loop alter the emission of bound [Lys7(NBD),Nle12]α-factor through direct interactions indicative proximity of these residues to ligand, is provided by the following observations: 1) In accordance with the screening paradigm, these substitutions do not significantly alter affinities for α-factor. In addition, most of them also do not alter receptor signaling function, making it unlikely that they alter receptor conformation. 2) We identified additional site-directed substitutions at positions F99, Y101, and Y128 that do, in fact, alter ligand binding (see Table 3). 3) The mutations in the first extracellular loop are the only substitutions recovered in the screen that are not located at, or near, previously-known sites of receptor-ligand interactions. 4) None of the sites of mutations recovered in the agonist screen reside at positions that are deep in the predicted transmembrane regions or at the intracellular surfaces of the receptor. If the mutations could induce changes in emission of NBD-labeled agonist via indirect effects (as was observed for the antagonist screen, see below) the recovered set of alleles would have been expected to include mutations at distant sites.
Three possible mechanisms for direct effects of mutations on the fluorescence of bound ligand all involve short distances between the altered residue and the ligand binding site: 1) Direct physical contact between the site of the mutation and the NBD-group of the bound ligand or its immediate site of attachment to ligand. Such contact could only occur if the mutated residue is within a few Å of the fluorophore or the linker. 2) Alteration of the electrostatic or dielectric environment of the bound fluorophores by substitutions at amino acid residues that are not in direct contact with fluorophore. The range of interactions capable of altering fluorescence emission of the bound ligand should be comparable to the range over which electrostatic effects are found to affect pKas of ionizable groups in proteins, generally less than about 10 Å 35; 36. 3) Alteration of the accessibility of the fluorophores to solvent. In structures of GPCRs in complex with small molecule ligands 2; 6; 7; 37; 38, the ligands are bound in cavities facing the extracellular surfaces of receptors. If this is also the case for peptide ligands, mutations affecting accessibility to solvent of groups on the ligand would be most likely to occur on surfaces lining these cavities.
The fluorescence changes observed on binding of [Lys7(NBD),Nle12]α-factor to receptors 19; 20, together with the fact that the screen was only able to recover mutations resulting in red-shifting of the NBD emission (indicative of increasing polarity) suggests that the side chain of Lys7 resides near a non-polar region when bound to wild-type Ste2p. Further indications of the polarity of the environments of different regions of bound ligand can be derived from comparisons of the behaviors of the two differentially labeled agonists. Mutations N205H, Q272P, G273S, and D275E in the predicted second and third extracellular loops of the receptor result in increased [Dap3(NBD),Arg7,Nle12]α-factor emission intensity compared with normal receptors (indicative of decreased polarity) whereas the same mutations cause decreased emission and red-shifting of bound [Lys7(NBD),Nle12]α-factor (indicative of increased polarity, perhaps accompanied by a decrease in the number of surface-exposed receptors). Together with additional differential interactions between particular mutated residues and the Dap3 and Lys7 positions on the ligand listed in Table 2, these observations support the existence of specific interactions between the substituted residues and the affected fluorophores and argue that the changes in emission are not due to mutation-induced global conformational changes in the receptor.
The environment of NBD in bound antagonists appears to differ significantly from that of NBD in bound agonist. Three different antagonists, all labeled with NBD at the equivalent of position Lys7 of normal α-factor, exhibit emission spectra upon binding to receptor that are considerably red-shifted (indicative of a shift to a polar environment) compared to the spectrum of similarly-labeled bound agonist (Table 1). Furthermore, in sharp contrast to the mutations affecting agonist fluorescence, the amino acid substitutions identified as causing changes in the fluorescence emission of receptor-bound antagonist all caused blue-shifting of the spectrum of NBD-labeled ligand, indicative of decreased polarity of the fluorophore environment. These changes in fluorescence appear to result from overall changes in the receptor conformation based on the following:
Most of the recovered mutations affecting [dTyr3,Lys7(NBD),Nle12]α-factor fluorescence were located at positions in the protein that are unlikely to interact directly with ligand, based on the predicted seven-transmembrane segment topology. This suggests that they are affecting fluorescence via a mechanism that does not involve direct contact with ligand.
None of the tested mutations that affect the emission of bound agonist had any significant effect on the emission spectrum of bound antagonist (Supplementary Table 3). Thus, either the NBD group of receptor-bound antagonist [dTyr3,Lys7(NBD),Nle12]α-factor does not interact with the regions of the receptor that appear to be in close proximity with agonist, or the NBD group of the antagonist resides in a highly solvent-exposed environment that can not be directly altered by mutations in the receptor.
None of the mutations that alter the emission spectrum of bound Lys7-labeled antagonist had any effect on the fluorophore of receptor-bound Lys7-labeled agonist (Supplementary Table 4). This implies either that these mutation-induced conformational changes do not alter the environment of bound agonist, or that these mutations do not alter the conformation of agonist-bound states.
All of the recovered mutations that alter the emission spectrum of receptor-bound antagonist [dTyr3,Lys7(NBD),Nle12]α-factor also confer on the mutant receptors the ability to signal in response to binding of ligands that act as antagonists of normal receptors (Table 4). In addition, most of the mutations affecting the [dTyr3,Lys7(NBD),Nle12]α-factor fluorescence also conferred constitutive signaling activity on the receptor. In contrast, none of the mutations affecting the emission of bound agonist resulted in significant (greater than 2-fold) constitutive activation of signaling with the exception of the I53F substitution, which exhibited an activation of 3.1 (± 0.8)-fold over normal receptors.
All but one of the mutations that alter the fluorescence emission of bound antagonist α-factor also resulted in enhanced binding affinity for antagonist (Table 4). In addition, some of these mutations also increased affinity for agonist (Supplementary Table 4). Thus, mutations of the receptor that directly alter the environment of the Lys7 position of antagonists without inducing wider conformational rearrangements that affect ligand affinity must be rare or non-existent. In contrast, receptor mutations affecting the fluorescence of bound agonist generally did not affect ligand binding affinity (Table 2).
When compared to the native agonist, α-factor, all known antagonists toward the α-factor receptor contain alterations in the N-terminal region of the peptide, such as deletion of extreme N-terminal amino acids, while maintaining the same C-terminal region as α-factor. Thus, the C-terminal region appears to mediate initial binding to receptor while the N-terminal region provides interactions necessary for receptor activation 26; 28 Viewed in this context, mutations that blue-shift the emission of bound antagonists may be seen as restoring agonist-type interactions with the N-terminal of the antagonist, driving the receptor into the conformation that it would normally have when bound to agonist and causing the antagonist to function as a weak agonist. Consistent with this, most such mutations enhance the receptor’s overall binding affinity for both antagonist and some increase the affinity for agonist (Table 4; Supplementary Table 4).
As a test of the idea that mutation-induced conformational shifts of the receptor moves the NBD fluorophore of bound antagonist into an environment similar to that of the fluorphore of bound agonist, we combined the mutations Q51H and D275E (which shift the fluorescence of bound agonist but do not, by themselves, affect the emission of bound antagonist) into alleles that also contain the constitutively activating mutation P258S (which does not affect the emission of bound agonist but shifts the emission of bound antagonist). The presence of the Q51H or D275E mutations in the same allele as the P258S substitution red-shifts the emission of the bound antagonist [dTyr3,Lys7(NBD),Nle12]α-factor compared to alleles containing the constitutive P258S mutation alone (Table 4). Thus, once the labeled region of the antagonist is induced (by the constitutively activating mutations) to interacting with receptor, its emission can be shifted by the same mutations that shift the emission of bound agonist.
The mutations recovered at positions M54 and F55 in the first transmembrane segment of Ste2p exhibit several properties indicative of direct interactions of these amino acids with N-terminal regions of the ligand that determine its efficacy: 1) The effects of mutations at these positions are ligand-specific: The F55S substitution reduces binding of both the antagonist [dTyr3,Lys7(NBD),Nle12]α-factor and the agonist [Dap3(NBD),Arg7,Nle12] α-factor to undetectable levels without significantly diminishing binding or signaling responses of the normal agonist [Lys7(NBD),Nle12] α-factor. 2) The F55L substitution is the only identified mutation that provides a spectral shift in the emission of bound antagonist without enhancing overall binding affinity or the EC50 of the antagonist. 3) The M54I and F55L substitutions shift the fluorescence emission of antagonist and enhance the maximal signaling responses to antagonists without causing any significant enhancement of constitutive signaling. These findings indicative of specific interactions between the first transmembrane segment of the receptor and the different N-terminal regions of the tested series of ligands observations are difficult to reconcile with previous evidence for direct contact between the extracellular regions of transmembrane segments 5–7 of the receptor and the N-terminal region of the ligand 26; 28. Taken together, the current and previous results suggest either that the N-terminal of the ligand interacts simultaneously with both the first transmembrane segment and segments 5–7 of the receptor, or that the mutations at M54 and F55 exert their effects via indirect effects on the conformation of nearby helices in Ste2p.
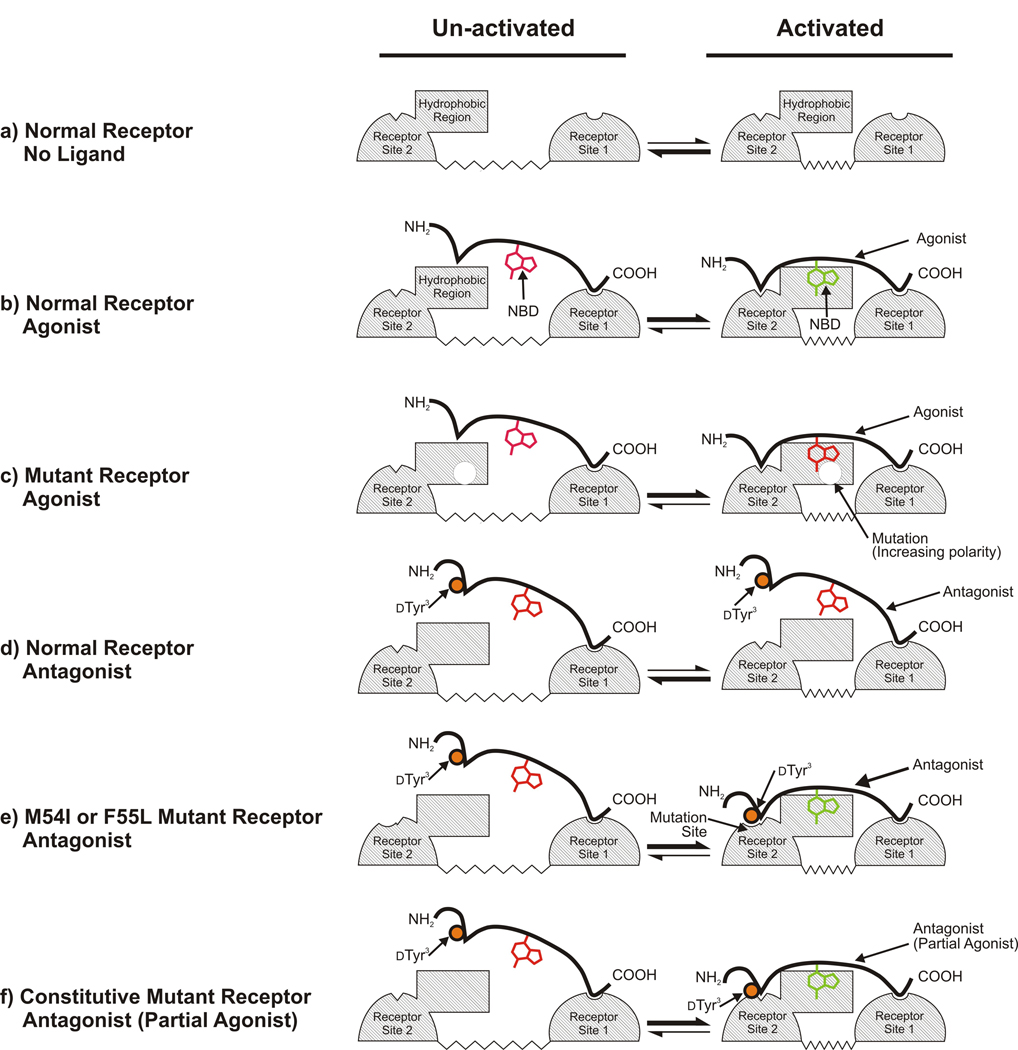
Overall analysis of the binding and signaling responses of normal and mutant receptors to agonists and antagonists is consistent with the mechanistic model of receptor action shown in Fig. 4. In the figure, the conformational change associated with activation is schematically represented as a close approach of two regions of the receptor that allows both the N- and C-termini of the agonist to make separate interactions with different portions of the ligand binding site (Fig. 4a). Under all conditions, the receptor undergoes transitions between the activated and un-activated states, however the relative populations of receptors in the different states is altered by ligands and mutations. The interaction between the C-terminal of the ligand and the receptor is the major contributor to initial binding. Weaker, but highly specific, binding between the activated-receptor conformation and the N-termini of agonists is required to maintain the receptor in the activated state (Fig. 4b). (If the ligand is held in position by binding to the C-terminal, avidity effects could allow the interaction with the N-terminal to be quite weak.) The interaction with the N-terminal region of the ligand stabilizes the active state of the receptor (close approach of two regions of the receptor in the model). When interactions with both the N- and C-termini of ligands occur, a fluorophore attached to Lys7 of α-factor is brought into a non-polar environment at the receptor-ligand interface, causing a blue-shift in emission. Mutations at the boundary between the predicted aqueous and transmembrane regions of each extracellular loop are capable of red-shifting the emission of bound ligand by increasing the polar nature of the environment of bound agonist (Fig. 4c).
Figure 4.
Model for the binding and activation of normal and mutant α-factor receptors by different ligands. Ligand-interacting surfaces on the receptor are indicated by cross-hatching. Site I interacts with the C-termini of ligands and is likely to consist of the extracellular end of the first transmembrane segment. Site 2 interacts with the N-termini of ligands and most likely consists of the extracellular end of the sixth transmembrane segment and the third extracellular loop of the receptor (see 13;14;15;16;17;18;32). The conformational equilibrium between inactive and activated states of the receptor is schematically represented as a change in the distance between Site 1 and Site 2. a) In the absence of ligand, the receptor favors the unactivated state with a large separation between ligand contact sites. b) Binding of agonist to normal receptors favors the activated state. c) Mutations affecting the emission spectrum of bound agonist change the environment of the fluorophore (red-shift) without altering receptor ligand binding or, generally, activation. d) Binding of ligands that act as antagonists toward normal receptors (labeled as “antagonist”) does not alter the receptor’s conformational equilibrium. Activation-specific interactions with the N-terminal of the ligand are blocked by the dTyr3 substitution (orange circle), leaving the NBD group attached at Lys7 exposed to solvent while the C-terminal region of the antagonist (which is identical to the corresponding region of agonist) maintains a high affinity interaction with receptor. e) Mutations that restore receptor binding to the altered N-terminal region of antagonist allow signaling responses to “antagonist” ligands. The altered binding mode to the mutant receptors allows the NBD group to be protected from solvent. f) Mutations that constitutively activate the receptor by favoring the activated conformation, enhancing the interaction of Site 2 of the receptor with antagonists that exhibit weak partial agonist activity. The altered binding mode allows the NBD group to be protected from solvent.
Viewed in the framework of the model, binding of true neutral antagonists to normal receptors would involve only the component of the receptor-agonist interactions involving the C-terminal of the ligand, leaving the fluorophore attached to Lys7 exposed to solvent and insensitive to effects of mutations that alter the environment of bound agonist (Fig. 4d). The loss of activating interactions of the N-terminal regions of antagonists with receptors may be overcome by mutations such as M54I and F55L in Ste2p that could directly enhance interactions between the N-terminal portions of agonists and the activated state of the receptor (Fig. 4e). Such mutations can alter the relative affinity of the ligand for the activated vs. un-activated states, converting a neutral antagonist to an agonist. Such a conversion could occur without any enhancement of the overall binding affinity for the ligand, as is observed for the F55L substitution.
Most of the mutations recovered from the antagonist screen provide simultaneous shifting of the emission spectrum, constitutive activation of receptors, enhanced overall binding affinity for ligands, and ability of receptors to be activated by ligands that serve as antagonists toward normal receptors (Table 4 and Supplementary Table 4). Based on the specific locations of these mutations, and their degree of dispersion throughout the sequence and predicted topology of the receptor, it is unlikely that they all could exert their affects by directly altering receptor-ligand interactions. Instead, it seems more likely that they act indirectly by altering the equilibrium of the receptor between activated and un-activated states as shown in Fig. 4f. Such an alteration would not be expected to alter the signaling responses to a true neutral antagonist, which, by definition, binds both states with equal affinity. Thus, the observed effects are most likely a result of the weak partial agonist activity of the antagonistic α-factor analogs, which has been reported previously 30, and can be seen in the dose response curves presented in Supplementary Fig. 5b and the notes to Table 4. Increased population of the activated state (which preferentially interacts with agonists), enhances the binding affinity of partial agonists for receptors, in addition to promoting constitutive signaling activity. Such behavior is consistent with an analytical expression describing the two-state model for receptor signaling (see 39), which predicts that constitutive signaling and signaling responses to weak partial agonists will exhibit similar dependencies on the equilibrium constant relating the populations of activated and un-activated ligand-free receptors (see supplementary text). However, it is surprising that substantial changes in the fluorescence emission of bound antagonist/partial agonist are observed for mutants that cause only small shifts in the population of activated receptors, as indicated by relatively weak constitutive activity. This suggests that, upon binding the weak partial antagonist, the mutant receptors may be efficiently converted to a conformation that is actually different from the normal antagonist-stabilized activated state. This new state could be intermediate between the normal activated and un-activated states, providing significant alteration of the environment of the bound ligand, but only inefficient activation of G protein.
The fluorescence-based screen described here provides a relatively rapid procedure for identifying receptor-ligand interactions that can complement existing biochemical approaches. The selectivity of the screen for mutant alleles that retain correct folding, efficient targeting to the cell surface, and high affinity ligand binding makes it more direct and specific than genetic approaches based on detection of loss of ligand binding or signaling. Since many mammalian GPCRs and other receptors can be expressed in functional form in yeast 10; 11, and since fluorescent ligand analogs are available for a variety of receptors 40, these procedures should be useful for characterizing ligand binding sites on a range of different receptors. Differential application of this approach to agonist and antagonist binding to the yeast α-factor receptor has provided a view of the distinct interactions of the two classes of ligands with receptors, leading to a mechanistic model for receptor responses to the different types of ligands.
Materials and Methods
Plasmid and Strains
The plasmids and strains used in this study are listed in Supplementary Table 5. Receptors for α-factor were expressed from multicopy plasmids with URA3 markers derived from plasmid pMD1230 19. STE2 in this plasmid contains a natural HpaI site at position 136 (numbered as base pairs from the beginning of the start codon) and XbaI, NheI, and KpnI restriction sites introduced as translationally silent mutations at positions 362, 617, and 822 in the predicted N-terminal region and extracellular loops 1, 2, and 3, respectively 41. The backbone of plasmid pMD1230 contains extra HpaI and XbaI sites that were removed as follows: To remove the extra HpaI site, plasmid pMD98519 was subjected to site directed mutagenesis to remove the HpaI site from its origin of replication, creating pMD1129. The STE2-containing SacI-SphI fragment was then excised from pMD1230 and then ligated into SacI-SphI-cut plasmid pMD1129, creating plasmid pMD1349. To remove both the extra HpaI and XbaI sites, plasmid pMD1383 was constructed by site-directed mutagenesis of plasmid pMD228 30. The SacI-SphI fragment from pMD1230 was then ligated into SacI-SphI-digested pMD1383, creating vector pMD1450.
Plasmid pMD228, lacking any STE2 insert, was used to transform negative control strains. Site-directed mutagenesis was performed as described previously42. The host Saccharomyces cerevisiae strain used for all studies was A57530, which has the genotype: MATa ste2-Δ far1-Δ bar1− cry1R ade2-1 his4-580 lys2oc trp1am tyr1oc SUP4-3ts leu2 ura3 FUS1∷p[FUS1-lacZ TRP1].
Library construction and screening
The starting allele of STE2 used for mutagenesis contained a truncation of cytoplasmic C-terminal residues 305–431. This deletion of the C-terminal tail of the receptor inhibits endocytosis, increasing the number of receptors at the cell surface and providing an enhanced fluorescence signal upon binding of [Lys7(NBD),Nle12]α-factor 19. C-terminally truncated receptors exhibit similar ligand binding affinities and hypersensitive responses to pheromone, compared to full-length versions 19; 43; 44.
Error-prone PCR was performed essentially as described previously 45 using plasmid pMD1230 encoding the truncated version of Ste2p as the template and four sets of primer pairs flanking the regions of interest in the STE2 gene (Supplementary Table 1 and Fig. 2). Following amplification, strain A575 30 was transformed with a mixture of the PCR product and linearized plasmid (Fig. 1), allowing homologous recombination between the PCR fragment and the plasmid. Transformants were cultured on synthetic medium lacking uracil (SD-ura). Approximately 10,000 transformants were obtained for each of the libraries except that covering the third extracellular loop, for which approximately 500 transformants were obtained. Co-transformations of linearized plasmid with PCR product resulted in 50 – 100 times the number of transformants obtained with linearized plasmid alone. The average mutagenesis rate ranged from 1.2 – 3.4 bases and 0.7 – 2.4 amino acids per plasmid.
Prior to cell sorting, the library of yeast strains containing mutagenized receptors was cultured in SD-Ura medium to an OD600 of approximately 1, then incubated in 15 mM MES buffer (pH 5.8) with 50 nM [Lys7(NBD),Nle12]α-factor for an hour in the dark on ice, essentially as described 19. The screen with [dTyr3,Lys7(NBD),Nle12]α-factor was performed in 15 mM sodium acetate buffer (pH 4.6) with 75 nM ligand. Cell sorting was conducted using a FACSVantage (Becton Dickinson) instrument with excitation at 488 nm, collecting cells exhibiting an increased ratio of emission in the FL2 channel (565–605 nm) to emission in the FL1 channel (515–545 nm) compared with an unmutagenized population. The collection parameters used for gating are represented by the rectangle in the two-dimensional dot plot in Fig. 1. For the N-terminal and EC3 libraries, two independent populations of potential mutants were collected, a “stringent” screen consisting of cells exhibiting the most significant change in FL1/FL2 ratio, and a “relaxed” screen for cells with more modest changes in channel ratio. Only a “relaxed” screen was performed for the other two libraries. Approximately 2 to 4 million cells were screened per library. For each library, approximately 600 – 5000 cells were collected, but only about 10% of these produced colonies when the cultures were plated on SD-ura.
Characterization of mutant alleles
In cases where the recovered alleles contained multiple substitutions, individual mutations were re-created by site-directed mutagenesis. In cases where substitutions of interest were originally recovered as single mutations in an allele, the sequence of the entire coding region of STE2 was obtained to verify the absence of additional substitutions. Plasmids encoding STE2 alleles with verified single amino acid substitutions of interest were re-transformed into a fresh yeast host for further testing. Approximately 50% of alleles failed to recapitulate the original phenotype following retransformation. The presence of such “false positive” isolates did not significantly affect the time required to isolate and characterize mutants.
Fluorescent Ligand Binding Assays
Normal α-factor was purchased from Bachem. All α-factor analogs were synthesized by solid phase synthesis, purified to near homogeneity, and characterized by MS, as described previously 46. Shifts in emission spectrum were quantitated based on equilibrium saturation binding curves for [Lys7(NBD),Nle12]-α-factor detected by analytical flow cytometry as described previously 19. 15 mM sodium acetate buffer (pH 4.6) was used instead of the MES buffer used during the screening process because ligand binding is more consistent and tighter at the lower pH. The mean fluorescence for each sample was measured using an Accuri C6 or Becton Dickinson FACSCalibur or FACScan flow cytometers. Autofluorescence of the yeast cells was determined in the absence of ligand for each independent yeast transformant and subtracted from each measurement. The levels of autofluorescence were generally 10–15 fold less than the maximal fluorescence seen with [Lys7(NBD),Nle12]α-factor-bound to normal receptor. Overall Bmax and Kd values were calculated by fitting the mean fluorescence per cell at each ligand concentration to an equation that allows for single-site specific binding as well as a non-specific component using the ligand-binding module of Sigmaplot (SPSS Inc.). Fluorescence in each channel, FL1 and FL2, was fit separately. The ratio of the Bmax values obtained from these fittings (referenced to the ratio for normal receptors measured in parallel on the same day) was taken as as the measure of the extent of shift in emission spectrum. Such ratios are expected to be relatively unaffected by experimental variations in the concentrations of ligand stocks or cell densities. Errors are reported as the standard error of the mean for assays conducted on three independent yeast transformants including, propagated contributions from assays with the wild-type receptors.
β-Galactosidase Assays
Assays of FUS1-lacZ induction in response to pheromone were performed as described 23. The EC50s and maximal induction levels were determined by fitting the relative β-galactosidase activity at each α-factor concentration to a sigmoidal dose-response equation using the ligand-binding module of Sigmaplot (SPSS Inc). The results are presented relative to the EC50 and maximal induction of the wild-type receptor in response to α-factor, determined in parallel. Errors are reported as the standard error of the mean for assays conducted on three independent yeast transformants, including, propagated contributions from assays of wild-type receptors. Assays with the antagonist were performed with the fluorescent analog.
Supplementary Material
Acknowledgements
We thank, Tim Bushnell, Peter Keng, and B. A. Warsop of the Flow Cytometry Core Facility of the University of Rochester and Randall Rossi for their assistance with flow cytometry. We also thank Jeffrey Zuber, Amir Taslimi, and Rajashri Sridharan for useful discussions and assistance in data analysis and preparation of this manuscript. This work was supported by NIH grants GM059357 to M.E.D. and GM22086 to F.N.
Abbreviations used
- GPCR
(G Protein Coupled Receptor)
- NBD
7-nitrobenz-2-oxa-1,3-diazol-4-yl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 2.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010 doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strader CD, Fong TM, Graziano MP, Tota MR. The family of G-protein-coupled receptors. Faseb J. 1995;9:745–754. [PubMed] [Google Scholar]
- 10.Brown AJ, Dyos SL, Whiteway MS, White JH, Watson MA, Marzioch M, Clare JJ, Cousens DJ, Paddon C, Plumpton C, Romanos MA, Dowell SJ. Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein alpha-subunit chimeras. Yeast. 2000;16:11–22. doi: 10.1002/(SICI)1097-0061(20000115)16:1<11::AID-YEA502>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Dowell SJ, Brown AJ. Yeast assays for G-protein-coupled receptors. Receptors Channels. 2002;8:343–352. [PubMed] [Google Scholar]
- 12.Son CD, Sargsyan H, Hurst GB, Naider F, Becker JM. Analysis of ligand-receptor cross-linked fragments by mass spectrometry. J Pept Res. 2005;65:418–426. doi: 10.1111/j.1399-3011.2005.00248.x. [DOI] [PubMed] [Google Scholar]
- 13.Son CD, Sargsyan H, Naider F, Becker JM. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry. 2004;43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 14.Lee BK, Khare S, Naider F, Becker JM. Identification of residues of the Saccharomyces cerevisiae G protein- coupled receptor contributing to alpha-factor pheromone binding. J Biol Chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 15.Naider F, Becker JM, Lee YH, Horovitz A. Double-mutant cycle scanning of the interaction of a peptide ligand and its G protein-coupled receptor. Biochemistry. 2007;46:3476–3481. doi: 10.1021/bi602415u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee BK, Lee YH, Hauser M, Son CD, Khare S, Naider F, Becker JM. Tyr266 in the sixth transmembrane domain of the yeast alpha-factor receptor plays key roles in receptor activation and ligand specificity. Biochemistry. 2002;41:13681–13689. doi: 10.1021/bi026100u. [DOI] [PubMed] [Google Scholar]
- 17.Umanah GK, Huang L, Ding FX, Arshava B, Farley AR, Link AJ, Naider F, Becker JM. Identification of residue-to-residue contact between a peptide ligand and its G protein-coupled receptor using periodate-mediated dihydroxyphenylalanine cross-linking and mass spectrometry. J Biol Chem. 2010 doi: 10.1074/jbc.M110.149500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umanah GK, Son C, Ding F, Naider F, Becker JM. Cross-linking of a DOPA-containing peptide ligand into its G protein-coupled receptor. Biochemistry. 2009;48:2033–2044. doi: 10.1021/bi802061z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajaj A, Celic A, Ding FX, Naider F, Becker JM, Dumont ME. A fluorescent alpha-factor analogue exhibits multiple steps on binding to its G protein coupled receptor in yeast. Biochemistry. 2004;43:13564–13578. doi: 10.1021/bi0494018. [DOI] [PubMed] [Google Scholar]
- 20.Ding FX, Lee BK, Hauser M, Davenport L, Becker JM, Naider F. Probing the binding domain of the Saccharomyces cerevisiae alpha-mating factor receptor with rluorescent ligands. Biochemistry. 2001;40:1102–1108. doi: 10.1021/bi0021535. [DOI] [PubMed] [Google Scholar]
- 21.Ding FX, Patri R, Arshava B, Naider F, Lee BK, Hauser M, Becker JM. Study of the binding environment of alpha-factor in its G protein- coupled receptor using fluorescence spectroscopy. J Pept Res. 2002;60:65–74. doi: 10.1034/j.1399-3011.2002.21004.x. [DOI] [PubMed] [Google Scholar]
- 22.Bajaj A, Connelly SM, Gehret AU, Naider F, Dumont ME. Role of extracellular charged amino acids in the yeast alpha-factor receptor. Biochim Biophys Acta. 2007;1773:707–717. doi: 10.1016/j.bbamcr.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celic A, Connelly SM, Martin NP, Dumont ME. Intensive mutational analysis of G protein-coupled receptors in yeast. Methods Mol Biol. 2004;237:105–120. doi: 10.1385/1-59259-430-1:105. [DOI] [PubMed] [Google Scholar]
- 24.Shah A, Marsh L. Role of Sst2 in modulating G protein-coupled receptor signaling. Biochem Biophys Res Commun. 1996;226:242–246. doi: 10.1006/bbrc.1996.1340. [DOI] [PubMed] [Google Scholar]
- 25.Dosil M, Giot L, Davis C, Konopka JB. Dominant-negative mutations in the G-protein-coupled alpha-factor receptor map to the extracellular ends of the transmembrane segments. Mol Cell Biol. 1998;18:5981–5991. doi: 10.1128/mcb.18.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abel MG, Zhang YL, Lu HF, Naider F, Becker JM. Structure-function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine-scanned analogs. J Pept Res. 1998;52:95–106. doi: 10.1111/j.1399-3011.1998.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 27.Eriotou-Bargiota E, Xue CB, Naider F, Becker JM. Antagonistic and synergistic peptide analogues of the tridecapeptide mating pheromone of Saccharomyces cerevisiae. Biochemistry. 1992;31:551–557. doi: 10.1021/bi00117a036. [DOI] [PubMed] [Google Scholar]
- 28.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 29.Konopka JB, Margarit SM, Dube P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci U S A. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommers CM, Martin NP, Akal-Strader A, Becker JM, Naider F, Dumont ME. A limited spectrum of mutations causes constitutive activation of the yeast alpha-factor receptor. Biochemistry. 2000;39:6898–6909. doi: 10.1021/bi992616a. [DOI] [PubMed] [Google Scholar]
- 31.Huang LY, Umanah G, Hauser M, Son C, Arshava B, Naider F, Becker JM. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry. 2008;47:5638–5648. doi: 10.1021/bi701866e. [DOI] [PubMed] [Google Scholar]
- 32.Henry LK, Khare S, Son C, Babu VV, Naider F, Becker JM. Identification of a contact region between the tridecapeptide alpha- factor mating pheromone of Saccharomyces cerevisiae and its G protein- coupled receptor by photoaffinity labeling. Biochemistry. 2002;41:6128–6139. doi: 10.1021/bi015863z. [DOI] [PubMed] [Google Scholar]
- 33.Akal-Strader A, Khare S, Xu D, Naider F, Becker JM. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J Biol Chem. 2002;277:30581–30590. doi: 10.1074/jbc.M204089200. [DOI] [PubMed] [Google Scholar]
- 34.Lin JC, Duell K, Konopka JB. A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein-coupled alpha-factor receptor. Mol Cell Biol. 2004;24:2041–2051. doi: 10.1128/MCB.24.5.2041-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antosiewicz J, McCammon JA, Gilson MK. The determinants of pKas in proteins. Biochemistry. 1996;35:7819–7833. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- 36.Bashford D, Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 38.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor P, Insel PA. The Molecular Basis of Pharmacological Selectivity. In: Pratt WBaTP., editor. Principles of Drug Action: The Basis of Pharmacology. New York, NY: Churchill Livingstone Inc.; 1990. [Google Scholar]
- 40.McGrath JC, Arribas S, Daly CJ. Fluorescent ligands for the study of receptors. Trends Pharmacol Sci. 1996;17:393–399. doi: 10.1016/s0165-6147(96)40004-9. [DOI] [PubMed] [Google Scholar]
- 41.Sommers CM, Dumont ME. Genetic interactions among the transmembrane segments of the G protein coupled receptor encoded by the yeast STE2 gene. J Mol Biol. 1997;266:559–575. doi: 10.1006/jmbi.1996.0816. [DOI] [PubMed] [Google Scholar]
- 42.Leavitt LM, Macaluso CR, Kim KS, Martin NP, Dumont ME. Dominant negative mutations in the alpha-factor receptor, a G protein-coupled receptor encoded by the STE2 gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:917–932. doi: 10.1007/s004380051039. [DOI] [PubMed] [Google Scholar]
- 43.Konopka JB, Jenness DD, Hartwell LH. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 44.Reneke JE, Blumer KJ, Courchesne WE, Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 45.Sommers CM, Dumont ME. Genetic approaches for studying the structure and function of G protein-coupled receptors in yeast. In: Wess J, editor. Structure-Function Analysis of G Protein-Coupled Receptors. New York: Wiley-Liss; 1999. pp. 141–166. [Google Scholar]
- 46.Naider F, Becker JM. Structure-activity relationships of the yeast alpha-factor. CRC Crit Rev Biochem. 1986;21:225–248. doi: 10.3109/10409238609113612. [DOI] [PubMed] [Google Scholar]
- 47.Abel MG, Lee BK, Naider F, Becker JM. Mutations affecting ligand specificity of the G-protein-coupled receptor for the Saccharomyces cerevisiae tridecapeptide pheromone. Biochim Biophys Acta. 1998;1448:12–26. doi: 10.1016/s0167-4889(98)00109-8. [DOI] [PubMed] [Google Scholar]
- 48.Martin NP, Celic A, Dumont ME. Mutagenic mapping of helical structures in the transmembrane segments of the yeast alpha-factor receptor. J Mol Biol. 2002;317:765–788. doi: 10.1006/jmbi.2002.5444. [DOI] [PubMed] [Google Scholar]
- 49.Lin JC, Parrish W, Eilers M, Smith SO, Konopka JB. Aromatic residues at the extracellular ends of transmembrane domains 5 and 6 promote ligand activation of the G protein-coupled alpha-factor receptor. Biochemistry. 2003;42:293–301. doi: 10.1021/bi026766o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.