Abstract
DREAM (downstream regulatory element antagonistic modulator) is a calcium-binding protein that regulates dynorphin expression, promotes potassium channel surface expression, and enhances presenilin processing in an expression level-dependent manner. However, no molecular mechanism has yet explained how protein levels of DREAM are regulated. Here we identified group I mGluR (mGluR1/5) as a positive regulator of DREAM protein expression. Overexpression of mGluR1/5 increased the cellular level of DREAM. Up-regulation of DREAM resulted in increased DREAM protein in both the nucleus and cytoplasm, where the protein acts as a transcriptional repressor and a modulator of its interacting proteins, respectively. DHPG (3,5-dihydroxyphenylglycine), a group I mGluR agonist, also up-regulated DREAM expression in cortical neurons. These results suggest that group I mGluR is the first identified receptor that may regulate DREAM activity in neurons.
Keywords: Calcium, DREAM, Group I mGluR, mGluR5, mGluR1, Pain
INTRODUCTION
Downstream regulatory element antagonistic modulator (DREAM) is a calcium-binding protein, and is variously known as calsenilin and KChIP3, named according to the original binding proteins when discovered independently [1]. DREAM acts as a transcriptional repressor of dynorphin, a subunit of potassium channels, and a modulator of presenilin [2-4]. DREAM binds to a dynorphin response element (DRE) upstream of the dynorphin gene, thereby suppressing its expression [2]. It also binds to presenilins and regulates the amount of fragmented presenilins [3]. DREAM interaction with the A-type potassium channel (carboxyl terminus) is reported to be essential for proper channel function [4]. Among these, the most studied mechanism of DREAM action is the regulation of nociception by modulating dynorphin expression. DREAM binding to DRE is inhibited by increased calcium, possibly due to its binding to the calcium-binding motifs of the protein, suggesting the presence of a calcium-mediated regulatory mechanism of DREAM activity [5]. DREAM knockout mice show increased expression of dynorphin in the spinal cord and, accordingly, decreased sensitivity to pain [2].
Most of the roles of DREAM have been explored based on overexpression and knockout experiments of the DREAM gene, and alteration in the levels of DREAM have been shown to result in changes in various cell functions [2-4]. However, very little information concerning the physiological regulatory mechanism of DREAM levels in cells is available. The only finding we have is that amyloid β (Aβ) increased expression of DREAM in cultured neurons [6]. Furthermore, in addition to the regulation of expression level of the molecule, DREAM must be localized to the nucleus to act as a repressor of dynorphin transcription. Calcium ionophore, thapsigargin, and serum starvation were reported to promote nuclear localization of DREAM [7,8]. However, little information is available about the signaling proteins or receptors that affect the subcellular localization of DREAM.
Metabotropic glutamate receptor 5 (mGluR5) and mGluR1 are members of the metabotropic glutamate receptor family of G protein-coupled receptors (GPCRs), which has 8 subtypes. mGluR5 has long been known to be implicated in the regulation of synaptic transmission and synaptic plasticity [9]. mGluR5 is also involved in neurological and psychiatric diseases such as anxiety, addiction, pain, and fragile X mental retardation syndrome [9-13]. At the molecular level, group I mGluR (comprised of mGluR1 and mGluR5) has been reported to interact or regulate calcium-binding proteins, such as the neuronal calcium sensors NCS-1 (neuronal calcium sensor-1) and VILIP-1 (visinin-like protein-1) [14]. We therefore hypothesized that mGluR1/5 may be a group of receptors that can relay extracellular signals to regulate the activity of DREAM, another member of the neuronal calcium sensor family of proteins.
Here we tested whether a functional interaction between mGluR1/5 and DREAM is present, and the consequence of this interaction. We report that group I mGluR up-regulates protein levels of DREAM and accumulates the protein in the nucleus and cytoplasm. These results suggest group I mGluR-initiated mechanism by which DREAM levels can be regulated.
METHODS
Materials
His-tagged rat DREAM was expressed in BL21 (DE3), purified with nickel-resin, and used for anti-DREAM antibody production. Commercial murine monoclonal anti-DREAM antibody (Millipore, Billerica, MA, USA) was also used. Monoclonal anti-His and anti-Myc antibodies were purchased from BD Biosciences (San Diego, CA, USA) and Sigma-Aldrich (St. Louis, MO, USA), respectively. EDTA-AM was purchased from Molecular Probes (Eugene, OR, USA). Unless otherwise noted, all other reagents were purchased from Sigma-Aldrich.
Constructs
Chicken β-actin promoter replaced the CMV promoter of the pcDNA3 vector (Invitrogen, Carlsbad, CA, USA), generating a pCBA vector. The coding region of rat DREAM was amplified with PCR and inserted into pcDNA3.1 His C and pCBA at the EcoRI site. The D150N, E186Q, and E234Q mutations were introduced for disabling Ca2+ binding to EF hand 2, 3, and 4, respectively, by site-directed mutagenesis with the primers: 5'-CCA TCC ACT TTG AGA ACT TTG TGG TTG GGC-3' and 5'-GCC CAA CCA CAA AGT TCT CAA AGT GGA TGG 3' for D150N, 5'-CAT CAC CAA AGA GCA GAT GCT GGC CAT C-3' and 5'-GAT GGC CAG CAT CTG CTC TTT GGT GAT G-3' for E186Q, and 5'-GGA GTA GTG ACT ATT GAT CAA TTT CTG GAG ACT TGT C-3' and 5'-GAC AAG TCT CCA GAA ATT GAT CAA TAG TCA CTA CTC C-3' for E234Q [15]. Multiple mutant constructs, QQ and NQQ, were made by sequential mutagenesis. The cDNA of mGluR5 was inserted into the pRK5. F767S mutant of mGluR5, which corresponds to F781S or Gαq protein binding mutant of mGluR1, and was made with 5'-AGA AAT GTT CCA GCC AAC TCT AAT GAG GCC AAA TAT ATT-3' and 5'-AAT ATA TTT GGC CTC ATT AGA GTT GGC TGG AAC ATT TCT-3' [16].
Cell culture
HEK293, HeLa, and SK-N-MC cells were obtained from ATCC (Manassas, VA, USA). All cells were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen, Carlsbad, CA, USA) containing 10 mM L-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 10% fetal bovine serum. Cells were maintained in 5% CO2/95% air atmosphere at 37℃.
Primary cortical neuron cultures are described elsewhere [17]. Briefly, the meninges were removed from brains of TP18 Sprague Dawley rats with fine forceps. Isolated cortices were minced, trypsinized, and triturated through a Pasteur pipette narrowed by flame. The resulting cell suspensions were counted and plated onto 6-well plates coated with poly-L-ornithine and fibronectin. Cultures were maintained in neurobasal medium (Invitrogen, Carlsbad, CA, USA) with B-27 supplement (Invitrogen, Carlsbad, CA, USA).
Cell fractionation
Cells were washed with PBS and pelleted by centrifugation followed by swelling in low salt buffer containing 20 mM HEPES pH 7.9, 1.5 mM MgCl2, and protease inhibitor cocktail. Swollen cells were lysed with syringe homogenization and centrifuged at 3,000 g for 3 min. Triton X-100 (1%) was added to the supernatant for cytoplasmic extraction. The resulting pellet was washed twice with low salt buffer and incubated in high salt buffer (low salt buffer supplemented with 420 mM KCl) for nuclear protein extraction.
Western blot analysis
Cells were plated onto 6-well plates at 1×105 to 2×105 per well for 18 h and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 36 h of incubation, cells were pelleted; lysed with denaturing lysis buffer containing 10 mM Tris, 2% SDS pH 7.4, and Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN, USA); boiled for 10 min; and cleared with centrifugation at 13,000 g for 10 min. Cell lysates were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membrane was blocked with 5% non-fat milk and probed with primary and horseradish peroxidase (HRP)-conjugated secondary antibodies followed by chemiluminescence detection with an ECL (Pierce, Rockford, IL, USA) in LAS-4000 CCD camera (GE Healthcare, Piscataway, NJ).
Immunostaining
pCBA-DREAM was co-transfected with pRK5 or pRK5-myc-mGluR5 into HEK293 cells, which were grown for 18 h after being plated onto cover slips. After further incubation for 36 h following transfection, cells were washed with PBS, fixed with 4% paraformaldehyde for 5 min, and blocked with goat serum. The cells on the cover slips were treated with rabbit anti-DREAM and anti-Myc antibodies, rinsed with PBS, and incubated with Alexa 488- and Alexa 568-conjugated secondary antibodies. Cells were observed under a Zeiss LSM710 confocal microscope (Jena, Germany).
RESULTS
Group I mGluR increases protein levels of DREAM
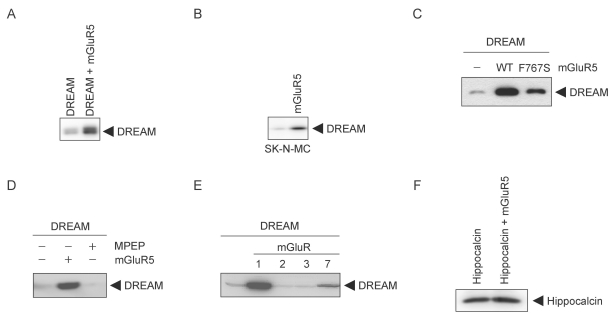
To investigate the possible functional interaction between mGluR5 and DREAM, HEK293 cells were transfected with mGluR5 and DREAM, and expression of DREAM was analyzed using Western blotting. The expression of DREAM was increased by mGluR5 (Fig. 1A). Transfection of mGluR5 in the SK-N-MC neuroblastoma cell line that expresses endogenous DREAM also promoted an increase in DREAM (Fig. 1B). Next we tested whether G protein coupled to mGluR5 is required for the effect of the receptor on the level of DREAM. HEK293 cells were transfected with DREAM and wild-type mGluR5 or mGluR5 F767S, a mutant incapable of G protein coupling [16]. Wild-type mGluR5 increased DREAM expression whereas the mGluR5 F767S mutant did not, suggesting that G protein mediated the effect of mGluR5 on the levels of DREAM (Fig. 1C). To confirm that mGluR5 activity is involved in the modulation of DREAM, we treated cells expressing mGluR5 and DREAM with MPEP (2-Methyl-6-(phenylethynyl)pyridine hydrochloride), a specific mGluR5 antagonist, for 15 h. MPEP inhibited the effect of mGluR5 on the levels of DREAM, indicating that constitutive activity of mGluR5 overexpressed in HEK293 cells increased the expression of DREAM (Fig. 1D). mGluRs are composed of eight subtypes and sub-classified into groups I, II, and III according to ligand specificity, sequence homology, and G protein couplings. The group I mGluRs (mGluR1 and mGluR5) couple to Gαq protein and the group II mGluRs (mGluR2 and mGluR3) and the group III mGluRs (mGluR4, 6, 7, 8) couple to Gαi/o proteins [9]. We examined whether the up-regulation of DREAM by a metabotropic glutamate receptor is specific to the mGluR5 subtype. Cells were co-transfected with DREAM and mGluR1, 2, 3, or 7 and changes in DREAM levels were measured using Western blot analysis. Among the mGluR subtypes tested, only mGluR1 profoundly increased DREAM expression, suggesting that only group I mGluRs can regulate the levels of DREAM (Fig. 1E). We also tested the effect of mGluR5 on hippocalcin, a protein belonging to the neuronal calcium sensor family [18], the same family as DREAM. mGluR5 did not increase the level of hippocalcin in cells co-expressing mGluR5 and hippocalcin (Fig. 1F), indicating that the effect of mGluR5 is not generalized toward all neuronal calcium sensor family proteins.
Fig. 1.
Group I mGluR increases DREAM protein expression. (A, C, and D~F) HEK293 cells were co-transfected with 500 ng of each plasmid expressing DREAM or hippocalcin and 50 ng of pRK5 empty vector, pRK5-mGluR subtypes, or pRK5-mGluR5 F767S (Gαq binding site mutant of mGluR5). Cell lysates were immunoblotted with rabbit anti-DREAM or anti-hippocalcin antibody. (B) pRK5 or pRK5-mGluR5 (200 ng) was transfected into SK-N-MC cells for 36 h before cell lysis. Immunoblotting using mouse anti-DREAM antibody was performed. (E) Cells were treated with MPEP for 15 h before lysis. Lysis was performed by boiling with denaturing lysis buffer after 36 h of cultivation. WT, wild-type.
Stabilization of DREAM by mGluR5
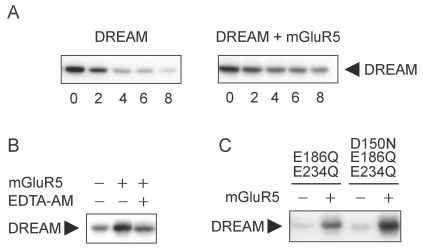
There are several potential mechanisms by which mGluR5 increases the level of DREAM in cells. Perhaps the simplest prediction is that mGluR5 could increase the stability of DREAM. Thus, we tested the hypothesis that mGluR5 enhanced the stability of DREAM. HEK293 cells were co-transfected with DREAM alone or mGluR5 and DREAM and pre-treated with cycloheximide for 8 h. The protein levels of DREAM at each time point (0, 2, 4, 6, and 8 h) were measured using Western blot analysis. Sustained levels of DREAM were observed over time in cells expressing both mGluR5 and DREAM compared to cells expressing DREAM alone (Fig. 2A). This result suggests that mGluR5 increases the levels of DREAM by enhancing the stability of the protein. Because DREAM is a calcium-binding protein and has typical calcium-binding motifs, we investigated whether calcium chelation or mutation of calcium-binding motifs had any effect on the stability of DREAM. We performed Western blotting in lysates from cells transfected with mGluR5 and DREAM, followed by administration of EDTA-AM (disodium ethyldiamine tetraacetic acid-acetoxymethyl ester) for 12 h. EDTA-AM decreased the level of DREAM, indicating that intracellular calcium is required for the mGluR5 signaling responsible for the regulation of DREAM levels (Fig. 2B). To determine the effects of mutations in calcium-binding motifs on the stability of DREAM, cells were transfected with mGluR5 and DREAM mutants (E186Q/E234Q mutant and D150N/E186Q/E234Q mutant), which have been reported to disrupt calcium binding [15]. We observed that no mutants abolished the effect of mGluR5 on their expression levels, suggesting no involvement of calcium-binding motifs of DREAM in mGluR5 regulation of the protein (Fig. 2C).
Fig. 2.
mGluR5 stabilizes DREAM protein. (A) pCBA-DREAM was co-transfected with pRK5 or pRK5-mGluR5 into HEK293 cells. Cells were lysed after 48 h of incubation. Cells were treated with cycloheximide (CHX) at 100 µg/ml for 0, 2, 4, 6, or 8 h before cell lysis. DREAM expression was analyzed by immunoblotting with rabbit anti-DREAM antibody. (B) HEK293 cells were transfected as indicated above. EDTA-AM (100 nM) was added to culture for 12 h to chelate intracellular Ca2+. Immunoblotting was performed using rabbit anti-DREAM antibody. (C) Lysates from cells expressing mGluR5 and DREAM (E186Q/E234Q) double mutant or DREAM (D150N/E186Q/E234Q) triple mutant were subjected to Western blotting.
mGluR5 increases DREAM protein in both the nucleus and cytoplasm
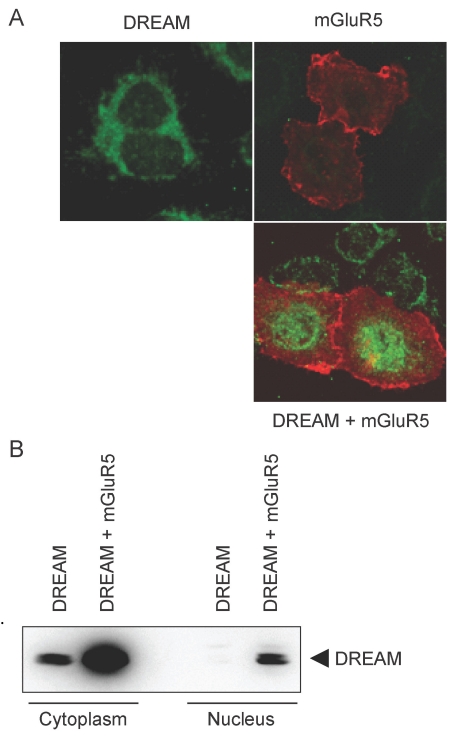
DREAM binding to the DRE region in the promoter of the dynorphin gene and consequent repression of dynorphin transcription is a well-known mechanism by which DREAM modulates nociception. To test whether the mGluR5-induced increase in DREAM expression caused accumulation of the protein in the nucleus, we transfected HeLa cells with mGluR5 and DREAM and performed immunofluorescence. We observed that mGluR5 promoted the nuclear localization of DREAM, as shown by a prominent increase in DREAM immunofluorescence in the nucleus (Fig. 3A). HEK293 cells were transfected with mGluR5 and DREAM and fractionated into the nucleus and cytoplasm. In Western blot analysis, mGluR5 increased the level of DREAM in both the cytoplasm and the nucleus of HEK293 cells expressing mGluR5 and DREAM (Fig. 3B). These results suggest that mGluR5 can promote the localization of DREAM in the nucleus, where it acts as a transcriptional repressor, and also increase the amount of DREAM in the cytoplasm, where it can interact with binding partners such as presenilin and channels.
Fig. 3.
Subcellular localization of increased DREAM by mGluR5. (A) pCBA-DREAM and/or pRK5-myc-mGluR5 were transfected into HeLa cells. Following 36 h incubation, cells were fixed with 4% paraformaldehyde, blocked with goat serum, and incubated with rabbit anti-DREAM and anti-myc antibody. After incubation, fixed cells were washed and treated again with Alexa 488 anti-rabbit and Alexa 568 anti-mouse antibodies. Images were obtained under a Zeiss LSM 710 confocal microscope. (B) HEK293 cells were cotransfected with pRK5-mGluR5 and pCBA-DREAM. Cytoplasmic and nuclear extracts were prepared from transfected cells. Western blotting was performed to determine the amount of DREAM in each fraction.
Increase in the protein level of DREAM by DHPG in primary cultured cortical neurons
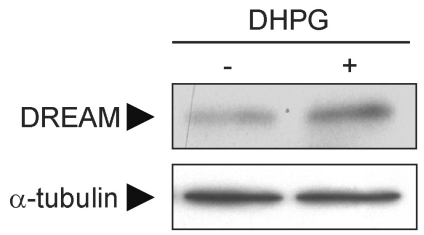
To investigate whether mGluR1/5-induced regulation of DREAM can also occur in primary cultured neurons, where group I mGluR and DREAM are naturally expressed, we prepared primary cultures of cortical neurons and treated cells with DHPG (3,5-dihydroxyphenylglycine), a group I mGluR agonist, for 12 h at day 12 in vitro. DHPG increased the level of DREAM in primary cultured cortical neurons (Fig. 4), suggesting that agonist-induced activation of endogenous group I mGluR in neurons contributes to the regulation of DREAM.
Fig. 4.
DHPG increases DREAM levels in primary cultured neurons. Primary cultures of rat cortical neurons were prepared from 17-day-old embryonic brains. At day 12 in vitro, neurons were treated with 200 µM DHPG for 12 h. Changes in DREAM level were measured using Western blot analysis.
DISCUSSION
The present investigation demonstrates that group I mGluR is a group of receptor that can regulate the protein level of DREAM. mGluR1/5 signaling is mainly mediated by Gαq protein-coupled mediators, although many G protein-independent signaling pathways have been increasingly reported [19,20]. The finding that the mGluR5 F767S mutant could not increase the level of DREAM protein suggests that DREAM stabilization by mGluR5 is mediated by Gαq protein-initiated signaling. This was further supported by the result that intracellular chelation of calcium, an important downstream second messenger of Gαq signaling, prevented the up-regulation of DREAM by mGluR5. mGluR1 is another group I mGluR coupled to Gαq protein. Unlike the group II and group III mGluRs, mGluR1 induced an increase in DREAM level, which also supports the involvement of a Gαq protein-mediated signaling mechanism. Although it is evident that mGluR5 regulates the stability of DREAM, the underlying mechanism remains unclear. mGluR5 up-regulates NCS-1 and VILIP protein levels in the hippocampus in vivo [14]. However, the protein stabilities or mRNA expressions were not measured. Interestingly, hippocalcin, a neuronal calcium sensor protein, was not affected by mGluR5 in our study. Thus, it seems that mGluR5-dependent regulation of neuronal calcium sensor proteins is not a universal phenomenon for all calcium-binding proteins that have characteristics of EF-hand calcium-binding domains in common. Domain study for DREAM suggests that the C-terminal portion of the protein is involved in the stabilization mechanism by mGluR5 (data not shown), suggesting that certain specific mechanisms might be present for the regulation of DREAM by calcium levels elicited by mGluR5. This finding suggests a need for further study.
The important roles of DREAM are implicated in the modulation of pain, regulation of neuronal excitability, and pathogenesis of Alzheimer disease [1,3,4,21]. The sites of actions of DREAM can be divided into two locations: near the membrane and inside the nucleus. DREAM regulation of Kv channels and gamma-secretase may occur near the membrane, and DREAM suppression of dynorphin expression takes place in the nucleus. The mGluR5-induced increase in DREAM in the cytoplasm, and more prominent increase in the nucleus, indicates the potential enhancement of DREAM functions in both regions. Probably, the specific cellular context of certain cell types stimulated by mGluR5 would be an important factor that determines the discernible phenotype of DREAM up-regulation.
mGluR5 has been implicated in various neurological and psychiatric diseases, including anxiety, depression, pain, and Alzheimer disease [9,10]. mGluR5 has also long been related to pain, since mGluR5 antagonists such as MPEP and MTEP (3-((2-Methyl-4-thiazolyl)ethynyl)pyridine) relieve pain in several animal models, and conditional mGluR5 knockout in the central nucleus of the amygdala results in the diminution of inflammation-induced hypersensitivity [22-24]. Interestingly, the analgesic effect of mGluR5 is reported to be mediated by its extracellular signal-regulated kinase-linked interaction with Kv4.2, the potassium channel regulated by DREAM [25]. DREAM regulation of dynorphin induction is one of the well-known mechanisms of pain regulation [2,21]. Taken together, these reports suggest an intriguing possibility that the mGluR5-mediated increase in DREAM in both the cytoplasm and nucleus may contribute to pronociceptive functions of mGluR5. We have demonstrated that mGluR5 increased the protein level of DREAM in both compartments. Hence, research toward a better understanding of the role of stabilized DREAM in the physiology of neurons and the pathophysiology of pain and Alzheimer disease may allow for the development of new treatments.
In conclusion, group I mGluR stabilize the DREAM protein, suggesting a potential link between mGluR5, DREAM, and neuronal dysfunctions in pain and Alzheimer disease.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (331-2008-1-E00056) to C.H. Kim. We thank the Yonsei-Carl Zeiss Advanced Imaging Center, Yonsei University College of Medicine, for technical assistance.
ABBREVIATIONS
- DREAM
downstream regulatory element antagonistic modulator
- mGluR
metabotropic glutamate receptor
References
- 1.Buxbaum JD. A role for calsenilin and related proteins in multiple aspects of neuronal function. Biochem Biophys Res Commun. 2004;322:1140–1144. doi: 10.1016/j.bbrc.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Cheng HY, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, Sarosi I, Woodgett JR, Oliveira-dos-Santos AJ, Ikura M, van der Kooy D, Salter MW, Penninger JM. DREAM is a critical transcriptional repressor for pain modulation. Cell. 2002;108:31–43. doi: 10.1016/s0092-8674(01)00629-8. [DOI] [PubMed] [Google Scholar]
- 3.Buxbaum JD, Choi EK, Luo Y, Lilliehook C, Crowley AC, Merriam DE, Wasco W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat Med. 1998;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 4.An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 5.Carrión AM, Link WA, Ledo F, Mellström B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 6.Jo DG, Lee JY, Hong YM, Song S, Mook-Jung I, Koh JY, Jung YK. Induction of pro-apoptotic calsenilin/DREAM/KChIP3 in Alzheimer's disease and cultured neurons after amyloid-beta exposure. J Neurochem. 2004;88:604–611. doi: 10.1111/j.1471-4159.2004.02159.x. [DOI] [PubMed] [Google Scholar]
- 7.Zaidi NF, Thomson EE, Choi EK, Buxbaum JD, Wasco W. Intracellular calcium modulates the nuclear translocation of calsenilin. J Neurochem. 2004;89:593–601. doi: 10.1046/j.1471-4159.2004.02362.x. [DOI] [PubMed] [Google Scholar]
- 8.Woo HN, Chang JW, Choi YH, Gwon AR, Jung YK, Jo DG. Characterization of subcellular localization and Ca2+ modulation of calsenilin/DREAM/KChIP3. Neuroreport. 2008;19:1193–1197. doi: 10.1097/WNR.0b013e3283089209. [DOI] [PubMed] [Google Scholar]
- 9.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro FM, Paquet M, Cregan SP, Ferguson SS. Group I metabotropic glutamate receptor signalling and its implication in neurological disease. CNS Neurol Disord Drug Targets. 2010;9:574–595. doi: 10.2174/187152710793361612. [DOI] [PubMed] [Google Scholar]
- 11.Dölen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Cho CH, Shin HK. Spinal metabotropic glutamate receptors (mGluRs) are involved in the melittin-induced nociception in rats. Korean J Physiol Pharmacol. 2008;12:237–243. doi: 10.4196/kjpp.2008.12.5.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krystal JH, Mathew SJ, D'Souza DC, Garakani A, Gunduz-Bruce H, Charney DS. Potential psychiatric applications of metabotropic glutamate receptor agonists and antagonists. CNS Drugs. 2010;24:669–693. doi: 10.2165/11533230-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 14.Brackmann M, Zhao C, Kuhl D, Manahan-Vaughan D, Braunewell KH. MGluRs regulate the expression of neuronal calcium sensor proteins NCS-1 and VILIP-1 and the immediate early gene arg3.1/arc in the hippocampus in vivo. Biochem Biophys Res Commun. 2004;322:1073–1079. doi: 10.1016/j.bbrc.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J Biol Chem. 2005;280:18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- 16.Francesconi A, Duvoisin RM. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J Biol Chem. 1998;273:5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Lee J, Choi KY, Hepp R, Lee JY, Lim MK, Chatani-Hinze M, Roche PA, Kim DG, Ahn YS, Kim CH, Roche KW. Calmodulin dynamically regulates the trafficking of the metabotropic glutamate receptor mGluR5. Proc Natl Acad Sci USA. 2008;105:12575–12580. doi: 10.1073/pnas.0712033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGarrigle D, Huang XY. GPCRs signaling directly through Src-family kinases. Sci STKE. 2007;2007:pe35. doi: 10.1126/stke.3922007pe35. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, McGarrigle D, Huang XY. When a G protein-coupled receptor does not couple to a G protein. Mol Biosyst. 2007;3:849–854. doi: 10.1039/b706343a. [DOI] [PubMed] [Google Scholar]
- 21.Costigan M, Woolf CJ. No DREAM, No pain. Closing the spinal gate. Cell. 2002;108:297–300. doi: 10.1016/s0092-8674(02)00640-2. [DOI] [PubMed] [Google Scholar]
- 22.Kolber BJ, Montana MC, Carrasquillo Y, Xu J, Heinemann SF, Muglia LJ, Gereau RW., 4th Activation of metabotropic glutamate receptor 5 in the amygdala modulates pain-like behavior. J Neurosci. 2010;30:8203–8213. doi: 10.1523/JNEUROSCI.1216-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker K, Bowes M, Panesar M, Davis A, Gentry C, Kesingland A, Gasparini F, Spooren W, Stoehr N, Pagano A, Flor PJ, Vranesic I, Lingenhoehl K, Johnson EC, Varney M, Urban L, Kuhn R. Metabotropic glutamate receptor subtype 5 (mGlu5) and nociceptive function. I. Selective blockade of mGlu5 receptors in models of acute, persistent and chronic pain. Neuropharmacology. 2001;40:1–9. doi: 10.1016/s0028-3908(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 24.Sevostianova N, Danysz W. Analgesic effects of mGlu1 and mGlu5 receptor antagonists in the rat formalin test. Neuropharmacology. 2006;51:623–630. doi: 10.1016/j.neuropharm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Hu HJ, Alter BJ, Carrasquillo Y, Qiu CS, Gereau RW., 4th Metabotropic glutamate receptor 5 modulates nociceptive plasticity via extracellular signal-regulated kinase-Kv4.2 signaling in spinal cord dorsal horn neurons. J Neurosci. 2007;27:13181–13191. doi: 10.1523/JNEUROSCI.0269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]