Abstract
The oxygen of a peptide bond has two lone pairs of electrons. One of these lone pairs is poised to interact with the electron-deficient carbon of the subsequent peptide bond in the chain. Any partial covalency that results from this n→π* interaction should induce pyramidalization of the carbon (C′i) toward the oxygen (Oi−1). We searched for such pyramidalization in 14 peptides that contain both α- and β-amino acid residues and that assume a helical structure. We found that the α-amino acid residues, which adopt the main chain dihedral angles of an α-helix, display dramatic pyramidalization but the β-amino acid residues do not. Thus, we conclude that Oi−1 and C′i are linked by a partial covalent bond in α-helices. This finding has important ramifications for the folding and conformational stability of α-helices in isolation and in proteins.
Keywords: α-helix, α/β-peptide, Bürgi–Dunitz trajectory, foldamer, n→π* interaction, protein folding, protein stability, stereoelectronic effect
Introduction
Electron delocalization is the source of partial covalency in many noncovalent interactions. For example, the partial covalency in a hydrogen bond stems from delocalization of the lone pair of electrons (n) of the hydrogen bond acceptor over the antibonding orbital (σ*) of the hydrogen bond donor.1–6 We have discovered another noncovalent interaction, termed the n→π* interaction, with partial covalency.7–11 In this interaction, the partial covalency arises due to overlap of the electron pair (n) of a donor group with the antibonding orbital (π*) of a carbonyl group.12–18 In common protein secondary structures 11 19 and peptoids, 20 21 the electron-pair donor is a proximal carbonyl oxygen. This interaction is the basis of many protein–ligand interactions 22 and is reminiscent of the Bürgi–Dunitz trajectory for nucleophilic additions to carbonyl groups.23
The partial covalency of the n→π* interaction should give rise to a distinctive structural signature. Specifically, an n→π* interaction, analogous to the approach of a nucleophile to a carbonyl group, 23 should engender pyramidalization of the acceptor carbon of the carbonyl group.10 In this pyramidalization, the acceptor carbon is displaced toward the donor group and away from the plane formed by its three pendant atoms (Fig. 1). Pyramidalization can be detectable in high-resolution crystal structures. Nonetheless, the magnitude of the pyramidalization is small, and its origin can be attributed to other forces, such as those involved in the formation of the crystal lattice.24 25 Here, we use α/β-peptides to search for the existence of this definitive signature for the n→π* interaction in residues with an α-helical conformation. Our findings provide new insight on this most renowned and common secondary structures.
Figure 1.
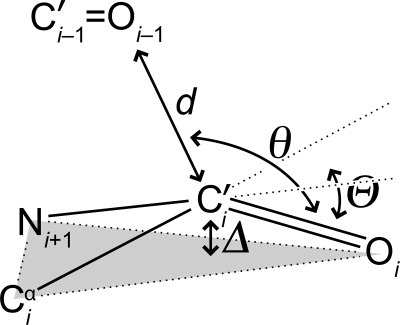
Pyramidalization of main-chain carbonyl groups due to the n→π* interaction showing the definition of distances: d and Δ and angles: θ and θ.
Results and Discussion
In an α-helix, the lone pair (n) of the oxygen (Oi−1) of the amide carbonyl group at position i − 1 overlaps with the antibonding orbital (π*) of the amide carbonyl group (C′i=Oi) at position i. This n→π* electron delocalization induces a short contact between these two carbonyl groups. It follows that the n→π* interaction could engender pyramidalization of the carbon of the acceptor carbonyl group, which would be observable in the high-resolution crystal structures of α-helices.
To establish that any observed pyramidalization arises from an n→π* interaction, an internal control is needed, wherein pyramidalization is absent when the n→π* interaction is absent. Gellman and coworkers 26–30 have determined crystal structures for 14 helical peptides containing both α- and β-amino acid residues (see Supporting Information). These structures were solved by direct methods rather than by experimental phasing or molecular replacement, and the atomic coordinates were deposited in the Cambridge Crystallographic Data Centre (CCDC). In these structures, the α-amino acid residues adopt the torsional angles φ (C′i−1–Ni–Ciα–C′i) and ψ (Ni–Ciα–C′i–Ni+1) that characterize an α-helix (Fig. 2).29 30 A close examination of these crystal structures revealed that carbonyl groups flanking the α-amino acid residues exhibited a C′i−1 =Oi−1···C′i distance of d < 3.22 Å, but analogous carbonyl groups flanking the β-amino acid residues had d > 3.22 Å, where 3.22 Å is the sum of the van der Waals radii of oxygen and carbon. The larger values of d for the β-amino acid residues stem from the additional carbon in their main chain. Hence, the carbonyl groups of the α-amino acid residues should experience a stronger n→π* interaction than the carbonyl groups of the β-amino acid residues. Accordingly, we reasoned that the carbonyl groups of the α-amino acid residues could likewise exhibit greater pyramidalization.
Figure 2.
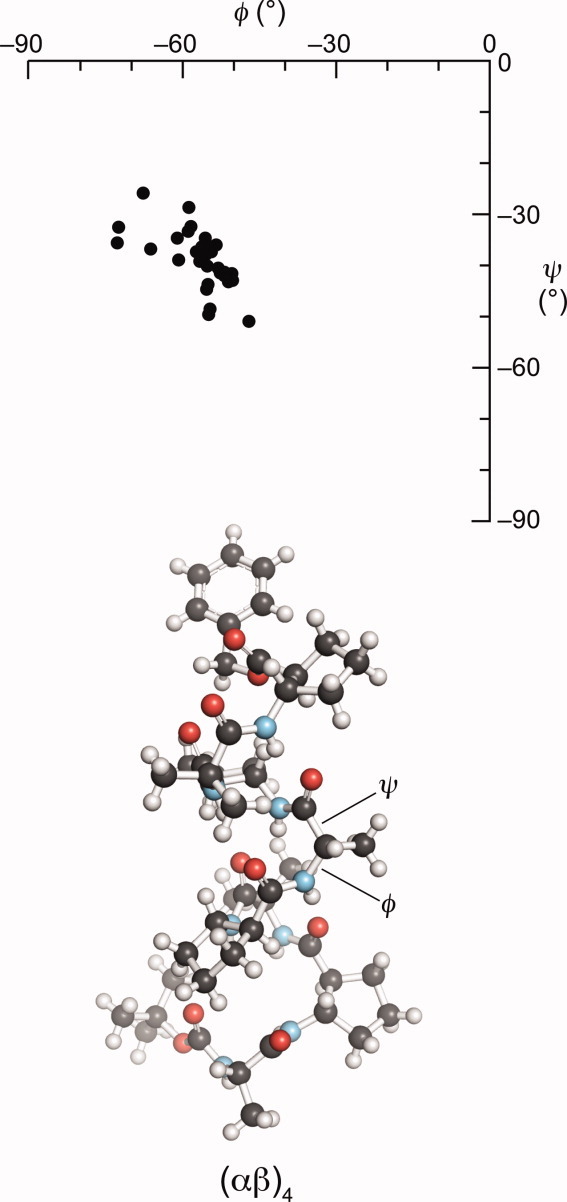
Ramachandran plot of α-amino acid residues in the 14 (αβ)n peptides analyzed herein. The ball-and-stick diagram depicts the structure with CCDC refcode OGAVAU.
The parameter θ reports on the extent of carbonyl pyramidalization (Fig. 1).10 In an α-helix, a value of θ > 0 indicates that C′i is closer to Oi−1 (and to the center of the α-helix) than the plane defined by Oi, Ciα, and Ni+1. Conversely, a value of θ < 0 indicates that C′i is farther from Oi−1 than that plane.
An examination of 14 crystal structures of oligopeptides with alternating α- and β-amino acid residues revealed that the acceptor carbonyl groups of the α-amino acid residues exhibit larger pyramidalization than those of the β-amino acid residues. Moreover, that pyramidalization has θ > 0, which is consistent with the partial covalency between Oi−1 and C′i. The mean value of θ for α-amino acid residues was (4.0° ± 1.1°), whereas that for β-amino acid residues was −(1.2° ± 1.9°) [Fig. 3(A)]. The first and the last residues were not included in this calculation and are also not depicted in Figures 2 and 3(A).
Figure 3.
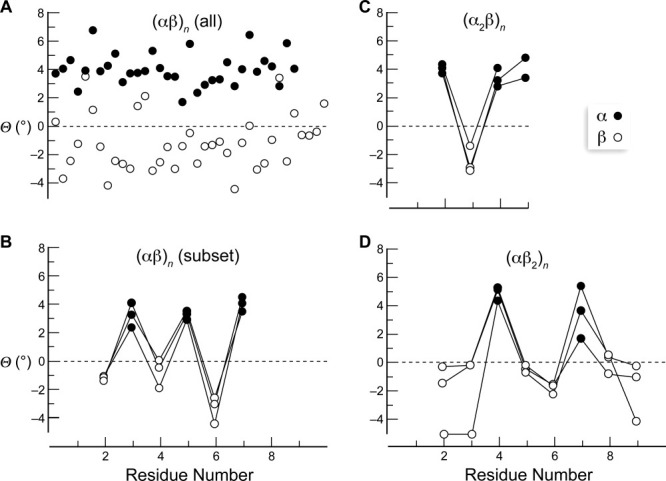
Pyramidalization in α/β peptides. The parameter θ is a measure of pyramidalization (Fig. 1). Data are from crystal structures in the CCDC.26–30 •, α-amino acid residue; ○, β-amino acid residue. A: (αβ)n (all 14); CCDC refcode OGATAS, CAXRID, OGASOF, OGASUL, OGATEW, OGATIA, COVFUP, OGATOG, OGATUM, OGAVAU, OGAVEY, OGAVIC, OGAVOI, and COVGAW. B: (αβ)n (subset); CCDC refcode COVFUP, OGATUM, and OGAVAU. C: (α2β)n; CCDC refcode PUCCIA and PUCCOG (two asymmetric units). D: (αβ2)n; CCDC refcode PUCCUM, PUCDEX, and PUCDUN.
It is noteworthy that the β-amino acid residues tend to have θ < 0. This inversion can be attributed to Pauli repulsion between the lone pair of the donor oxygen and the π orbital of the carbonyl group (Fig. 4).31 The slight negative pyramidalization of the acceptor carbonyl carbon obviates this Pauli repulsion. Similar closed shell repulsions had been invoked previously to explain small carbonyl pyramidalization.24 25 We also note that the oxygen donor for a β-amino acid residue blocks the si face of the acceptor carbonyl group, as the donor oxygen is close to the carbon acceptor (d ≈ 3.5–4.0 Å) but not close enough to induce a positive pyramidalization. The re face of the carbonyl group is exposed to short contacts from solvent and other molecules in the crystal lattice that could induce a small negative pyramidalization.
Figure 4.
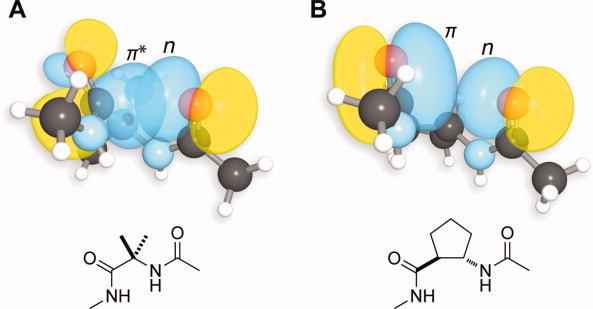
Typical α- and β-amino acid residues in α/β peptides. A: Favorable overlap between n- and the π*-orbital in an α-amino acid residue. B: Unfavorable overlap between n- and the π-orbital in a β-amino acid residue. Orbital images were obtained using computational methods described previously 11 on residues in CCDC refcode OGAVAU capped with acetyl and N-methylamino groups.
Quite strikingly, the carbonyl pyramidalization follows a distinct sawtooth pattern in helical peptides with alternating α- and β-amino acid residues [Fig. 3(B)]. To ascertain that the sawtooth pattern for alternating α- and β-amino acid residues was dependent on residue type, we analyzed crystal structures of helical peptides with α2β- and αβ2-amino acids as repeating motifs. In all of these sequences, we found that the pyramidalization pattern was indeed dependent on the type of residue [Fig. 3(C,D)]. The α-amino acid residues showed consistent positive pyramidalization, and the β-amino acid residues showed either negligible or slightly negative pyramidalization.
These findings lend strong credence to the existence of n→π* interaction in α-helices. Consistent with our findings, Mazzarella and coworkers reported carbonyl pyramidalization in α-helices of high-resolution protein crystal structures.32 Likewise, using average isotropic chemical shifts and X-ray crystallography, Lario and Vrielink 33 reported that the π-electron clouds of carbonyl groups are more polarized in α-helices than that in β-sheets. In α-helices, the adjacent carbonyl groups are poised for a strong n→π* interaction, but they are too far apart for any significant n→π* interaction in β-sheets. Accordingly, the polarization of the π-bond, which is a natural outcome of the n→π* interaction, is apparent in α-helices but not β-sheets. Such polarization of the π-bond could strengthen the canonical i→i + 4 hydrogen bond by making the carbonyl oxygen a better hydrogen-bond acceptor.
The existence of the n→π* interaction in α-helices has bearing on their folding and conformational stability. The short contact effected by the n→π* interaction between adjacent carbonyl groups fortifies a compact structure that aligns the distal hydrogen-bond donor and acceptor groups for a strong hydrogen bond. In an α-helix, the s-rich carbonyl lone pair participates in i→i+4 hydrogen bond. The other lone pair, which is p-rich, is engaged in the n→π* interaction between adjacent carbonyl groups.11 The n→π* interaction not only engages this lone pair for intramolecular association but also prevents it from participating in structure-disrupting hydrogen bonds with other molecules. In addition to contributing to conformational stability, the n→π* interaction could contribute to the folding of an α-helix. The nucleation of an α-helix involves the formation of its first turn, which is disfavored by both entropy and enthalpy.34–36 The n→π* interaction, which operates between the adjacent carbonyl groups, can compensate for these energetic penalties.
Acknowledgments
We thank Prof. S.H. Gellman, C.N. Bradford, B.R. Caes, Prof. G.R. Krow, and Dr. M.D. Shoulders for contributive discussion.
Supplementary material
References
- 1.Pimentel GC, McClellan AL. The hydrogen bond. San Francisco, CA: W.H. Freeman; 1960. [Google Scholar]
- 2.Reed AE, Curtiss LA, Weinhold F. Intermolecular interactions from a natural bond orbital, donor–acceptor viewpoint. Chem Rev. 1988;88:899–926. [Google Scholar]
- 3.Pervushin K, Ono A, Fernandez C, Szyperski T, Kainosho M, Wüthrich K. NMR scalar couplings across Watson–Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc Natl Acad Sci USA. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaacs ED, Shukla A, Platzman PM, Barbiellini DR, Tulk CA. Covalency of the hydrogen bond in ice: a direct x-ray measurement. Phys Rev Lett. 1999;82:600–603. [Google Scholar]
- 5.Weinhold F. Resonance character of hydrogen-bonding interactions in water and other H-bonded species. Adv Protein Chem. 2005;72:121–155. doi: 10.1016/S0065-3233(05)72005-2. [DOI] [PubMed] [Google Scholar]
- 6.Khaliullin RZ, Cobar EA, Lochan RC, Bell AT, Head-Gordon M. Unravelling the origin of intermolecular interactions using absolutely localized molecular orbitals. J Phys Chem A. 2007;111:8753–8765. doi: 10.1021/jp073685z. [DOI] [PubMed] [Google Scholar]
- 7.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. Collagen stability: insights from NMR spectroscopic and hybrid density functional computational investigations of the effect of electronegative substituents on prolyl ring conformations. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
- 8.Hinderaker MP, Raines RT. An electronic effect on protein structure. Protein Sci. 2003;12:1188–1194. doi: 10.1110/ps.0241903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodges JA, Raines RT. Energetics of an n→π* interaction that impacts protein structure. Org Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary A, Gandla D, Krow GR, Raines RT. Nature of amide carbonyl–carbonyl interactions in proteins. J Am Chem Soc. 2009;131:7244–7246. doi: 10.1021/ja901188y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. n→π* interactions in proteins. Nat Chem Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. Conformational stability of collagen relies on a stereoelectronic effect. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins CL, Lin G, Duo J, Rapolu D, Guzei IA, Raines RT, Krow GR. Substituted 2-azabicyclo[2.1.1]hexanes as constrained proline analogues: implications for collagen stability. J Org Chem. 2004;69:8565–8573. doi: 10.1021/jo049242y. [DOI] [PubMed] [Google Scholar]
- 14.Horng J-C, Raines RT. Stereoelectronic effects on polyproline conformation. Protein Sci. 2006;15:74–83. doi: 10.1110/ps.051779806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonntag L-S, Schweizer S, Ochsenfeld C, Wennemers H. The “azido gauche effect”—implications for the conformation of azidoprolines. J Am Chem Soc. 2006;128:14697–14703. doi: 10.1021/ja0654938. [DOI] [PubMed] [Google Scholar]
- 16.Shoulders MD, Guzei IA, Raines RT. 4-Chloroprolines: synthesis, conformational analysis, and incorporation in collagen triple helices. Biopolymers. 2008;89:443–454. doi: 10.1002/bip.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotch FW, Guzei IA, Raines RT. Stabilization of the collagen triple helix by O-methylation of hydroxyproline residues. J Am Chem Soc. 2008;130:2952–2953. doi: 10.1021/ja800225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoulders MD, Satyshur KA, Forest KT, Raines RT. Stereoelectronic and steric effects in side chains preorganize a protein main chain. Proc Natl Acad Sci USA. 2010;106:559–564. doi: 10.1073/pnas.0909592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fufezan C. The role of Bürgi–Dunitz interactions in the structural stability of proteins. Proteins. 2010;78:2831–2838. doi: 10.1002/prot.22800. [DOI] [PubMed] [Google Scholar]
- 20.Gorske BC, Bastian BL, Geske GD, Blackwell HE. Local and tunable n→π* interactions regulate amide isomerism in the peptoid backbone. J Am Chem Soc. 2007;129:8928–8929. doi: 10.1021/ja071310l. [DOI] [PubMed] [Google Scholar]
- 21.Gorske BC, Stringer JR, Bastian BL, Fowler SA, Blackwell HE. New strategies for the design of folded peptoids revealed by a survey of noncovalent interactions in model systems. J Am Chem Soc. 2009;131:16555–16567. doi: 10.1021/ja907184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulini R, Müller K, Diederich F. Orthogonal multipolar interactions in structural chemistry and biology. Angew Chem Int Ed. 2005;44:1788–1805. doi: 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- 23.Bürgi HB, Dunitz JD, Shefter E. Chemical reaction paths. IV. Aspects of O···C=O interactions in crystals. Acta Cryst B. 1974;30:1517–1527. [Google Scholar]
- 24.Rondan NG, Paddon-Row MN, Caramella P, Houk KN. Nonplanar alkenes and carbonyls: a molecular distortion which parallels addition stereoselectivity. J Am Chem Soc. 1981;103:2436–2438. [Google Scholar]
- 25.Jeffrey GA, Houk KN, Paddon-Row MN, Rondan NG, Mitra J. Pyramidalization of carbonyl carbons in asymmetric environments: carboxylates, amides, and amino acids. J Am Chem Soc. 1985;107:321–326. [Google Scholar]
- 26.Schmitt MA, Choi SH, Guzei IA, Gellman SH. Residue requirements for helical folding in short α/β-peptides: crystallographic characterization of the 11-helix in an optimized sequence. J Am Chem Soc. 2005;127:13130–13131. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt MA, Choi SH, Guzei IA, Gellman SH. New helical foldamers: heterogeneous backbones with 1:2 and 2:1 α:β-amino acid residue patterns. J Am Chem Soc. 2006;128:4538–4539. doi: 10.1021/ja060281w. [DOI] [PubMed] [Google Scholar]
- 28.Choi SH, Guzei IA, Gellman SH. Crystallographic characterization of the α/β-peptide 14/15-helix. J Am Chem Soc. 2007;129:13780–13781. doi: 10.1021/ja0753344. [DOI] [PubMed] [Google Scholar]
- 29.Choi SH, Guzei IA, Spencer LC, Gellman SH. Crystallographic characterization of helical secondary structures in α/β-peptides with 1:1 residue alternation. J Am Chem Soc. 2008;130:6544–6550. doi: 10.1021/ja800355p. [DOI] [PubMed] [Google Scholar]
- 30.Choi SH, Guzei IA, Spencer LC, Gellman SH. Crystallographic characterization of helical secondary structures in 2:1 and 1:2 α/β-peptides. J Am Chem Soc. 2009;131:2917–2924. doi: 10.1021/ja808168y. [DOI] [PubMed] [Google Scholar]
- 31.Jakobsche CE, Choudhary A, Raines RT, Miller SJ. n→π* Interaction and n)(π Pauli repulsion are antagonistic for protein stability. J Am Chem Soc. 2010;132:6651–6653. doi: 10.1021/ja100931y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esposito L, Vitagliano L, Zagari A, Mazzarella L. Pyramidalization of backbone carbonyl carbon atoms in proteins. Protein Sci. 2000;9:2038–2042. [PMC free article] [PubMed] [Google Scholar]
- 33.Lario PI, Vrielink A. Atomic resolution density maps reveal secondary structure dependent differences in electronic distribution. J Am Chem Soc. 2003;125:12787–12794. doi: 10.1021/ja0289954. [DOI] [PubMed] [Google Scholar]
- 34.Perczel A, Angyan JG, Kajtar M, Viviani W, Rivail JL, Marcoccia JF, Csizmadia IG. Peptide models. 1. Topology of selected peptide conformational potential energy surfaces (glycine and alanine derivatives) J Am Chem Soc. 1991;113:6256–6265. [Google Scholar]
- 35.Yang A-S, Honig B. Free energy determinants of secondary structure formation: I. α-helices. J Mol Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- 36.Cantor CR, Schimmel PR. Biophysical chemistry: Part III: the behavior of biological macromolecules. New York: Freeman and Company; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


