Abstract
Few proteins have had such a strong impact on a field as the lac repressor and λ repressor have had in Molecular Biology In bacteria, the genes required for lactose utilization are negatively regulated; the lac repressor binds to an upstream operator blocking transcription of the enzymes necessary for lactose utilization. A similar switch regulates the virus life cycle; λ repressor binds to an operator site and blocks transcription of the phage genes necessary for lytic development. It is now 50 years since Jacob and Monod first proposed a model for gene regulation, which survives essentially unchanged in contemporary textbooks1. This model provides a cogent depiction of how a set of genes can be coordinately transcribed in response to environmental conditions and regulates metabolic events in the cell. A historical perspective is presented that illustrates the role these two repressor molecules played and their contribution to our understanding of gene regulation.
Introduction
The discovery of the operon marked a pivotal point in science - the discovery not only provided a model for gene regulation, but also established a new era in science with the emergence of molecular biology. By the 1940's, it had been established that bacteria metabolize sugars for energy production. Most bacteria are versatile and can meet their energy demands by oxidizing a variety of sugars. Glucose, however, is the preferred sugar source, and it is used almost exclusively as an energy source even if other sugars are present. Only when the glucose supplies are exhausted will alternate sugars, such as lactose, be taken up and oxidized. This ability of the bacteria to switch from one metabolite to another was described by Jacques Monod as diauxic growth,2 a pattern observed when metabolites are used sequentially rather than simultaneously. Seeking to understand the molecular basis of this phenomenon provides the backdrop for what became the paradigm for gene regulation.
Monod developed his ideas about gene regulation in response to a prevailing hypothesis of the day that tried to describe diauxic growth as a consequence of “enzymatic adaptation”, i.e. the conversion of a preexisting enzyme from a non-functional to a functional form in response to particular metabolites. The ability of the enzyme to adapt was believed to benefit the organism, so these enzymes were called “adaptive”. Fascinated by the concept of diauxic growth, Leo Szilard hypothesized that constitutive enzyme synthesis was not necessarily stimulated by the presence of some inducer; but could also be due to the absence of product, which he described as a classic case of feedback control3. As a consequence, Szilard, Monod and others abandoned the term “enzyme adaptation” in favor of “enzyme induction.” This prompted Monod to identify and characterize the enzyme that splits the disaccharide lactose into galactose and glucose. By studying the kinetics of this enzyme's induction, Monod found that the addition of a metabolite to E. coli cells actually increased the production of the enzyme, now called β-galactosidase. 2; 4 Over the next several years, Monod explored the biochemistry and genetics of lactose metabolism. He determined that the natural inducer of β-galactosidase is allolactose, an analog of lactose that is created by a side reaction of this enzyme. Several effector molecules were synthesized that mimic the natural inducer and affect the synthesis of β-galactosidase. A particularly effective gratuitous inducer is 1-isopropyl-β-D-thiogalactoside (IPTG). Monod also identified bacterial strains that altered the synthesis of β-galactosidase. Many altered strains abolished the ability of the bacteria to make an active enzyme, while others changed the inducible character of enzyme synthesis by constitutively producing β-galactosidase, i.e. the de novo synthesis of the enzyme proceeded in the absence of an inducer. The fundamental questions that confronted Monod were: what is responsible for inducible and constitutive enzyme synthesis; how do inducers increase the production of β-galactosidase; and how does this relate to diauxie?
While Monod was focusing on lactose metabolism in one corridor of the Pasteur Institute, Andre Lwoff was exploring the mechanism of phage development down the hall. Lwoff noted that in some instances a virus can infect its host and replicate, producing hundreds of viral progeny. While at other times, the virus takes up residence in the host, presumably integrating its DNA into the host chromosome, establishing a prophage. Under normal conditions, these lysogenic phage are highly stable and replicates passively along with the bacteria. However, in response to specific environmental stimuli or stress, the virus can switch from lysogenic to lytic development in a process referred to as prophage induction.5 Lwoff wanted to determine how phage can live quiescently with its host and then reappear when confronted with environmental change. Although it was not readily apparent at the time, the fundamental question being asked by Lwoff was in fact similar to that being asked by Monod: how are cellular events turned on or off?
Francois Jacob entered Lwoff's laboratory to work on phage development and the mechanism of lysogeny. Jacob with Elie Wollman exploited the recently discovered phenomenon of bacterial conjugation to explore the mechanism of prophage induction.6 When a chromosome bearing a λ prophage enters a nonlysogenic recipient cell, half of the bacteria lyse and produced phage particles. In contrast, when a nonlysogenic donor is crossed with a lysogenic recipient, none of the bacteria release viral progeny. Jacob and Wollman concluded that the immunity of the lysogenic bacteria is caused by a cytoplasmic factor.7
Bacterial conjugation also proved useful for exploring bacterial functions and the genetics of lactose metabolism. Having isolated a number of mutants that affected the production of β-galactosidase, Monod reckoned that mutants could be crossed in various combinations and the relationship between the inducible and constitutive nature of enzyme synthesis could be determined. Published in 1959, the PaJaMa experiments (named for Pardee,Jacob,and M(a)onod) demonstrated that inducers function by relieving inhibition, consistent with the hypothesis of Szilard8. Moreover, there appeared to be molecules in the cytosol that are responsible for regulating the expression of the structural genes. As a consequence, the formerly accepted “induction model” of enzyme synthesis was cast aside in favor of a “repression model”.
Prophage induction and galactosidase production are at first glance two unrelated phenomena, yet both processes could be controlled in fundamentally similar ways. Jacob recognized that the synthesis of the genes of the lac system and the synthesis of genes required for lytic development might be regulated by a common mechanism. In both systems, a molecule in the cell appears to block gene expression and this repression could be reversed by a signal, either a metabolite in the case of lac or UV irradiation in the case of λ. These cytosolic molecules were called “repressors”.
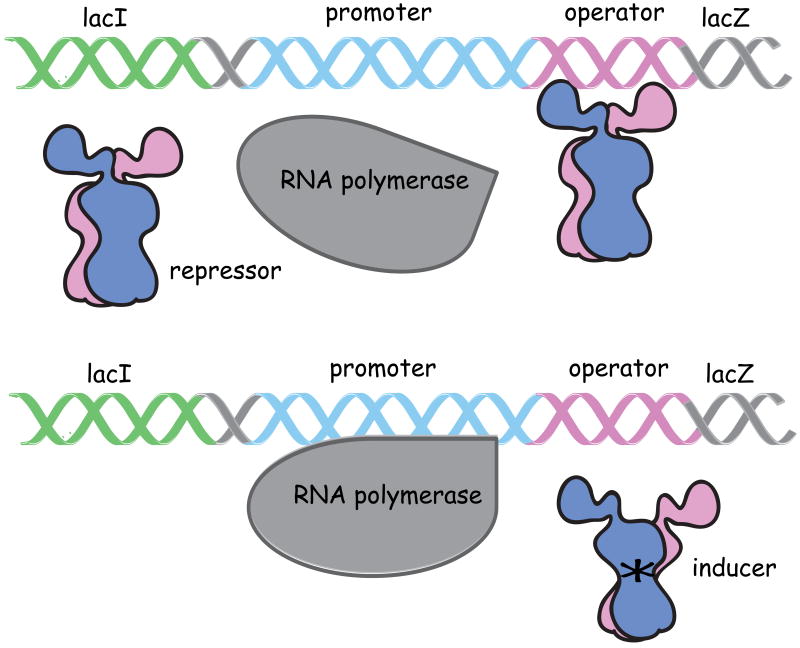
In 1961, JMB published Jacob and Monod's manuscript entitled “Genetic Regulatory Mechanisms in the Synthesis of Proteins” that outlined how a set of genes might be coordinately regulated, providing a model for gene regulation1. Jacob and Monod described an operon as a simple control circuit where a collection of structural genes could be regulated in a coordinated fashion. These structural genes code for a group of proteins that are responsible for a particular metabolic function. Regulating the ensemble of proteins requires a master switch, called the repressor molecule, which is produced from a regulator gene (lacI coding for the lac repressor and the cI gene for λ repressor). When the regulator binds to a specific site on the genome, an operator, it blocks transcription of the structural genes. The operator, a small fragment of DNA, was originally identified by Monod as cis-acting constitutive (oc) mutants. These mutants were located between the end of the gene that codes for the repressor (lacI) and the beginning of the structural genes lacZ, lacY, and lacA, which code for three proteins involved in lactose metabolism: β-galactosidase, lac permease, and a transacetylase. Repression is relieved when a regulatory protein binds a metabolite or an inducer, which lowers or eliminates the affinity of the repressor for the operator (Figure 1). A similar model could be used to explain the life cycle of the phage. A repressor molecule, produced from a cI regulator gene, binds to an operator site and blocks transcription of the phage genes necessary for lytic development. In this instance, repression is relieved by an environmental stimulus, such as UV irradiation rather than a metabolite. The model for gene regulation proposed by Jacob and Monod was met with criticism, for it was equally plausible that the whole operator is transcribed as part of the mRNA and the repressor would regulate translation of the message into protein. Moreover, the molecular composition of the repressor molecule was unknown: it could be either protein or nucleic acid. To get to the answer, the regulatory gene products had to be isolated and their mode of action established, but isolating these repressors would not be a simple feat.
Figure 1.
Diagram of the operon model of Jacob and Monod. The repressor molecule which was produced by the LacI gene binds to operator site and blocks transcription of a set of structural genes (Lac Z) that follows the operator site. The repressor when bound to an inducer (marked by a yellow star) decreases the affinity of the repressor for operator and allows transcription of the set of structural genes.
Isolation and Characterization of the Repressors and Their Operators
The number of repressor molecules in the bacterial cell is small and constitutes a minor fraction of the total cellular protein, thus making it difficult to purify. Nevertheless, both lac and λ repressors were purified by the mid 60's, albeit using very different approaches. Benno Muller Hill, working with Walter Gilbert, identified a potential lac repressor protein by isolating cell fractions that bound a radioactively labeled inducer.9 Since there were no known inducer molecules of λ repressor, Mark Ptashne developed an alternate strategy.10 He differentially labeled two cultures of bacteria, one infected with a λ strain that produced a functional repressor and another infected with a λ strain that was incapable of making repressor. The cell cultures were mixed together, sonicated, and the soluble extract was fractionated. Only one fraction appeared that was singly labeled and presumably contained the repressor molecule.
Once these proteins were isolated, the critical experiments could be performed. Ptashne demonstrated that the selectively labeled protein fraction contained a molecule that co-sediments with the wild type λ operator DNA but not with DNA that contains a defective operator, a λ virulent mutation.11 Similarly, Gilbert and Muller-Hill observed that the isolated fraction, which contained a molecule that binds to an inducer, co-sediments with lac operator DNA but with the DNA containing an operator constitutive mutation. Moreover, the addition of a gratuitous inducer prevented this molecule from sedimenting with the operator DNA.12 Gilbert and Ptashne had isolated the two repressors molecules, demonstrating that at least part of the operon model proposed by Jacob and Monod was indeed correct. Attention now focused on exploring the finer details of gene regulation.
The first step was to study the biochemical properties of these proteins. However, the amount of protein initially recovered was too small for further characterization. Using a variety of clever genetic manipulations, Muller-Hill created altered bacterial strains that produced large amounts of lac repressor.13 The overexpressed protein was purified and used initially for amino acid sequencing 14; 15 and structural characterization.16 The lac repressor contains 360 amino acids that self-associate, forming a tetrameric assembly. Each repressor monomer has a modular structure; when treated with trypsin it is cleaved into two domains: a small N-terminal domain, which was referred to as the headpiece, and a larger C-terminal fragment, called the core. The headpiece recognizes the operator and the core binds to effector ligands. 16 Once the gene for λ repressor was cloned and over expressed, the repressor protein could also be purified and characterized. At 236 amino acids, λ repressor is smaller than lac repressor, but it also self-associates, forming higher order oligomers. Each λ repressor monomer is composed of two domains; like the lac repressor, these domains are tethered by a protease sensitive linker. 17 The amino terminal domain binds to DNA and the carboxy-terminal domains allow the λ repressor to self-associate.
Attention not only focused on characterizing the repressor, but also on their site of action, the operator. Cis-acting constitutive (Oc) mutations were originally used to map the location of the lac operator between the end of the LacI gene and the beginning of the LacZ gene.18 Roughly ten years later, the operator site was isolated19 allowing Gilbert and Maxam20 to sequence a 27-base pair section of the double stranded DNA. The lac operator sequence is pseudo-symmetric, possessing an approximate dyad axis about a central G·C base pair. The minimal size of operator required for specific binding was later shown to be only 17 base pairs and corresponds to the center of the originally found 27-base pair sequence.21 The regulatory region on the phage genome is considerably more complex; two operator regions exist, OR and OL, that are separated by ∼2.4 kilo base pairs.22 Once the operators were isolated and sequenced, Ptashne established that OR and OL are both roughly 85 base pairs and contain three DNA binding sites that occur in tandem. The OR region consists of operator sites OR1, OR2, and OR3 with a similar arrangement of three operators for OL. All six operators are ∼17 base pairs and contain similar but not identical sequences with a pseudo two-fold axes of symmetry.23
How do these repressors recognize a specific operator sequence of DNA? Seeman et al. suggested that sequence-specific DNA recognition could be achieved by having the amino acid side chains hydrogen bond to the exposed bases in the major and minor grooves of the DNA.24 Unfortunately, despite considerable effort, the three-dimensional architecture of these molecules remained elusive for many years. As a consequence, a fallback position was taken and attention was focused on the structures of the isolated DNA binding domains. The amino terminal domain (NTD) of λ repressor adopts a globular structure that contains 5 α-helices. 25 A bi-helical unit, now referred to as a ‘helix-turn-helix’ motif (HTH), protrudes from the surface of the molecule. In the crystals, the NTD self-associates, albeit weakly, forming a dimer. As a consequence, two HTH motifs, one from each monomer, are oriented to potentially fit into successive grooves of the operator. Later, the crystal structure of the NTD bound to a synthetic operator verified that the amino terminal domain of λ repressor uses the HTH motif to recognize a 17 base pair nucleotide containing an operator sequence. 26; 27 The two-fold axis of the protein dimer is aligned with the pseudo two-fold axis of the operator. Amino acid side chains on the HTH motif recognize the operator by forming hydrogen bonds to the bases in the major grove of each half site, and recognition is achieved by a handful of specific interactions. The headpiece domain of lac repressor bound to DNA was determined in solution. It is a small, compact globular structure that contains a HTH motif.28 As was seen with λ repressor, the side-chains that protrude from the HTH motif make sequence specific interactions with the two half sites of the operator. In addition, hydrophobic interactions contribute to the specificity and the binding affinity of the complex. As it turned out, this HTH motif is a characteristic of many DNA binding proteins.29
After seeing how the repressor recognizes its operator, attention turn to the C-terminal domains (CTD) of the repressors and the regulation of these molecular switches. The core domain of the lac repressor is composed of two subdomains that each contain a six stranded parallel β-sheet sandwiched between four α-helices.30 The quaternary structure of the repressor is unique; the molecule self-associates into an unusual tetramer that appears roughly as a V-shaped dimer of dimers. The CTD of λ repressor crystallized readily and tends to self-associate, forming dimers as well as tetramers and octamers.31 A monomeric unit folds into a highly twisted 7-stranded anti-parallel β-sheet followed by a single turn of 310-helix32
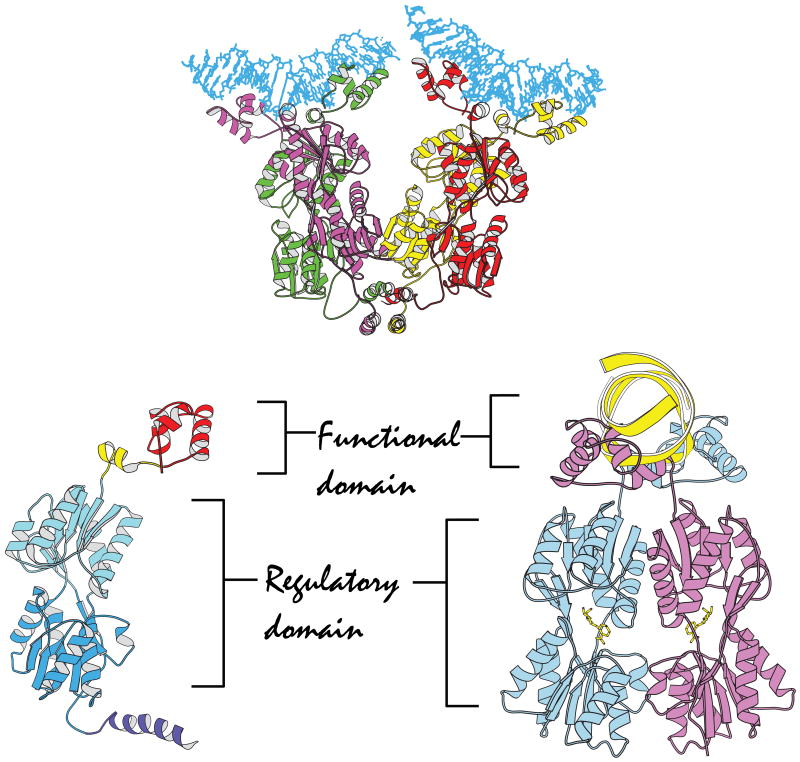
The structures of these domains provided only fragmental information as to how these repressors function. It was therefore imperative to determine the architecture of the intact molecules as well as these molecules bound to various ligands, effectors and operators. Eventually, the structure of the intact lac repressor was solved both in the presence and absence of its operator.33 The protein scaffold of the intact lac repressor contains four discrete functional units: the headpiece, a hinge region, a sugar binding domain, and a C-terminal helix33 (Figure 2). The headpiece, which binds to the DNA, is connected to the core by a stretch of polypeptide chain called the hinge region. In the absence of DNA, the hinge region is disordered and the headpiece domain is highly mobile. In contrast, when the headpiece is bound to the operator, the hinge region adopts a helical conformation that also makes sequence specific interactions with the operator and recognizes bases in the minor groove. The hinge helices pry open the minor grove and dramatically bend the DNA; deforming the center of the operator from a canonical B form conformation. The significance of this deformation and bending, however, remains unclear. The core of the repressor consists of two structural domains; the C-terminal subdomain is responsible for dimerization while the N-terminal subdomain performs the important role of communicating to the headpiece whether a metabolite is present. The lac repressor terminates with an α-helix that self-associates, forming a four helix bundle to create the biologically observed tetrameric repressor.
Figure 2.
A ribbon diagram of the quaternary structure of the lac repressor complexed to DNA. Lac repressor is a tetrameric structure where each monomer is drawn in color. Each dimeric repressor binds to a 21 base pair duplex deoxyoligonucleotides. The observed lac tetramer appears to be a tethered dimer of dimers. Below is a view of the lac repressor monomer from the complex with DNA. Four functional domains occur in repressor monomer: starting from the N-terminus (1) the DNA binding domain or headpiece (residues 1--45) is colored red; (2) the hinge region (residues 46--62) is yellow; (3) the ligand binding domain or core (residues 63--329) which has distinct N- and C-terminal subdomains are colored in two shades of blue; and (4) the tetramerization helix (residues 340--357) is purple. The monomers associate into a functional dimer that binds to the operator. Dimer is shown bound to the anti-inducer ONPF
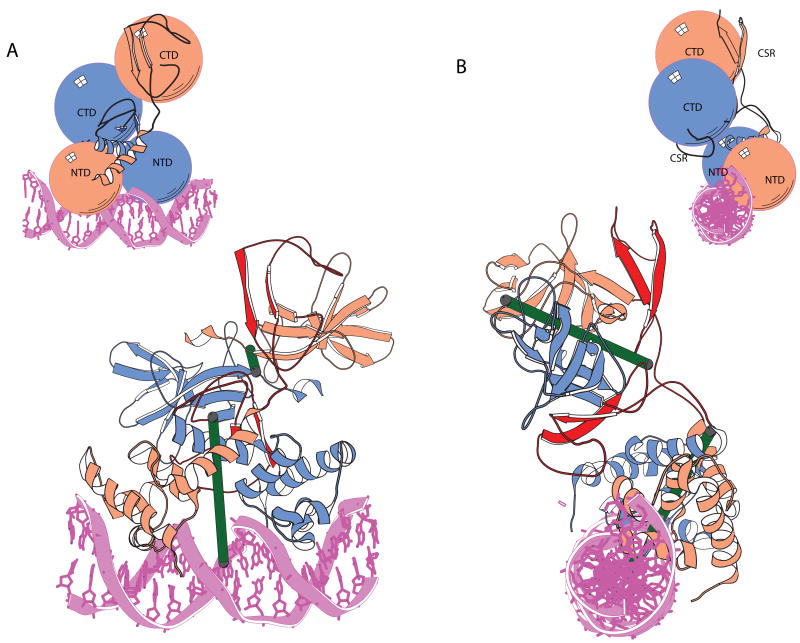
The full-length repressor λ repressor bound to its operator DNA confirmed that the two distinct domains of known structure are tethered by a short segment of polypeptide.34 (Figure 3). Although this linker was previously assumed to be flexible and void of structure, it is an integral part of the CTD, forming a pair of antiparallel β strands that drape across the surface of this domain. The most remarkable aspect of the intact dimeric structure is its lack of global symmetry. The dimeric repressor does not have a single axis of symmetry; the two-fold axes that relate the NTD and CTD dimers are substantially skewed and are nearly orthogonal to one another.
FIGURE 3.
Orthogonal views of the λ repressor-operator complex. One subunit of the dimer (salmon colored) adopts an extended conformation and the other adopts a compact conformation (blue). The radii of the spheres were determined on the basis of the actual size of the NTD and CTD. The NTD and CTD are connected by a pair of antiparallel β-strands and a connecting loop that contains the site of RecA-mediated autocleavage are highlighted in red. The DNA is shown in pink. The symmetry axes of the individual domains are shown as green rods, and are nearly perpendicular to one another.
Lambda-Cooperativity and Gene Regulation from a Structual Perspective
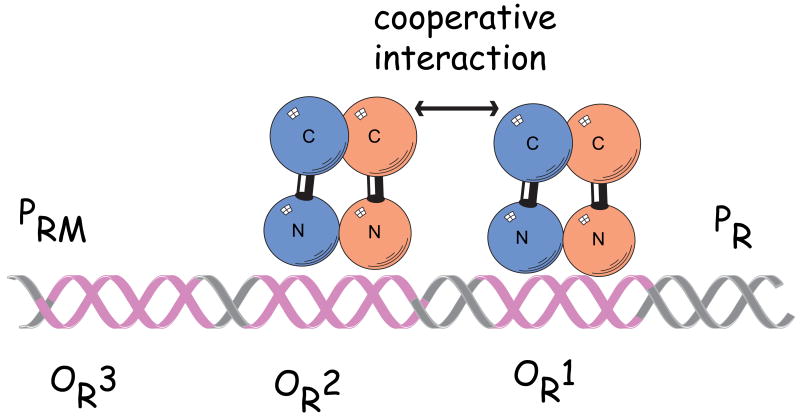
Regulation of the virus life cycle requires a control mechanism that is more elaborate than a simple bi-stable switch. At the heart of the system is the repressor protein, which unexpectedly functions as both a repressor and an activator of transcription. When λ repressor binds to its operators, it not only blocks transcription of the viral genes necessary for lytic growth but also stimulates transcription of its own gene. Figure 4 is a schematic representation of the right operator region, illustrating the binding of the λ repressor to operators in a λ lysogen.
FIGURE 4.
The interactions of λ repressor at the right operator regions in a λ lysogen. The repressor dimers bound cooperatively to adjacent operator sites in OR and OL. The repressor monomer are shown in blue and cyan. Each subunit of the dimer consists of an N-terminal DNA-binding domain (N), a C-terminal oligomerization domain(C), and a linker region (black) connecting the two. The dimer pair bound cooperatively at OR1 and OR2 represses transcription from PR and the dimer bound at OR2 also activates transcription from PRM. Dimer pair bound also binds cooperatively at OL1 and OL2 represses transcription from PL. (not shown)
When λ repressor binds to adjacent sites on the DNA the proteins interact cooperatively. Cooperativity is a consequence of a linked equilibrium, which manifests itself by a left shift of a binding isotherm. The affinity of the repressor for OR1 is greater than OR2 or OR3, as a consequence cooperativity facilitates or increases the binding affinity of a second dimer to the lower-affinity site OR2.35 Binding to OR2 increases the level of repression of PR. The physiological consequence of the cooperative interaction is to decrease in the amount of repressor needed to maintain a lysogen.36 Cooperativity is not only essential for lysogeny, it also amplifies the sensitivity of the switch that controls prophage induction. 37 Mutations that disrupt cooperativity, while having no effect on the binding of a dimer to a single operator site, were found within the CTD and lie near the interface formed when two repressor dimers associate into a tetramer.38;39 A structural model of cooperativity (Figure 5) was created by superimposing the intact dimer on to the structure of the CTD tetramer. 34 The model illustrates that when a cooperativity complex is established the repressor dimers bind to the lateral sides of the operator while the CTD's are positioned medially.
FIGURE 5.
Structural modeling depicting interaction of adjacently bound λ repressor dimers. (a) The pairwise cooperativity complex was modeled by superimposing the CTDs of two intact DNA-bound dimers onto the structure of the CTD tetramer. This superimposition brings the DNA of one repressor-operator complex into rough alignment with that of the second complex. Also shown are the C-terminal α-helices of the NTDs, which mediate formation of a weak NTD dimer. (b) The pairwise cooperativity complex is rotated by 90° to illustrate that the NTDs bind on opposite sides of the DNA with the CTD tetramer below. (c) The model of the cooperativity complex is rotated by 180° with respect to A in order to view the cooperativity interface.
The repressor concentration ultimately dictates whether the wild type phage will be maintained as a lysogen or enters lytic growth. Binding of a repressor at OR2 not only increases repression of PR, it also stimulates transcription of the cI gene, which codes for the λ repressor, by increasing the affinity of RNA polymerase for the promoter, PRM.40 Mutant repressors that bind to the DNA normally, but fail to activate transcription, have amino acid substitution on the HTH motif albeit distinct from amino acids required for operator binding 41. By activating transcription of its own gene the phage ensures there is sufficient repressor for maintaining the lysogen. Of course an excessive amount of repressor would impede the ability of the phage to enter lytic growth. Consequently, the repressor maintains a steady state level of its own gene product by binding to the operator OR3 and blocking transcription from PRM. This auto-regulation requires communication between OR and OL.42
Cooperativity at a distance requires communication between the left and right operators. During lysogenic growth, binding of λ repressor to OR1 and OR2 and to OL1 and OL2 represses transcription of the early lytic genes by physically blocking access of the polymerase to the promoter. A cooperatively bound pair of dimers at OR1 and OR2 can interact with a cooperatively bound pair of dimers at OL1 and OL2 by forming a higher-ordered octameric complex. When this octamer forms, the intervening 2.4 kilo-bases of DNA form a loop that juxtaposes OL3 and OR3. The close proximity of OL3 and OR3 promotes a pair of dimers to bind cooperatively to these two operators. As a consequence, OR3 is occupied at lower concentrations of repressor than would be predicted simply by its intrinsic affinity for this operator.42 Mutation of OL3, in the context of an otherwise wild-type λ phage, results in a substantial defect in prophage induction due to loss of negative autoregulation 42. Figure 6A is a model of cooperativity at a distance that was created by superimposing the dimeric repressor onto the crystal structure of the CTD octamer. This assembly brings the operators OR3 and OL3 into close proximity. Figure 6B illustrates how two repressor dimers can associate and negatively regulate transcription of PRM. The model was created by superimposing the intact dimer onto the tetrameric CTD.
FIGURE 6.
A model depicting higher-order looped complex that facilitates negative autoregulation. The cooperatively bound λ repressor dimer pair at OR1 and OR2 interacts with the cooperatively bound repressor at OL1 and OL2, forming an octameric complex that loops 2.4 kb of intervening DNA. (a) The model was created by superimposing the CTDs of four DNA-bound dimers onto the structure of the CTD octamer. In this case, the superimposition was performed by first constructing two pairwise cooperativity complexes. (b). Formation of the looped complex juxtaposes OL3 and OR3, allowing another pair of repressor dimers to bind cooperatively to these sites. A model depicting interaction of nonadjacently bound repressor dimers shows how two repressor dimers might look when bound cooperatively to OL3 and OR3 in the context of the looped complex.
Prophage induction or the entry into lytic growth occurs with the destruction of the repressor, allowing RNA polymerase access to the promoters PR and PL and transcription of the lytic genes. The repressor self-destructs by an auto-cleavage reaction.43 The cleavage hydrolyzes a peptide bond, which physically separates the NTD from the CTD.44 The amino acids that perform this catalysis are a pair of conserved residues, serine and lysine, that are located within a shallow groove on the surface of the CTD. The mechanism for the self-cleavage is analogous to the serine proteases 45; the lysine residue, in an unprotonated form, removes a proton from the serine residue, thereby activating a nucleophile that attacks the carbonyl carbon atom of the peptide bond in the linker region. 43 Quite unexpectedly, these catalytic amino acids are located at the cooperativity interface. Consequently, the quaternary structure of the repressor must be disrupted for the active site residues to be exposed and find its substrate. The cleavage reaction occurs in response to stress. DNA damage activates the SOS response and the cellular RecA protein.46 How RecA facilitates the cleavage reaction is not well understood. Mutations that inhibit dimerization have an increased susceptibility to RecA-mediated cleavage and conversely repressor that form more stable dimers are resistant to RecA mediated cleavage. It appears that RecA may associate with the repressor and alter the dimer- dimer equilibrium. Mutant repressors that prevent the RecA-mediated cleavage render the lysogen non-inducible.47
Lac - Allostery and Gene Regulation from a Structual Perspective
At first glance the regulatory circuit that controls the lac operon appears to be a simple bi-phasic system; binding of lac repressor to its operator physically denies RNA polymerase access to a promoter. In 1963 Monod, Changeaux, and Jacob published in JMB a model that describes how metabolites might play a pivotal role in gene regulation. 48 The fundamental tenet of their model is that activity of a protein can be modulated by altering its conformation. Effector molecules (metabolites) when bound to a protein have the potential to alter the structure and consequently the protein's ability to perform a specific function at a distal site. Building on this model, Monod, Wyman, and Changeaux published two years later a rigorous description for how a conformational transition in multimeric proteins alters the ability of the molecule to perform a given function. 49 Although this theory of allosteric transitions, which became known as the MWC model, did not explicitly mention the lac operon, it would be safe to assume that they anticipated that the lac operon would be allosterically regulated.
The repressor adopts two distinct conformational states; one bound to the operator and another when bound to inducer. This was first illustrated by observing that crystals of the repressor - operator complex when exposed to an allosteric inducer immediately shattered.50 This is, of course, reminiscent of the shattering of crystals of deoxyhemoglobin when exposed to air. 51 The two distinct conformational states of the repressor, R and R* (designated as R and T by MWC), are related by an equilibrium constant KRR*. The linked equilibria that describe the binding of the repressor to its operator and to the inducer are illustrated in Figure 7. There are 12 repressor species that contribute to the total repressor concentration. The amount of repressor that is able to bind to the operator, R, depends upon the conformational equilibrium, the inducer concentration, and the inducer binding affinities to the R and R* conformation. These three parameters are responsible of establishing the relative amount of active and inactive repressor. For the lac repressor, the conformational equilibrium constant is close to unity and in solution the two conformations are readily interchangeable with the inactive conformation (R*) being modestly favored over the active conformation. 52
FIGURE 7.

A diagram illustrating the linked equibria. Kro is the equilibrium constant that describes the binding of the repressor (R) to the operator (O). The equilibrium constant Krr* is an inherent property of the repressor and dictates the ratio of the repressor in the active and inactive conformation. The inducer binds to the repressor with affinities dictated by KIr and Kir*; the binding of the inducer to these two conformational states. Below is a schematic representation of the dimeric repressor in the active and the inactive conformations.
The role of the inducer molecule is to shift the apparent equilibrium, KRR*, driving the repressor toward the inactive or R* conformation. At high concentrations of inducer, the apparent value of KRR* increases by several orders of magnitude and a larger fraction of the total repressor concentration adopts the R* conformation; incapable of binding to the operator. Inducers shift the apparent equilibrium by forming an intricate hydrogen bonding network that cross links the NTD and the CTD. Figure 8 illustrates the extensive hydrogen bonding between the inducer and the repressor. All of the oxygen atoms of the galactose ring form hydrogen bonds either by directly bonding to the amino acid side-chains or mediated through water molecules. Anti-inducers bind to the repressor like inducers, but form a different hydrogen bonding network. As seen in Figure 9 the anti-inducer binds to the repressor in the presence and the absence of operator by making alternate interactions. In general, inducers have a C6 hydroxyl, which is used to establish a water-mediated hydrogen bonding network. While this hydroxyl is a necessary prerequisite for induction, it is not sufficient. The functional groups attached at the C1 position also play a crucial role in discriminating inducers from non-inducers and in determining the potency of the inducer. In order for a galactoside containing the C6 hydroxyl to be a potent inducer, it must have a C1 substituent group which sufficiently increases the binding energy while maintaining conformational flexibility. 53
FIGURE 8.
Illustrates the binding of the inducer (IPTG) to the repressor. The dark blue portions of the structure correspond to residues in the C-terminal domain while the light blue corresponds to the N-terminal portion of the structure. The inducer and the water mediate hydrogen bonds that stabilize this conformation of the repressor.
FIGURE 9.
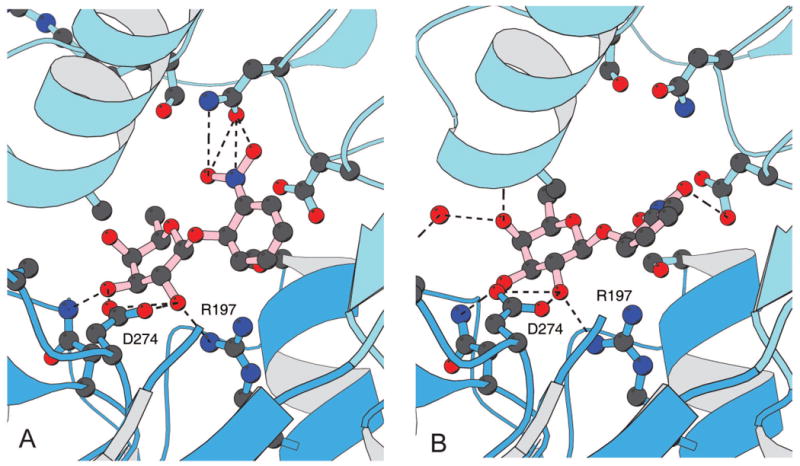
The binding of the anti-inducer, ONPF, the repressor in the absence and the presence of the operator. (a) In the presence of DNA, the anti-inducer forms a ternary complex with the repressor primarily by establishing hydrogen bonds between the O2 and O3 hydroxyls of the fucoside and residues R197, N246 and D274 of the repressor and the nitrophenyl group hydrogen bonds to N146. (b) In absence of DNA, the anti-inducer is also bound to the repressor by hydrogen bonding to the fucoside but the nitrophenyl group does not appear to be ordered or adopt the same conformation.
The allosteric signal is transmitted through the dimer interface. Over 4,000 single amino acid substitutions of the repressor were created and some of which resulted in mutant repressor molecules, classified by an Isphenotype, that bind to operator DNA with wild type affinity but are incapable of induction.54. Over half of these ls point mutations appear in the effector binding pocket and directly alter the affinity of the inducer for the repressor. Other mutations that produce the Is phenotype cluster at the monomer-monomer interface between the N-terminal subdomains (Figure 10). From the position of these mutations, it appears that a signal is transmitted from the effector site, through the dimer interface to the hinge helices and the DNA binding domains. The inducer stabilizes a conformation of the repressor that precludes the headpiece and the hinge helices from efficiently binding to the operator.
FIGURE 10.
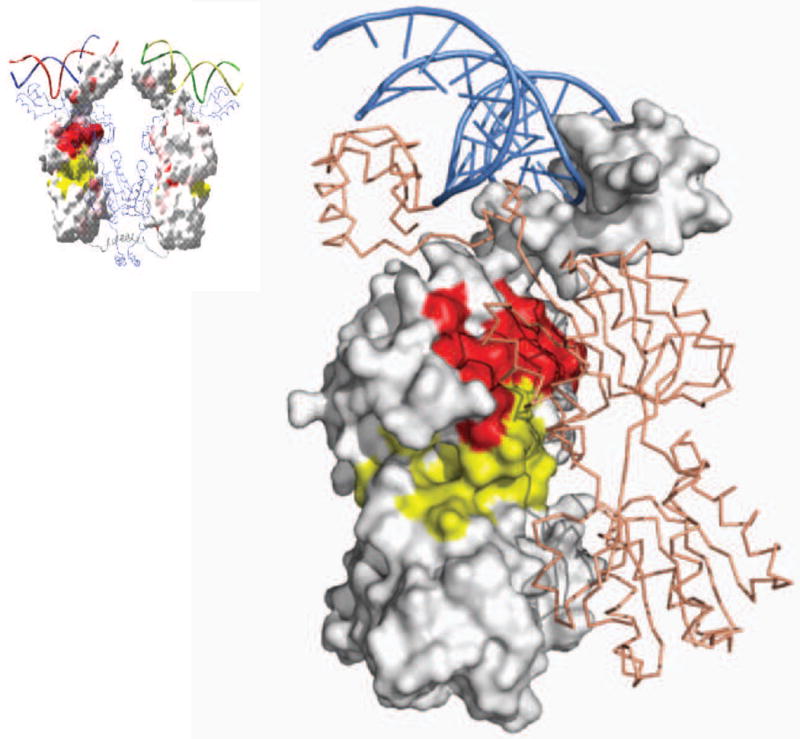
A summary of the genetic data for mutations that produce the Is phenotype. Each amino acid replacement was tested for β-galactosidase activity in the presence of IPTG. The genetic data is mapped on the surface structure of lac repressor. Locations of mutants are shown in yellow and red. Yellow areas are within 8 Å of the IPTG molecule.
How can a bi-phasic switch produce levels of β-galactosidase that increase linearly as a function of inducer concentration? This was an issue that greatly concerned Monod. 3 The answer was arrived at by Jacob after watching one of his sons playing with his electric trains. An on-or-off switch can function like a rheostat by oscillating between the on and off states with different frequencies.3 The fraction of time the promoter is accessible and the duration of time it is free are both relevant for effective transcription. The kinetics of transcriptional initiation of the lac operon have been well characterized. 55 RNA polymerase binds to the promoter region forming a ‘closed’ complex, which then undergoes an ‘isomerization’ to establish the open (strand separated) initiating complex; a process that takes ∼60 seconds. In the absence of an inducer, the repressor binds to the operator and is released in a stochastic fashion. On average the amount of time the repressor is bound to the operator greatly exceeds the amount of time is free. Based on the kinetic data of Riggs et al 56, the operator and the repressor form a stable complex that has a lifetime of ∼1600 seconds. Of course the complex dissociates but the operator is only vacant for ∼6 seconds before the two re-associate. As a consequence there is not enough time for the polymerase to bind to the promoter and initiate transcription and the switch is effectively turned off.
The inducer decreases the affinity of the repressor for the operator by three orders of magnitude, which could be a consequence of either decreasing the on rate, an increase in the off rate, or a combination of the two. If the inducer only altered the dissociation rate of the repressor, then the mean occupancy time would be significantly reduced; however the mean time the operator is free of repressor would be unchanged, and therefore on average the time frame would still be too short for transcription to initiate. In contrast, if the inducer altered the equilibrium by depressing the on rate, then the average time the DNA is free of repressor would increase sufficiently, allowing transcription to initiate. At high inducer concentrations- , the off rate was estimated to increase ∼10-fold 57, which implies that there is a substantial decrease in the on rate. Altering the association rate rather than the dissociation rate allows polymerase to initiate transcription and produce levels of β-galactosidase that increase linearly as a function of inducer concentration.
Cooperativity at a distance may also play a role in regulating the lac operon. On the bacterial genome there are two stretches of DNA resemble the lac operator; one of the sequences, O2, is 401 base pairs downstream of the primary operator, and the other, O3, is 92 base pairs upstream of O1. Although these two sequences resemble the primary operator, constitutive mutations were never found. On the other hand, these pseudo-operator sites increase repression of the operon.58; 59 The tetrameric repressor, in principal, is ideally suited to bind simultaneously two of these operators and create repression loops that may play a role in regulation.60 Models for looping the DNA are consistent with the architecture of the tetrameric lac repressor which may act like a clamp; bringing two operators that are separated in linear sequence close together63.
Conclusion
Regulating gene expression is a fundamental process of life and is essential for controlling metabolic events, development, and disease. Although the specific details of regulation can be extraordinarily diverse and complex, the novel concepts put forward by Jacob and Monod were revolutionary and have served as the foundation for understanding gene regulation. Over the past half century, the details of the operon have been elucidated using genetic, biochemical, and structural techniques, yet the principles that were originally put forward have only been slightly altered or refined. The structures of these two repressors provide detailed molecular models that help us understand how proteins recognize DNA and how the synthesis of proteins is regulated.
Many transcription regulators, in both prokaryotes and eukaryotes, activate and repress the transcription of genes by recruitment.64 The structures of λ repressor beautifully illustrate how a protein bound to one site on the DNA can recruit a second protein to bind another site. When λ repressor binds to an operator, a second repressor is recruited to an adjacent site. The recruitment is driven by adhesive forces that are created by protein-protein interactions. A repressor bound to the adjacent operator can then recruit polymerase to bind its promoter. The same weak interactions that are responsible of bringing repressor molecules together at adjacent sites are also responsible for recruitment of repressors bound to operators that are widely spaced along the genome. Recruitment of transcriptional regulators is essential for maintaining the functionality of this switch. Both positive and negative regulation is achieved by simple adhesive interactions. Thermodynamically, recruitment and cooperativity are a consequence of linked equilibrium.
The diauxic growth of bacterial cultures Monod described in his doctoral thesis could not be explained by the initial operon model. 65 Not included in the original model is that transcription of the operon is positively activated by a cyclic AMP-dependent catabolite gene regulator protein (CAP).66 In glucose starved cells, the level of cyclic AMP increases dramatically which in turn binds to CAP. Once CAP is activated, the protein binds to the DNA; increasing the affinity of RNA polymerase for its promoter and transcription of the structural genes of the lac operon. 67 Transcription of the enzymes necessary for lactose metabolism occurs when the glucose levels in the cell are low and lactose is elevated. Although the detailed mechanism for alternate metabolite oxidation is likely to be more complex, to a first approximation this combination of repression and activation can account for the diauxie.
The model of the operon has molded our understanding of gene regulation and opened up new and exciting areas of research. The regulatory components of the lac operon have been used to control transcription of the tyrosinase gene in a mouse. 68 In the absence of the inducer the tyrosinase gene is repressed and the mice have a white coat color. When the mice were fed IPTG in their drinking water, repression of tyrosinase was relieved, resulting in mice with brown eyes and fur. This simple bacterial switch successfully controlled a gene in a mammalian system. In the future it is certainly plausible that specific loci could be switched on and off repeatedly to create reversible models of human disease and normal development. As the mouse is the most widely used experimental system to model human disease and development, the ability to regulate genes could greatly broaden the range of biological questions that can be addressed experimentally. Monod would not have been at all surprised to see lac in mice, as he once wrote, “anything that is true of E. coli must be true of elephants, except more so.”
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacob F, Monod J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Monod J, Paris H, editors. These Doctorat es Sciences. 1942. Recherches sur la croissance des cultures bacteriennes. [Google Scholar]
- 3.Judson HF. The eighth day of creation : makers of the revolution in biology. Simon and Schuster; New York: 1979. [Google Scholar]
- 4.Cohn M. Contributions of studies on the b-galactosidase of E-coli to our understanding of enzyme synthesis. Bacteriol Rev. 1957;274:765–769. doi: 10.1128/br.21.3.140-168.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lwoff A. Lysogeny. Bacteriol Rev. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 7.Jacob F, Wollman E. Spontaneous induction of the development of bacteriophage lambda during genetic recombination in Escherichia coli K12. C R Hebd Seances Acad Sci. 1954;239:317–9. [PubMed] [Google Scholar]
- 8.Pardee AB, Jacob F, Monod J. The genetci control and cytoplasmic expression of inducibility in the synthesis of b-galactosidase in E Coli. J Mol Biol. 1959;1:165–178. [Google Scholar]
- 9.Gilbert W, Muller-Hill B. Isolation of the Lac Repressor. Proc Natl Acad Sci USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptashne M. ISOLATION OF THE lambda PHAGE REPRESSOR. Proc Natl Acad Sci U S A. 1967;57:306–13. doi: 10.1073/pnas.57.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ptashne M. Specific binding of the lambda phage repressor to lambda DNA. Nature. 1967;214:232–4. doi: 10.1038/214232a0. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W, Muller-Hill B. The lac operator is DNA. Proc Natl Acad Sci U S A. 1967;58:2415–21. doi: 10.1073/pnas.58.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller-Hill B, Crapo L, Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968;59:1259–64. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyreuther K, Adler K, Geisler N, Klemm A. The amino-acid sequence of lac repressor. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3576–80. doi: 10.1073/pnas.70.12.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beyreuther K. Revised sequence for the lac repressor. Nature. 1978;274:767. [Google Scholar]
- 16.Geisler N, Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977;16:938–43. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- 17.Pabo CO, Sauer RT, Sturtevant JM, Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979;76:1608–12. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob F, Monod J. On the Regulation of Gene Activity. Cold Spring Harbor Symp Quant Biol. 1961;26:193–211. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 19.Bourgeois S, Riggs AD. The lac repressor-operator interaction. IV. Assay and purification of operator DNA. Biochemical & Biophysical Research Communications. 1970;38:348–54. doi: 10.1016/0006-291x(70)90719-9. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert W, Maxam A. The Nucleotide Sequence of the lac Operator. Proc Natl Acad Sci USA. 1973;70:3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahl CP, Wu R, Stawinsky J, Narang SA. Studies on the lactose operon. Minimal length of the lactose operator sequence for the specific recognition by the lactose repressor. Proc Natl Acad Sci USA. 1977;74:966–970. doi: 10.1073/pnas.74.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ptashne M, Hopkins N. The operators controlled by the lambda phage repressor. Proc Natl Acad Sci U S A. 1968;60:1282–7. doi: 10.1073/pnas.60.4.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniatis T, Ptashne M. Multiple repressor binding at the operators in bacteriophage lambda. Proc Natl Acad Sci U S A. 1973;70:1531–5. doi: 10.1073/pnas.70.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–8. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pabo CO, Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982;298:443–7. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- 26.Beamer LJ, Pabo CO. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992;227:177–96. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- 27.Jordan SR, Pabo CO. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988;242:893–9. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- 28.Chuprina VP, Rullmann JA, Lamerichs RM, van Boom JH, Boelens R, Kaptein R. Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics. Journal of Molecular Biology. 1993;234:446–62. doi: 10.1006/jmbi.1993.1598. [DOI] [PubMed] [Google Scholar]
- 29.Sauer RT, Yocum RR, Doolittle RF, Lewis M, Pabo CO. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982;298:447–51. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- 30.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–7. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 31.Bell CE, Lewis M. Crystal structure of the lambda repressor C-terminal domain octamer. Journal of Molecular Biology. 2001;314:1127–36. doi: 10.1006/jmbi.2000.5196. [DOI] [PubMed] [Google Scholar]
- 32.Bell CE, Frescura P, Hochschild A, Lewis M. Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101:801–11. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- 33.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–54. doi: 10.1126/science.271.5253.1247. see comment. [DOI] [PubMed] [Google Scholar]
- 34.Stayrook S, Jaru-Ampornpan P, Ni J, Hochschild A, Lewis M. Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding. Nature. 2008;452:1022–5. doi: 10.1038/nature06831. [DOI] [PubMed] [Google Scholar]
- 35.Johnson AD, Meyer BJ, Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979;76:5061–5. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson N, Adams C, Youderian P. Genetic selection for mutations that impair the cooperative binding of lambda repressor. Mol Microbiol. 1994;11:567–79. doi: 10.1111/j.1365-2958.1994.tb00337.x. [DOI] [PubMed] [Google Scholar]
- 37.Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci U S A. 1982;79:1129–33. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckett D, Burz DS, Ackers GK, Sauer RT. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry. 1993;32:9073–9. doi: 10.1021/bi00086a012. [DOI] [PubMed] [Google Scholar]
- 39.Whipple FW, Hou EF, Hochschild A. Amino acid-amino acid contacts at the cooperativity interface of the bacteriophage lambda and P22 repressors. Genes Dev. 1998;12:2791–802. doi: 10.1101/gad.12.17.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ptashne M, Backman K, Humayun MZ, Jeffrey A, Maurer R, Meyer B, Sauer RT. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–61. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- 41.Hochschild A, Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–7. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- 42.Dodd IB, Egan JB. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol Microbiol. 2002;45:697–710. doi: 10.1046/j.1365-2958.2002.03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slilaty SN, Little JW. Lysine-156 and serine-119 are required for LexA repressor cleavage: A possible mechanism. Proc Natl Acad Sci U S A. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer RT, Ross MJ, Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J Biol Chem. 1982;257:4458–62. [PubMed] [Google Scholar]
- 45.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–52. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 46.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 47.Gimble FS, Sauer RT. Mutations in bacteriophage lambda repressor that prevent RecA-mediated cleavage. J Bacteriol. 1985;162:147–54. doi: 10.1128/jb.162.1.147-154.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monod J, Changeux JP, Jacob F. Allosteric Proteins and Cellular Control Systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 49.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 50.Pace HC, Lu P, Lewis M. lac repressor: crystallization of intact tetramer and its complexes with inducer and operator DNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1870–3. doi: 10.1073/pnas.87.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haurowitz F. Hoppe-Seyler's Z Physiol Chem. 1938;254:268–274. doi: 10.1515/bchm2.1963.331.1.67. [DOI] [PubMed] [Google Scholar]
- 52.Daber R, Sharp K, Lewis M. One is not enough. Journal of Molecular Biology. 2009;392:1133–44. doi: 10.1016/j.jmb.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daber R, Stayrook S, Rosenberg A, Lewis M. Structural analysis of lac repressor bound to allosteric effectors. Journal of Molecular Biology. 2007;370:609–19. doi: 10.1016/j.jmb.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. Journal of Molecular Biology. 1994;240:421–33. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 55.Schlax PJ, Capp MW, Record MT., Jr Inhibition of transcription initiation by lac repressor. Journal of Molecular Biology. 1995;245:331–50. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 56.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. Journal of Molecular Biology. 1970;53:401–17. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 57.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 58.Mossing M, Record MT., Jr Upstream operators enhance repression of the lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 59.Oehler S, Eismann ER, Kramer H, Muller-Hill B. The three operators of the lac operon cooperate in repression. EMBO Journal. 1990;9:973–9. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kania J, Muller-Hill B. Construction, isolation and implications of repressor-galactosidase - beta-galactosidase hybrid molecules. European Journal of Biochemistry. 1977;79:381–6. doi: 10.1111/j.1432-1033.1977.tb11819.x. [DOI] [PubMed] [Google Scholar]
- 61.Flashner Y, Gralla JD. DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell. 1988;54:713–21. doi: 10.1016/s0092-8674(88)80016-3. [DOI] [PubMed] [Google Scholar]
- 62.Bellomy GR, Mossing MC, Record MT., Jr Physical Properties of DNA in Vivo As Probed by the Length Dependence of the lac Operator Looping Process. Biochemistry. 1988;27:3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 63.Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: the influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci U S A. 2006;103:9879–84. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci. 2005;30:275–9. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Makman RS, Sutherland EW. Adenosine 3′,5′-Phosphate in Escherichia Coli. J Biol Chem. 1965;240:1309–14. [PubMed] [Google Scholar]
- 66.Ullmann A, Monod J. Cyclic AMP as an antagonist of catabolite repression in Escherichia coli. FEBS Lett. 1968;2:57–60. doi: 10.1016/0014-5793(68)80100-0. [DOI] [PubMed] [Google Scholar]
- 67.Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabloite-sensitive genes: A postive control. Proc Natl Acad Sci. 1970;66:104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes & Development. 2001;15:1506–17. doi: 10.1101/gad.892001. see comment. [DOI] [PMC free article] [PubMed] [Google Scholar]