Abstract
Interferon response factor 5 (IRF5) regulates innate immune responses to viral infection. IRF5 genetic variants have been shown to be strongly associated with risk for systemic lupus erythematosus (SLE). Functional roles of IRF5 exon 6 structural variants that occur as part of a SLE risk-associated haplotype, including a 30-bp in/del (in/del-10) and a 48-bp splice-site variant (SV-16), have not been established. In this study, we used IRF5 deficient cells overexpressing human IRF5 variants to investigate the roles of exon 6 in/del-10 and SV-16 in regulation of the apoptosis response, nuclear translocation, and ability to transactivate IRF5 responsive cytokines. We found that expression of IRF5 isoforms including either SV-16 or in/del-10 confers ability of IRF5 to impair the apoptotic response and correlates with reduced capacity for IRF5 nuclear translocation in MEFs after a DNA-damaging stimulus treatment. Interestingly, the presence or absence of both SV-16 and in/del-10 results in abrogation of both the anti-apoptotic and enhanced nuclear translocation effects of IRF5 expression. Only cells expressing IRF5 bearing SV-16 show increased IL-6 production upon LPS stimulation. MEFs expressing hIRF5 variants containing in/del-10 showed no significant difference from the control; however, cells carrying hIRF5 lacking both SV-16 and in/del-10 showed reduced IL-6 production. Our overall findings suggest that exon 6 SV-16 is more potent than in/del-10 for IRF5-driven resistance to apoptosis and promotion of cytokine production; however, in/del-10 co-expression can neutralize these effects of SV-16.
Keywords: SLE, IRF5 variants, exon 6, apoptosis, nuclear translocation
INTRODUCTION
Interferon regulatory factor 5 (IRF5) is a transcription factor that regulates innate immune responses downstream of Toll-Like Receptors (TLR) and following viral infections [1–3]. Association between variants in the IRF5 locus and risk of human systemic lupus erythematosus (SLE) has been recently established [4,5], although the precise mechanism by which these risk variants influence autoimmunity is still unclear. SLE associated risk in the IRF5 locus is carried on a complex haplotype with multiple variants that potentially influence the function and/or expression of IRF5.
IRF5 has important roles in both the gene-regulatory networks in the host innate immune system and the regulation of oncogenesis. IRF5 primarily resides in the cytoplasm in the absence of any activating stimulus, and is phosphorylated upon viral infection, toll-like receptor (TLR) dependent signaling, and DNA damage [3,6,7]. Phosphorylated IRF5 translocates to the nucleus [3,6,8,9] where it regulates TLR-dependent induction of proinflammatory cytokine genes by binding to MyD88 [1], and induces type I interferons and proinflammatory cytokines [8]. The induction of proinflammatory cytokines, such as IL-6, IL-12, and TNFα, is impaired in IRF5 deficient (IRF5−/−) spleen-derived dendritic cells (DCs) and macrophages after stimulation with certain TLR agonists [1]. However, this impairment is cell type and stimulus dependent [10,11]. In IRF5−/− bone-marrow-derived DCs and macrophages, proinflammatory cytokine production is normal after stimulation with LPS, however, following Poly (I:C) stimulation, DCs but not macrophages show reduced cytokine production [10]. The role of IRF5 in cytokine production by other cell types, such as embryonic fibroblasts (MEF), has not been reported.
IRF5 also functions as a tumor suppressor gene, presumably through its effects on apoptosis [12,13]. Much less is known of IRF5 apoptotic signaling pathways. p53, a tumor suppressor gene and apoptosis regulator, can transactivate the IRF5 gene [12]. However, IRF5 acts in an apoptotic pathway distinct from that for p53 [2,6,9]. Recent data indicated that IRF5 is a mediator of the death receptor-induced apoptotic signaling pathway [7]. IRF5 inhibits the growth of tumor cells both in vitro and in vivo [13], and can sensitize tumor cells to DNA damage-induced apoptosis by irinotecan (CPT-11) [6]. As with its role in regulating TLR-driven cytokine responses, IRF5 function in apoptosis is also cell type dependent. Couzinet et al reported IRF5 was required for death receptor induced apoptosis in DCs and hepatocytes, but not in thymocytes and MEFs [14].
Efforts to map the structural basis for the enhanced risk of SLE conferred by IRF5 alleles have resulted in a complex genetic picture. Graham et al described a risk haplotype defined by 3 variants: a SNP (rs2004640) that is located in the 5’UTR, a splice junction of an alternative exon 1B that permits expression of exon 1B transcripts, a 3’ UTR polyadenylation site SNP (rs10954213) that results in a truncated mRNA isoform that demonstrates a longer half-life, and a 30-bp insertion/deletion (in/del-10) in exon 6 in the IRF5 PEST domain (proline (P), glutamate (E), serine (S) and threonine (T) [4,15]. More recently, a pentanucleotide (CGGGG) repeat in the IRF5 promoter has been shown to be associated with SLE [5]. Conditional analyses suggest that the 4X CGGGG allele explains most of the genetic risk attributable to variants in the 5’ UTR of IRF5 [5]. Differential binding of SP1 to the sequence produced by 4X CGGGG has been proposed as a potential functional mechanism for this in/del [5,16].
Of the potential functional polymorphisms carried on IRF5 SLE associated risk haplotype, little is known about the ability of the exon 6 in/del-10 to alter function of IRF5. Adding to the complexity is the observation that exon 6 can be spliced at an alternative site 48-bp (SV-16) downstream of the canonical splice junction. While the exon 6 in/del-10 in isolation does not demonstrate association with SLE risk [4], its presence on risk haplotypes is likely to influence the function of IRF5. Herein, we describe the results of a reductionist approach to defining functional effects of the exon 6 in/del-10 and SV-16 within IRF5. We employ murine embryonic fibroblasts from IRF5 deficient mice stably transfected with human IRF5 (hIRF5) molecules representing each possible combination of the exon 6 in/del-10 and SV-16. We evaluate potential roles of these variant IRF5 features in regulation of the apoptosis response, nuclear translocation, and ability to transactivate IRF5 responsive cytokines.
METHODS
Cells and Reagents
IRF5 deficient murine embryonic fibroblasts (IRF5−/−-MEF) were developed as described [17] and obtained from Dr. Tak Mak, University of Toronto, Ontario, Canada. IRF5−/−-MEF were immortalized by retroviral transduction of SV40 large T antigen using Phoenix cells as the viral packaging system [18]. Cells were maintained in DMEM (Gibco Invitrogen, Carlsbad CA) with 10% FBS, L-glutamine (2mM), penicillin and streptomycin (100 units/mL). CPT-11 was provided by Dr. Ameeta Kelekar (University of Minnesota), and also purchased from Sigma-Aldrich (Saint Louis, MO). Leptomycin B and lipopolysaccharide (LPS) were purchased from Sigma-Aldrich.
Genes, Plasmid Constructs and Retroviral Gene Transfer
cDNAs of human IRF5 (hIRF5) variants V1 and V2, were purchased from Origene (Rockville, MD). Isoforms V5 and V10 were constructed with QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), using the manufacturer’s recommended protocol and the following mutagenesis primers: V10_sense, 5’-tgtcaagtggccgcctactctgcagccgcc-3’; V10_antisense, 5’-ggcggctgcagagtaggcggccacttgaca-3’; and V5_sense, 5’-gcctgagcctcacagatgcagtgcagtctggcccccacatgacaccctattctttactcaaagaggatgtcaagtggc-3’, V5_antisense, 5’-gccacttgacatcctctttgagtaaagaatagggtgtcatgtgggggccagactgcactgcatctgtgaggctcaggc-3’. cDNAs encoding GFP fused to the N’-terminus of hIRF5 isoforms (V1, V2, V5, V10) were inserted into a retroviral vector, pMSCVneo (Clontech, Mountain View, CA). The Calphos Mammalian Transfection Kit (Clontech) was used to co-transfect pMSCVneo with Gag-pol and VSVG helper plasmids (gifts of Dr. Nick Somia, University of Minnesota) into 293T cells. 48 hour supernatants containing replication-deficient retroviruses encoding GFP-hIRF5s or GFP alone were collected. IRF5−/−-MEF cells were infected according to protocols described in the Retroviral Gene Transfer and Expression User Manual (Clontech).
FACS Analysis and Fluorescence Microscopy
Cells were analyzed on a FACSCalibur flow cytometer (BD, Franklin Lakes, NJ) for GFP-hIRF5 expression. For fluorescence microscopic imaging, cells seeded on cover-slips were stimulated and then fixed with 4% paraformaldehyde. ProLong Gold antifade reagent with nuclear dye DAPI (Invitrogen) was used to make the slides as the mounting medium. The slides were then directly visualized with a Zeiss Axioskop fluorescence microscope.
Apoptosis Assay
Cells were plated into 6-well plates with 5 ×104 per well. After overnight attachment and growth, cells were washed once with fresh growth medium, and replaced with fresh growth medium or medium with 2uM CPT-11. After 24 hours of treatment, the cells were harvested (adherent + suspended = total population). After labeling apoptotic cells with Annexin V-Phycoerythrin (PE) from the PE Annexin V apoptosis detection kit (BD Biosciences, San Jose, CA), cells were analyzed on a FACSCalibur flow cytometer (BD).
Imaging Scanning Flow Cytometry
We used imaging flow cytometry (AMNIS, Seattle, WA) to study nuclear translocation of GFPhIRF5s. Cells were seeded into 10cm2 dish and allowed to reach approximately 80% confluence. Cells were detached with TrypLE Express (Gibco) and resuspended with 2uM CPT-11 medium and incubated for 3 hours. Cells were then fixed in 2% paraformaldehyde for 20 minutes washed twice with PBS and stained overnight with 7AAD at 4°C. Treated and untreated cells were assayed for nuclear co-localization of GFP-hIRF5 and 7-AAD using the AMNIS cytometer.
Western Blot, and Antibodies
Cells were washed with cold PBS and lysed in cell lysis buffer (Cell Signaling, Boston, MA) with protease inhibitor (Sigma-Aldrich). After brief sonication (5 seconds, twice on ice), insoluble material was removed by centrifugation at 13,000g, 4°C. For SDS-PAGE, 20ug of total protein was resolved on 4–12% Bis-Tris gels (Bio-Rad, Hercules, CA), and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked at room temperature for one hour with 5% dry non-fat milk in TBST before overnight incubation with polyclonal goat anti-IRF5 (Abcam, Cambridge, MA) at 4°C. The membranes were subsequently incubated at room temperature for one hour with horseradish peroxidase-conjugated anti-goat secondary (Abcam), and signal was visualized by addition of ECL substrate (Amersham, GE Healthcare, Buckinghamshire, UK) followed by autoradiography.
Measurement of Cytokine Production
Cells were seeded into 12-well plates at 5 × 105 per well. The next day, cells were washed once with fresh medium exposed to stimuli for 24 hrs. Cytokine concentrations in the culture supernatants were measured with Mouse Cytokine 20-Plex Panel (Invitrogen). Trypan Blue dye (Sigma-Aldrich) -excluding viable cell numbers were quantified after trypsin-mediated detachment (TrypLE Express, Gibco).
Statistics
Analysis was performed using GraphPad Software, PRISM for Mac. Data were expressed as mean ± SEM. Student’s t-test was used to compare the means of the different variables between the IRF5−/−-MEF control and a particular hIRF5 isoform overexpressed in IRF5−/−-MEFs. A p value < 0.05 was considered significant.
RESULTS
Generation of Stable Cell Lines Expressing Human IRF5 Variants
Graham et al. originally described a three variant SLE risk-conferring IRF5 haplotype consisting of a 3’ UTR variant that influences mRNA stability, an exon 1B splice donor variant and a 10 amino acid in/del in exon 6 (in/del-10) [4,19]. Of these three variants, no specific functional role has yet been attributed to the variably expressed polypeptide regions within IRF5 exon 6. In addition to in/del-10, exon 6 is capable of alternative splicing 48-bp down stream of the canonical splice site resulting in the loss of 16 amino acids (SV-16). We therefore explored whether or not the presence or absence of either exon 6 SV-16 or in/del-10 influences IRF5 cell biological function.
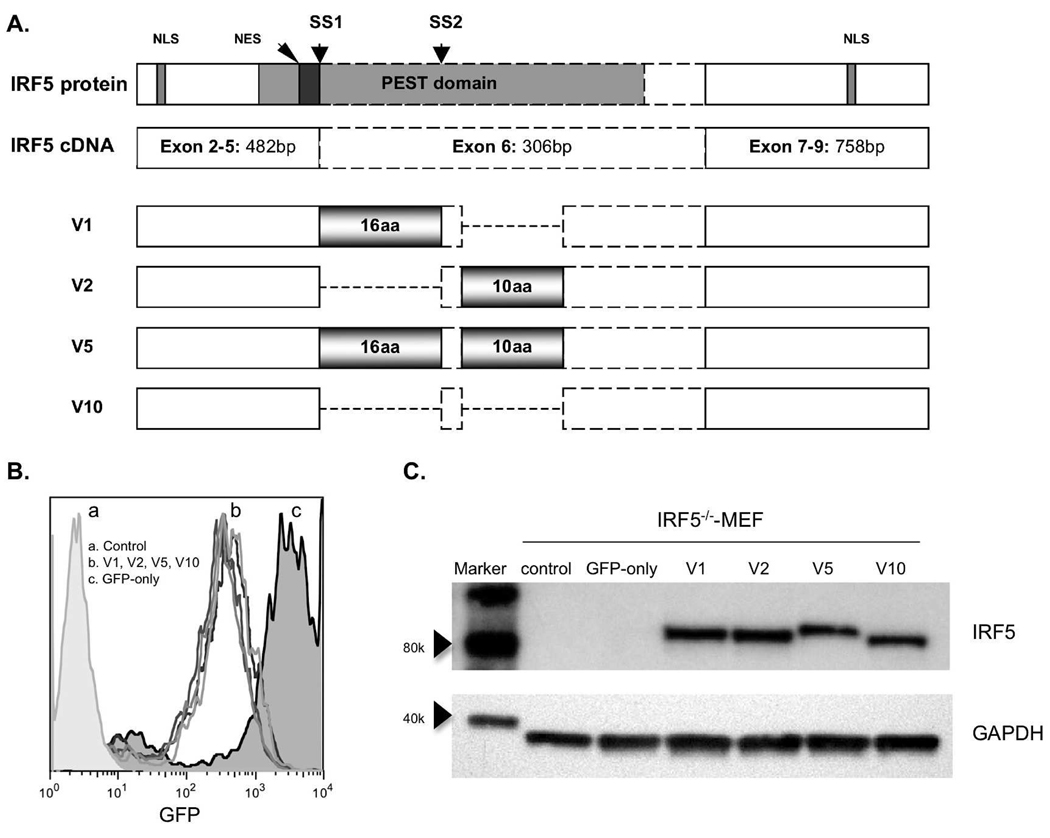
We constructed green fluorescent protein (GFP)-tagged full length human IRF5 (hIRF5) cDNA that comprise all possible combinations of exon 6 SV-16 and in/del-10, termed IRF5 V1, V2, V5, V10 according to the nomenclature of GenBank™ (U51127, BC004139, AY6936655, DQ277633) [19,20] (Figure 1A). cDNA encoding GFP-hIRF5 chimeric proteins were cloned into the retroviral vector, and stable hIRF5 expressing cell lines were prepared in IRF5−/−-MEFs. IRF5−/− cells were employed to prevent difficulties with data interpretation stemming from potential interactions between human and murine IRF5. FACS analysis showed high and approximately equivalent GFP expression levels in all transduced MEF cell lines (Figure 1B). GFP expression levels in the studied cell lines were stable over several weeks of continuous cell culture (data not shown). Equivalent expression levels of GFP-hIRF5 proteins in IRF5 isoform-transduced cell lines were confirmed by Western blot for IRF5 (Figure 1C).
Figure 1.
A. Schematic displaying exon 6 structural variants that comprise four isoforms of human IRF5 (not to scale). Gray horizontal gradient boxes indicate the two areas of structural variation: one is a 48-bp splice-site variant (SV-16) between the splice site 1 (SS1) and SS2; the other one is 30-bp in/del (in/del-10). Two nuclear localization signals (NLS) are located 449bp upstream and 480bp downstream from exon 6; PEST domain expanding from exon 3 to exon 6 encloses both the two structural variation areas; one nuclear export signal (NES) locates inside the PEST domain, immediately N’-terminal of exon 6. B, FACS analysis shows equivalent levels of GFP signal in cell lines stably transduced with hIRF5 variants. C, Western analysis of cell lysates derived from IRF5−/−-MEFs stably expressing GFP-hIRF5.
IRF5 V1 and V2 Resist CPT-11-induced Apoptosis
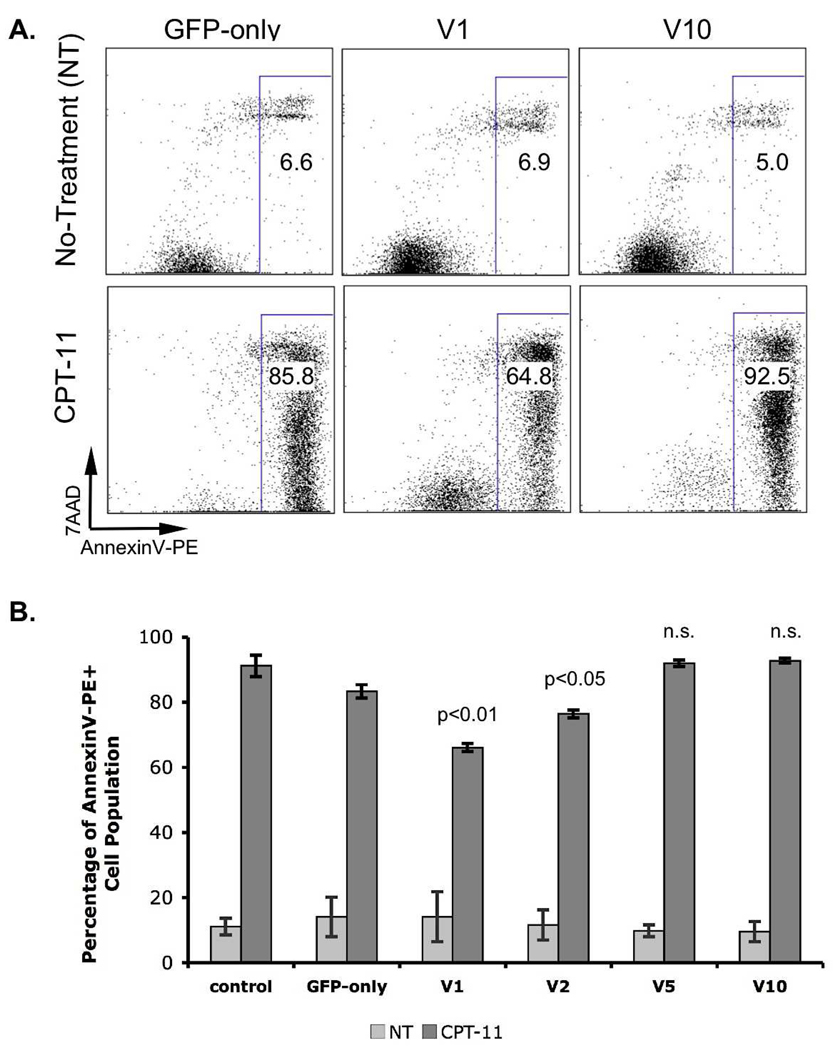
Recent studies had demonstrated that IRF5 was a mediator of cell cycle arrest and cell death [2,6]. IRF5 was required for DNA damage-induced apoptosis [6]. To determine whether exon 6 structural variants might alter the apoptosis-modulating effect of IRF5, we studied stable IRF5 variant expressing cell lines described above. Accordingly, we exposed hIRF5-expressing IRF5−/−-MEFs to the DNA damaging and apoptosis-inducing agent irinotecan (CPT-11) and measured Annexin V binding after 22 hours, the time after which maximal induction of apoptosis was observed based on preliminary time course experiments (data not shown). Increase in Annexin V binding capacity was one of the earliest indicators of impending cell death [21]. We observed that a high fraction of IRF5−/−-MEFs exhibited increased Annexin V binding after CPT-11 (Figure 2A). Expression of either IRF5 isoform V5 (containing both SV-16 and in/del-10) or V10 (lacking both SV-16 and in/del-10) resulted in no change in the fraction of IRF5−/−-MEFs cells exhibiting high Annexin V binding after CPT-11 treatment (Figure 2A and B). In contrast, IRF5−/−-MEFs expressing V1 or V2 hIRF5 isoforms (containing either SV-16 or in/del-10 within exon 6) displayed reduced reactivity with Annexin V following exposure to CPT-11 (V1; 66.1% Annexin V positive cells, p < 0.01, V2; 76.5%, p < 0.05). These results suggested that isoforms harboring either exon 6 SV-16 or in/del-10 conferred ability of IRF5 to impair the apoptotic response in MEFs. Intriguingly, the presence of both exon 6 SV-16 and in/del-10 (V5) resulted in abrogation of any CPT-11-induced apoptosis-regulating effect.
Figure 2.
CPT-11 induced apoptosis. IRF5−/−-MEF cell lines were treated with 2uM CPT-11 for approximately 22 hours. Cells were stained with AnnexinV-PE/7AAD. A, FACS analysis of 7AAD- and Annexin V-PE-labeled cells. Total apoptotic cell (AnnexinV-PE+ populations; rectangular gate) fraction of total is indicated on FACS plots. B, Bar graph comparing the fraction of AnnexinV-PE+ cells in IRF5-expressing cell lines. p-values relative to the control indicated on the graph, n.s. = not significant. Standard error of the mean was calculated based on values from three independent experiments.
Translocation of GFP-hIRF5s in IRF5−/−-MEF
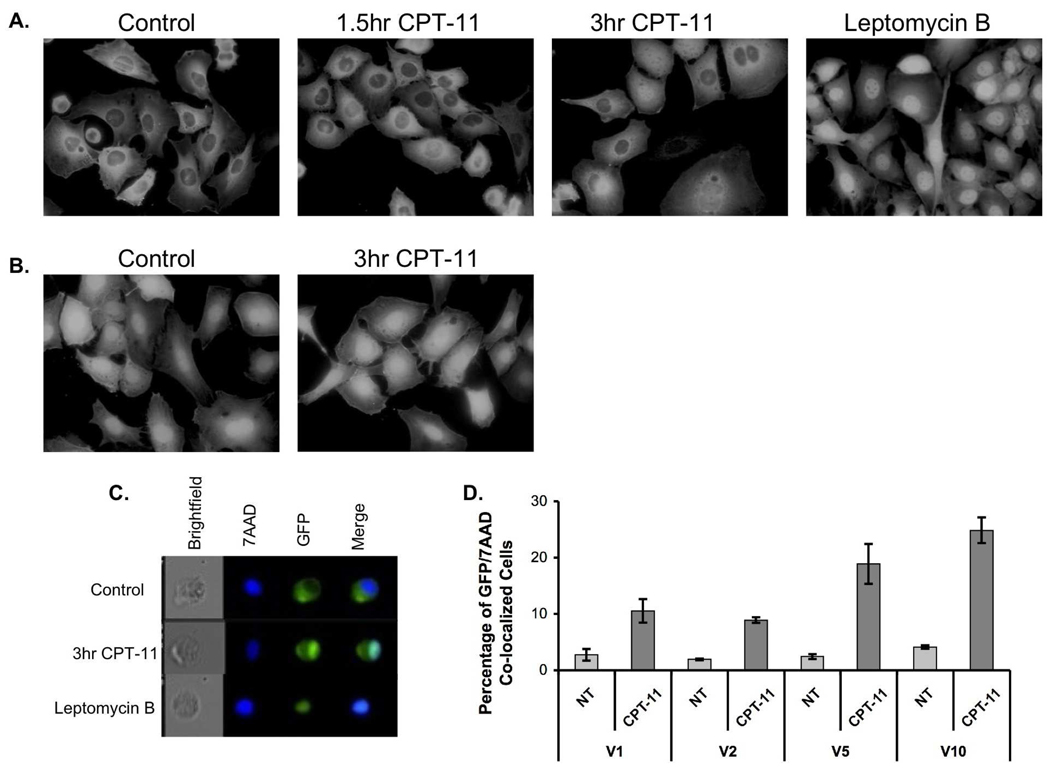
Transactivation of numerous pro-inflammatory genes occurs following nuclear translocation of IRF5 [6]. A nuclear localization signal (NLS) sequence resides near the structural variants within exon 6 [22] (Figure 1A). To determine whether IRF5 exon 6 structural variants differentially regulate IRF5 nuclear translocation, we stimulated GFP-hIRF5-expressing MEFs with CPT-11 and monitored GFP translocation using fluorescence microscopy. Cell lines expressing IRF5 isoforms were evaluated at various time points following CPT-11 exposure. Leptomycin B (nuclear export inhibitor) treatment served as a positive control for nuclear translocation. In the absence of stimulation, IRF5 was primarily located in the cytoplasm. Following leptomycin B treatment, we observed expected IRF5 nuclear accumulation (Figure 3A). In cells treated with CPT-11, nuclear accumulation of GFP-hIRF5 in IRF5−/−-GFPV1 MEFs was readily detectable after 3 hours. Cell lines carrying V2, V5, and V10 isoforms showed the same patterns of GFP-hIRF5 translocation (data not shown). In the GFP alone-expressing IRF5−/−-MEF control cell line, the fluorescent signal localized in cytoplasm and nucleus approximately equally, and CPT-11 treatment did not change signal distribution (Figure 3B).
Figure 3.
GFP-hIRF5 undergoes nuclear translocation following an apoptotic stimulus. A, Fluorescent micrographs show GFP-hIRF5 V1 distribution in untreated cells, in Leptomycin B-treated cells, and in cells following CPT-11 stimulation for indicated times. B, Distribution of GFP signal in cells stably expressing GFP only. As a nuclear exporter inhibitor, Leptomycin B (LMB) showed robust GFP-hIRF5 accumulation in the nucleus. C, Image scanning flow cytometry (AMNIS) images show fluorescence distribution of GFP-hIRF5 V1 (green), and nuclear dye 7-AAD (deep blue). Confluence of the 2 fluorescent signals on the Merge panel is indicated by light-green/blue coloration. D, Image scanning flow cytometry was used to determine the percentage of cells with co-localization of GFP-hIRF5s and 7AAD. Standard error of the mean was calculated based on means of co-localization values from three independent experiments.
To quantitatively assess the effects of exon 6 SV-16 and in/del-10 expression on nuclear translocation, we used image scanning flow cytometry (AMNIS) to measure cell specific IRF5 nuclear translocation. We stimulated GFP-hIRF5 variant-expressing IRF5−/−-MEFs with CPT-11 for 3 hours, then fixed the cells and stained them with the nuclear dye, 7AAD, overnight (Figure 3C). The fraction of cells exhibiting overlay/merging of fluorescent signals derived from GFP and 7-AAD was calculated to determine the proportion of the cells exhibiting IRF5 nuclear translocation (Figure 3D). We observed no differences in nuclear translocation levels between the four GFP-hIRF5-expressing cell lines in the absence of treatment. Following 3 hours of CPT-11 treatment, cells expressing hIRF5 V1 (10.53%) and V2 (8.89%) isoforms demonstrated reduced nuclear translocation compared with hIRF5 V5 (18.87%) and V10 (24.85%) isoforms. Together, these results suggest that inclusion of either the SV-16 or in/del-10 IRF5 exon 6 structural variant (V1 and V2 isoforms) associates with reduced capacity for IRF5 nuclear translocation after CPT-11 treatment. When both IRF5 exon 6 SV-16 and in/del-10 are present (V5), or when both are absent (V10), CPT-11-induced nuclear translocation is enhanced.
Cytokine Production in The hIRF5-overexpressing MEFs
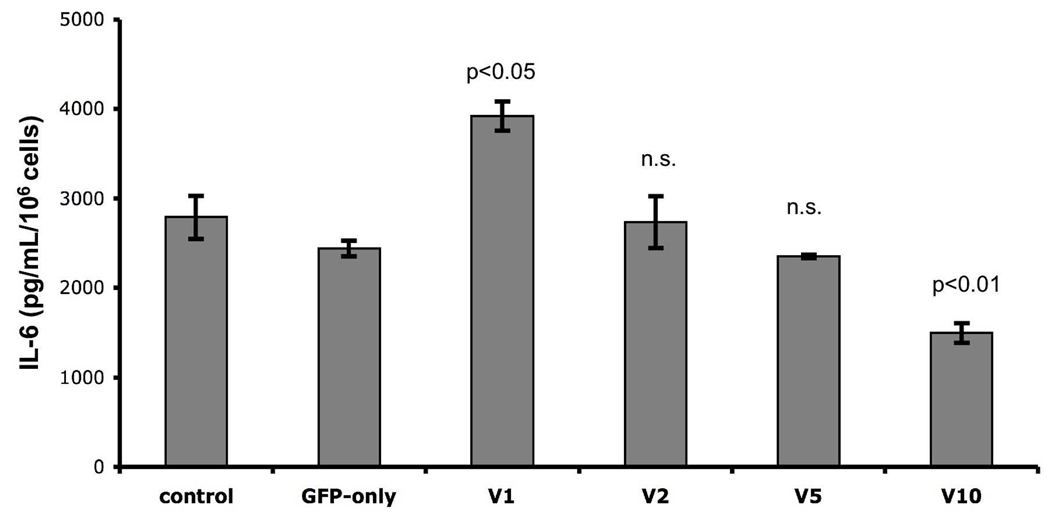
IRF5 promotes production of proinflammatory cytokines after TLR engagement in several cell types [1]. To determine whether the structural variants of IRF5 exon 6 regulate cytokine production, we stimulated GFP-hIRF5-expressing MEFs with 100ng/mL of LPS and measured levels of secreted cytokines regulated by IRF5, including IL-6, IL-12b, and TNFα, by ELISA [1]. After LPS stimulation, all cell lines, including the IRF5−/−-MEF and GFP-only controls, released IL-6 (Figure 4). Interestingly, MEFs expressing hIRF5 V1 produced more IL-6 (3.9 ng/mL/106 cells) compared to GFP-only expressing control MEFs (2.4 ng/mL/106 cells). In contrast, MEFs expressing IRF5 variants containing in/del-10 (V2, V5) showed no significant difference from controls in LPS-stimulated IL-6 production. Cells carrying IRF5 lacking both exon 6 SV-16 and in/del-10 (V10) showed reduced (p<0.01) IL-6 production compared to controls. LPS did not stimulate significant IL-12β or TNFα production by any MEF cell line. Together, these findings suggest that expression of IRF5 SV-16 can promote LPS-stimulated cytokine production, but only when in/del-10 is absent.
Figure 4.
IL-6 production after 24 hrs of treatment with 100 ng/mL of LPS. Bar graph shows IL-6 concentration in pg/mL/106 cells of LPS treated MEFs. p-values relative to control indicated on the graph, n.s. = not significant. Standard error of the mean was calculated from triplicate values arising from one experiment. Similar results were observed in an additional independent experiment.
DISCUSSION
In this study, we investigated potential cell biological and functional effects of IRF5 exon 6 variants. Recent large genetic association studies defined risk and protective IRF5-based haplotypes for human systemic lupus [4], several of which encode structural variants within IRF5 exon 6. Expression of variant IRF5 isoforms correlate with elevated disease-risk (Figure 1A), with protection from disease, or with neutrality. Although at least 10 IRF5 isoforms have been described in human cells [20], the four IRF5 isoforms studied here were selected because they differ only in the presence or absence of short insertions within exon 6 and because they may influence the risk of SLE [19]. Our findings suggest that the alternative splice site-specified IRF5 variant (SV-16) is more potent than the polymorphic exon 6 in/del (in/del-10) for IRF5-driven resistance to apoptosis and promotion of cytokine release. However, in/del-10 co-expression appears capable of neutralizing these effects of SV-16.
Although IRF5 has been characterized as an inducer of Type I Interferon-driven genes, additional key reported functions of IRF5 include the regulation of apoptosis induction [7] and promotion of pro-inflammatory cytokine secretion [22]. Nuclear localization sequences within IRF5 are critical for movement of phosphorylated IRF5 from cytoplasm to nucleus, and for IRF5 transactivating capacity [22], strongly suggesting that major IRF5 functional properties depend upon nuclear translocation.
Our finding that IRF5 variants can act as negative regulators of apoptosis is surprising, since other reports suggest that IRF5 promotes cell death induced by DNA damaging agents [2,6,9]. Potential explanations for the discrepancy between our findings and published reports include cell-type specificity of the apoptotic response and variation in apoptosis-inducing stimuli employed. Barnes et al found that overexpression of human IRF5 in the BJAB lymphoma cell line caused cell-cycle arrest and spontaneous apoptosis [9]. However, it is not clear whether these overexpression studies employed IRF5 variants carrying or lacking the SV-16 exon 6 regions. Later, Hu et al reported enhanced apoptosis of p53-deficient tumor cells after overexpression of hIRF5 V3 (lacking both SV-16 and in/del-10) [6]. Our findings are consistent with the latter, since overexpression of hIRF5 V10 (like V3, harboring neither in/del-10 nor SV-16) failed to impair high rates of CPT-11-induced apoptosis observed in IRF5-deficient MEFs (figure 2). While Yanai et al studied IRF5−/−-MEFs, similar to the cells used in experiments reported here, they examined apoptosis in response to viral infection and to gamma irradiation in the presence of overexpressed HA-Ras [2], rather than chemotherapy (irinotecan)-induced death. Both of the former modes of apoptosis may occur through activation of death receptor signaling cascades, which Hu et al have recently demonstrated are positively regulated by IRF5 [7]. Thus it is formally possible that IRF5 might serve as an inhibitor of apoptosis induced by topoisomerase I inhibition (irinotecan), while also having ability to promote cell death after DNA damage (such as would occur after irradiation). Our results suggest that the IRF5 SV-16 is required for the protective effect of hIRF5 expression from cell death in MEFs, although the mechanism whereby co-expression of in/del-10 abrogates SV-16 effects remains unclear. Further studies examining the role of IRF5 exon 6 SV-16 and in/del-10 in the apoptosis response of primary innate immune cells to stimuli like viral infection will clarify whether and how much IRF5 exon 6 variants regulate apoptosis relevant to immune-mediated processes.
Overexpresssed IRF5 associates with transactivation of proapoptotic genes such as Bax, Bak and p21 [9]. This observation is commensurate with our finding that protection from CPT-11-induced apoptosis correlates with reduced IRF5 nuclear translocation (observed in experiments with IRF5−/−-MEFs expressing the V1 and V2 IRF5 isoforms, figures 2, 3). The mechanism whereby either IRF5 exon 6 SV-16 or in/del-10 modulates nuclear translocation is not known. A conformational change associated with presence of either insertion may hinder transporter protein access to either of two IRF5 nuclear localization sequences that dynamically regulate IRF5 shuttling between nucleus and cytoplasm [13,22]. Lin et al have documented that constitutive cytoplasmic localization of IRF5 depends upon dominant function of an IRF5 nuclear export sequence (NES) [23]. Interestingly, the N-terminal portion of exon 6 harboring both in/del-10 and SV-16 is located immediately carboxy-terminal to the IRF5 NES [23]. Thus, the presence of either exon 6 SV-16 or in/del-10 could enhance access of the nuclear exporting transport protein CRM1 to IRF5, and sustain IRF5 presence in the cytoplasm. The observations that the presence of both exon 6 sv-106 and in/del-10, or of neither, is associated with enhanced nuclear localization (figure 3), suggest that regulation of IRF5 subcellular localization is multifactorial and complex.
Data from IRF5 structure/function analyses with respect to promotion of cytokine secretion are limited. Mancl et al showed that overexpression of hIRF5 lacking both exon 6 SV-16 and in/del-10 (V3) had diminished capacity to activate a reporter element derived from the IFNα gene promoter [20]. Recently, Yang et al showed that IRF5 DNA binding capacity is required for LPS-induced IL-6 secretion by a macrophage cell line [24]. The role of IRF5 exon 6 structural variants in regulating TLR-driven cytokine production has not been examined, however. Our finding that expression of IRF5 carrying the SV-16, but not in/del-10 (V1), results in enhanced LPS-induced IL-6 production in cell culture (figure 4) may be related to augmentation of reported IRF5 interaction with molecular mediators (e.g. TRAF6, MyD88) of the TLR signaling pathway leading to proinflammatory cytokine production [1]. Biochemical studies examining the IRF5 structural requirements for protein-protein associations with TLR pathway mediators will shed further light on this potential mechanism.
Expression of an IRF5 exon 6 that is associated with increased risk of SLE (V2; absent SV-16, present in/del-10) correlated with weak IRF5 capacity to prevent CPT-11-induced apoptosis, and no ability to augment TLR-dependent IL-6 production in MEFs (figure 4). These findings of comparatively weaker functional effects of in/del-10 relative to SV-16 are compatible with findings of recent genetic studies suggesting that expression of the in/del-10 exon 6 motif in isolation is not functional or deleterious with respect to human SLE risk [5,25]. These studies suggest instead that in/del-10 may be in linkage disequilibrium with additional disease risk-modifying polymorphisms of the IRF5 gene. Indeed, a 5 base pair in/del sequence element located 5’ to the first untranslated exon of IRF5 appears to explain the apparent contributions of both in/del-10 and SV-16 to increased risk of SLE [5].
Acknowledgements
We are grateful to Drs. Yoji Shimizu, Brandon Burbach and Jason Mitchell at the University of Minnesota for technical assistance with AMNIS and FACS analysis and to Mr. Thuan Nguyen from Oklahoma Medical Research Foundation for assisting with the cytokine assays.
Disclosure of funding sources:
This research was supported by NIH grants, AR052125 and AI063274, to P.G.
Footnotes
Declaration of interests:
The authors report no conflicts of interest. The authors alone are responsible for the content of the paper.
REFERENCES
- 1.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 2.Yanai H, Chen H, Inuzuka T, Kondo S, Mak TW, Takaoka A, Honda K, Taniguchi T. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng T, Brzostek S, Ando O, Scoy SV, Kumar KP, Reich NC. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J Immunol. 2006;176:7462–7470. doi: 10.4049/jimmunol.176.12.7462. [DOI] [PubMed] [Google Scholar]
- 4.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LRL, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, Bauer JW, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigurdsson S, Goring HHH, Kristjansdottir G, Milani L, Nordmark G, Sandling JK, Eloranta ML, Feng D, Sangster-Guity N, Gunnarsson I, et al. Comprehsive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872–881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 6.Hu G, Mancl ME, Barnes BJ. Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 2005;65:7403–7412. doi: 10.1158/0008-5472.CAN-05-0583. [DOI] [PubMed] [Google Scholar]
- 7.Hu G, Barnes BJ. IRF-5 is mediator of the death receptor-induced apoptotic signaling pathway. J Biol Chem. 2009;284:2767–2777. doi: 10.1074/jbc.M804744200. [DOI] [PubMed] [Google Scholar]
- 8.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of novel interferon regulatory fator, IRF-5, results in the induction of distinct interferon α genes. J Biol Chem. 2001;276:23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 9.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63:6424–6431. [PubMed] [Google Scholar]
- 10.Yasuda K, Richez C, Maciaszek JW, Agrawal N, Akira S, Marshak-Rothstein A, Rifkin IR. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178:6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- 11.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci USA. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H. Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene. 2002;21:2914–2918. doi: 10.1038/sj.onc.1205459. [DOI] [PubMed] [Google Scholar]
- 13.Barnes BJ, Field AE, Pitha-Rowe PM. Virus-induced heterodimer formation between IRF-5 and IRF-7 modulates assembly of the IFNA enhanceosome in vivo and transcriptional activity of IFNA genes. J Biol Chem. 2003;278:16630–16641. doi: 10.1074/jbc.M212609200. [DOI] [PubMed] [Google Scholar]
- 14.Couzinet A, Tamura K, Chen H, Nishimura K, Wang ZC, Morishita Y, Takeda K, Yagita H, Yanai H, Taniguchi T, Tamura T. A cell-type-specific requiredment for IFN regulatory factor (IRF5) in Fas-induced apoptosis. Proc Natl Acad Sci USA. 2008;105:2556–2561. doi: 10.1073/pnas.0712295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A, Meraro D. ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J Interferon Cytokine Res. 2002;22:153–160. doi: 10.1089/107999002753452764. [DOI] [PubMed] [Google Scholar]
- 16.Dideberg V, Kristjansdottir G, Milani L, Libioulle C, Sigurdsson S, Louis E, Wiman AC, Vermeire S, Rutgeerts P, Belaiche J, et al. An insertion-deletion polymorphism in the interferon regulatory factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Human Mol Genetics. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 17.Samuelson LC, Metzger JM. Isolation and freezing of primary mouse embryonic fibroblasts (MEF) for feeder plates. Cold Spring Harb. Protoc. 2006 doi: 10.1101/pdb.prot4482. [DOI] [PubMed] [Google Scholar]
- 18.Van Der Haegen BA, Shay JW. Immortalization of human mammary epithelial cells by SV40 large T-antigen involves a two step mechanism. In Vitro Cell Dev Biol. 1993;29A:180–182. doi: 10.1007/BF02634177. [DOI] [PubMed] [Google Scholar]
- 19.Graham RR, Kozyrev SV, Baechler EC, Linga Reddy PMV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nature genetics. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 20.Mancl ME, Hu G, Sangster-Guity N, Olshalsky SL, Hoops K, Fitzgerald-Bocarsly P, Pitha PM, Pinder K, Barnes BJ. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. J Biol Chem. 2005;280:21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 21.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis: Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labeled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 22.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Molecular and Cellular Biology. 2002;22(16):5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R, Yang L, Arguello M, Penafuerte C, Hiscott J. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J Biol Chem. 2005;280(4):3088–3095. doi: 10.1074/jbc.M408452200. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Zhao T, Shi X, Nakhaei P, Wang Y, Sun Q, Hiscott J, Lin R. Functional analysis of a dominant negative mutation of interferon regulatory factor 5. PLoS ONE. 2009;4(5):e5500. doi: 10.1371/journal.pone.0005500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyzyrev SV, Lewen S, Linga Reddy PMV, Pons-Estel B, Witte T, Junker P, Laustrup P, Gutierrez C, Suarez A, Martin J, et al. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis & Rheumatism. 2007;56(4):1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]