Abstract
Objectives
Subtypes of juvenile idiopathic arthritis (JIA) share phenotypic features with other autoimmune disorders. We investigated several genetic variants associated with rheumatoid arthritis (RA) and other autoimmune disorders for association with JIA, to test the hypothesis that clinically distinct phenotypes share common genetic susceptibility factors.
Methods
Cases were 445 children with JIA, and controls were 643 healthy adults. Eight single nucleotide polymorphisms (SNPs) in 7 loci [TNFAIP3 (rs10499194 and rs6920220), RSBN1 (rs6679677), C12ORF30 (rs17696736), TRAF1 (rs3761847), IL2RA (rs2104286), PTPN2 (rs2542151), and STAT4 (rs7574865)] were genotyped by the TaqMan assay. Alleles and genotypes were analyzed for association with JIA and JIA subtypes. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated.
Results
The strongest associations were observed for TNFAIP3 variants rs10499194 (OR: 0.74 (0.61-0.91); p <0.004), and TNFAIP3 rs6920220 (OR: 1.3 (1.05-1.61); p <0.02). We also observed associations between JIA and STAT4 (OR: 1.24 (1.02-1.51); p <0.03) and C12ORF30 (OR: 1.2 (1.01-1.43); p <0.04) variants. The PTPN2 variant rs2542151 deviated from Hardy-Weinberg equilibrium and was excluded from analyses. Variants in IL2RA, TRAF1, and RSBN1 were not associated with JIA. After stratification by JIA subtype, TNFAIP3 and C12ORF30 variants were associated with oligoarticular JIA, while the STAT4 variant was associated primarily with polyarticular JIA.
Conclusions
We have demonstrated associations between JIA and variants in TNFAIP3, STAT4 and C12ORF30 regions that have previously shown associations with other autoimmune diseases, including RA and systemic lupus erythematosus. Our results suggest that clinically distinct autoimmune phenotypes share common genetic susceptibility factors.
Keywords: JRA, genetics, autoimmune, association, juvenile idiopathic arthritis, rheumatoid arthritis
Juvenile idiopathic arthritis (JIA) comprises a collection of chronic childhood arthritides (1). Although associations between JIA and variants encoding human leukocyte antigens (HLA) have been consistently described, the HLA-DR locus has been estimated to account for only ~17% of JIA susceptibility (2). This suggests that non-HLA loci likely play significant roles in predisposition to JIA. To date, most investigators have utilized candidate-gene association studies to find genes predisposing to JIA susceptibility. More than 100 genetic loci have been tested for associations with JIA, most of which have not demonstrated meaningful associations (3). Candidates have often been chosen ad hoc, and many studies have suffered from lack of statistical power. However, convincing associations have been demonstrated with several non-HLA variants such as PTPN22, MIF, WISP3 using relatively large cohorts of children with JIA (3).
Children with JIA have an increased prevalence of other autoimmune disorders(4). Furthermore, relatives of children with JIA have an increased prevalence of autoimmune disorders (5). These observations suggest that clinically distinct autoimmune phenotypes may have common genetic susceptibility factors. Recently variants in genes encoding PTPN22 and CTLA4 have been shown to predispose to clinically distinct autoimmune phenotypes. Our objective was to test the hypothesis that genetic variants predisposing to multiple autoimmune phenotypes could also underlie susceptibility to JIA. We investigated 8 single nucleotide polymorphisms (SNPs) in seven loci associated with rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), type 1 diabetes (T1D) and inflammatory bowel disease (IBD) in our cohort of children with JIA and healthy controls.
Patients and methods
Cases were 445 children with JIA from the Intermountain States Database of Childhood Rheumatic Diseases. Patients were diagnosed according to the ILAR criteria (1). The median onset age was 5.6 years, and 67% of the subjects were female. There were 201 children with oligoarticular JIA, 100 with rheumatoid factor (RF)-negative polyarticular JIA, 40 with enthesitis related arthritis, 35 with RF-positive polyarticular JIA, 36 with systemic JIA, and 33 with other subtypes. Controls were 643 healthy adults (60 % female) screened for several common autoimmune diseases ascertained from the same geographic region. A questionnaire was used to screen controls for autoimmune disorders. Controls who reported an autoimmune disorder, or had a first-degree relative with an autoimmune disorder were excluded. Cases and controls were very similar in ethnicity, and predominantly (>90%) were of Northern European ancestry._Subjects were enrolled under protocols approved by the Institutional Review Board at the University of Utah.
Genotyping
DNA was isolated from blood using the Puregene DNA purification kit from Qiagen (Valencia, CA). Subjects were genotyped for the eight SNPs in seven loci including rs10499194 and rs6920220 in theTNFAIP3 locus, rs6679677 in the RSBN1 locus, rs17696736 in the C12orf30 locus, rs3761847 in the TRAF1/C5 locus, rs2104286 in the IL2RA locus, rs7574865 in the STAT4 locus, and rs2542151 in the PTPN2 locus. These variants were chosen based on their reported associations with other autoimmune phenotypes including RA, SLE, T1D and IBD (6-9). Each of these variants has been shown to have an association with at least two autoimmune phenotypes. Genotyping was performed using Taqman Pre-designed SNP genotyping assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocols. For quality control twelve samples were typed in duplicate on different plates; the concordance was 100% for all samples.
Analysis
All variants were evaluated for deviation from Hardy-Weinberg equilibrium (HWE) by comparing observed and expected frequencies of genotypes. Alleles and genotypes were investigated for association with JIA. Since rs10499194 and rs6920220 were both in the TNFAIP3 locus, haplotypes were also analyzed for an association with JIA. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated for alleles, as well as genotypes using dominant and recessive models. Since JIA is a collection of several heterogeneous subtypes, analyses were repeated after stratifying subjects into onset types and gender. The polyarticular subtypes were analyzed both individually and together to improve statistical power.
The STAT4 variant rs7574865 has been reported to be associated with several autoimmune phenotypes including RA(9-16), SLE (9-11, 15, 17), T1D (18, 19), Sjogren’s syndrome (20), and IBD(18). Published case-control association studies of STAT4 rs7574865 were identified using a PUBMED search. Allele frequency data were obtained from these studies. We performed meta-analyses of STAT4 associations with RA and SLE using a fixed-effects model, weighting studies by number of subjects. Mantel-Haenszel Chi-square tests were performed for the individual studies and for the studies combined. The Breslow-Day test was performed for heterogeneity among studies with significance set at p <0.05. All analyses were carried out using SAS 9.1.3. We did not perform meta-analyses of the other variants since their associations were mainly reported in genome-wide association studies, and there were only one or two published studies of these other variants.
RESULTS
The genotyping call rates ranged from 96.8 to 99.8% for the eight SNPs. SNP rs2542151 at the PTPN2 locus deviated significantly from HWE in the controls (p <0.01) and hence was not analyzed further. The allele and genotype frequencies of the seven remaining SNPs in cases and controls are shown in Table 1. The minor allele frequencies (MAF) among our controls did not differ by more than ~10% from MAF reported for individuals of European descent. The ancestral alleles were the major allele at SNP rs10499194, rs6920220, rs6679677, rs17696736, rs3761847, and rs2104286. However at rs7574865 in the STAT4 locus, the ancestral allele, T, was the minor allele (MAF = 0.24 among controls).
Table 1.
Case-control analysis of autoimmunity associated variants and JIA.
| Locus | Variant | Alleles | Controls | Cases | OR (95%CI) | P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 2 | N | 1/1 | 1/2 | 2/2 | MAF | N | 1/1 | 1/2 | 2/2 | MAF | ||||
| TNFAIP3 | rs10499194 | C T | 621 | 0.52 | 0.39 | 0.09 | 0.29 | 433 | 0.58 | 0.37 | 0.05 | 0.23 | 0.74 (0.61-0.91) | 0.004 |
| TNFAIP3 | rs6920220 | G A | 619 | 0.67 | 0.29 | 0.04 | 0.19 | 441 | 0.59 | 0.37 | 0.04 | 0.23 | 1.30 (1.05-1.61) | 0.015 |
| RSBN1 | rs6679677 | C A | 642 | 0.81 | 0.18 | 0.01 | 0.10 | 444 | 0.77 | 0.21 | 0.02 | 0.13 | 1.28 (0.98-1.68) | 0.070 |
| C12orf30 | rs17696736 | A G | 631 | 0.36 | 0.48 | 0.16 | 0.41 | 443 | 0.30 | 0.51 | 0.20 | 0.45 | 1.20 (1.01-1.43) | 0.041 |
| TRAF1/C5 | rs3761847 | G A | 641 | 0.34 | 0.49 | 0.18 | 0.42 | 440 | 0.34 | 0.47 | 0.19 | 0.42 | 1.02 (0.86-1.21) | 0.818 |
| IL2RA | rs2104286 | A G | 634 | 0.55 | 0.39 | 0.06 | 0.25 | 438 | 0.53 | 0.41 | 0.05 | 0.26 | 1.04 (0.85-1.27) | 0.693 |
| STAT4 | rs7574865 | G T | 641 | 0.56 | 0.39 | 0.04 | 0.24 | 443 | 0.51 | 0.43 | 0.07 | 0.28 | 1.24 (1.02-1.51) | 0.029 |
MAF: Minor allele frequency.
Odds ratio for the carriage of allele 2 are shown.
At the TNFAIP3 locus, the T allele at TNFAIP3 SNP rs10499194 demonstrated a significant protective effect against JIA in the entire cohort (OR 0.74 (0.61-0.91), p <0.004). A recessive model significantly improved the OR (0.49 (0.30-0.83), p <0.006). The effect was also pronounced in female subjects (0.71 (0.55-0.91), p <0.007). When analyses were repeated after stratification by subtypes, the association was significant in the oligoarticular JIA cohort (Table 2). In contrast, the minor allele (A) at TNFAIP3 SNP rs6920220 conferred risk to the JIA cohort as a whole (OR 1.30 (1.05-0.61), p < 0.02). At this locus, a dominant model improved the OR (1.44 (1.12-1.86, p <0.004). The association was also pronounced in female subjects (1.42 (1.09-2.66), p <0.009). Similar to the other variant, when analyses were repeated after stratification by subtypes, the association was significant in the oligoarticular JIA cohort (Table 2).
Table 2.
Case-control analysis of autoimmunity associated variants and JIA subtypes
| Locus | Variant | Phenotype | N | MAF | OR | 95%CI | p |
|---|---|---|---|---|---|---|---|
| TNFAIP3 | rs10499194 | Controls | 621 | 0.29 | |||
| All JIA | 433 | 0.23 | 0.75 | 0.61-0.91 | 0.004 | ||
| Oligoarticular | 198 | 0.22 | 0.71 | 0.55-0.93 | 0.012 | ||
| All poly | 131 | 0.23 | 0.75 | 0.55-1.02 | 0.065 | ||
| Polyarticular | 98 | 0.24 | 0.75 | 0.53-1.07 | 0.116 | ||
| Polyarticular RF+ | 33 | 0.23 | 0.72 | 0.40-1.30 | 0.279 | ||
| ERA | 37 | 0.19 | 0.57 | 0.32-1.04 | 0.064 | ||
| Systemic | 35 | 0.30 | 1.05 | 0.62-1.78 | 0.844 | ||
| Other | 32 | 0.26 | 0.82 | 0.46-1.46 | 0.500 | ||
| TNFAIP3 | rs6920220 | Controls | 619 | 0.19 | |||
| All JIA | 441 | 0.23 | 1.30 | 1.05-1.61 | 0.017 | ||
| Oligoarticular | 199 | 0.24 | 1.39 | 1.05-1.82 | 0.019 | ||
| All poly | 133 | 0.23 | 1.34 | 0.97-1.84 | 0.072 | ||
| Polyarticular | 99 | 0.23 | 1.33 | 0.93-1.92 | 0.116 | ||
| Polyarticular RF+ | 34 | 0.24 | 1.35 | 0.76-2.04 | 0.300 | ||
| ERA | 40 | 0.20 | 1.10 | 0.63-1.92 | 0.738 | ||
| Systemic | 36 | 0.19 | 1.06 | 0.58-1.92 | 0.841 | ||
| Other | 33 | 0.22 | 1.19 | 0.65-2.17 | 0.581 | ||
| C12orf30 | Rs17696736 | Controls | 631 | 0.41 | |||
| All JIA | 443 | 0.45 | 1.20 | 1.01-1.43 | 0.041 | ||
| Oligoarticular | 201 | 0.47 | 1.28 | 1.02-1.60 | 0.033 | ||
| All poly | 133 | 0.44 | 1.17 | 0.90-1.53 | 0.244 | ||
| Polyarticular | 98 | 0.44 | 1.17 | 0.87-1.59 | 0.302 | ||
| Polyarticular RF+ | 35 | 0.44 | 1.17 | 0.72-1.90 | 0.529 | ||
| ERA | 40 | 0.39 | 0.93 | 0.58-1.48 | 0.758 | ||
| Systemic | 36 | 0.42 | 1.05 | 0.65-1.70 | 0.843 | ||
| Other | 33 | 0.47 | 1.38 | 0.84-2.27 | 0.198 | ||
| STAT4 | rs7574865 | Controls | 641 | 0.28 | |||
| All JIA | 443 | 0.28 | 1.24 | 1.02-1.5 | 0.031 | ||
| Oligoarticular | 201 | 0.26 | 1.09 | 0.85-1.42 | 0.485 | ||
| All polyarticular | 134 | 0.32 | 1.50 | 1.13-2.00 | 0.005 | ||
| Polyarticular | 99 | 0.31 | 1.45 | 1.04-2.00 | 0.026 | ||
| Polyarticular RF+ | 35 | 0.34 | 1.66 | 1.0-2.76 | 0.050 | ||
| ERA | 39 | 0.29 | 1.33 | 0.80-2.20 | 0.268 | ||
| Systemic | 36 | 0.26 | 1.14 | 0.66-1.95 | 0.637 | ||
| Other | 33 | 0.28 | 1.19 | 0.68-2.08 | 0.538 |
Since both rs10499194 and rs6920220 are in the TNFAIP3 region, we investigated haplotype combinations of these two SNPs for association with JIA. The haplotype carrying the major alleles at both SNPs (CG) was neutral, while haplotype TG was protective (case frequency 0.23, control frequency 0.28, p <0.006) and haplotype CA conferred a risk (case frequency 0.22, control frequency 0.18, p <0.01).
An association was also observed between JIA and the G allele of rs17696736 (allelic OR 1.2, p <0.04) at the C12ORF30 locus. A recessive model improved the OR slightly in the entire JIA cohort (1.3 (1.00-1.68), p <0.05). This association was also pronounced in the oligoarticular JIA subset (Table 2). The minor allele (A) at SNP rs6679677 in the RSBN1 locus demonstrated a marginal association with JIA in the entire JIA cohort (OR 1.28 (0.98-1.68), p <0.07). When analyses were repeated after stratification, there was significant risk conferred to male subjects (146 cases, 257 controls; OR = 1.9 (1.20-2.97), p <0.006). The variants at TRAF1 and IL2RA did not show an association either with JIA or any of the JIA subtypes in our cohorts.
Finally, the T allele at the STAT4 SNP rs7574865 demonstrated a significant association with JIA (OR 1.24 (1.02-1.51), p <0.03). The association was pronounced in the polyarticular JIA subtypes (Table 2). When the RF-negative and RF-positive polyarticular JIA subsets were combined to improve power, the magnitude of the association with the STAT4 variant improved (OR 1.5 (1.13-2.0), p <0.005).
Meta-analyses of STAT variant rs7574865
The rs7574865 variant has been investigated for an association with an autoimmune phenotype in 18 case-control comparisons (Figure 1). Statistically significant associations had been reported between rs7574865 and RA (OR 1.17-1.9), SLE (OR 1.55-1.71), T1D (OR 1.36-1.94) and Sjogren’s syndrome (OR 1.47), as well as IBD (OR 1.29) (9-17, 19, 20). The subjects in these studies were predominantly of European ancestry, and most of the remainder were of Asian ancestry. There were 8 case-control associations of RA and STAT4. The pooled analysis confirmed an association between RA and STAT4 with an OR of 1.26 (1.22-1.31), p <10−7. When all 8 studies were included, the Breslow-Day statistic was significant (p <0.01), suggesting heterogeneity between studies. This was mainly due to the study by Zervou et al., which included a genetically homogeneous population from the Greek island of Crete(16). When we repeated the analysis excluding this study, there was no heterogeneity (Breslow-Day p < 0.17), and the OR estimate changed minimally, with an OR of 1.25 (1.21-1.30) p <10−7. The pooled OR for the 5 investigations of SLE was 1.59 (1.49-1.70), p <10−7. The Breslow-Day statistic for heterogeneity was not significant (p <0.97).
Figure 1. Case-control association results of STAT4 variant rs7574865.
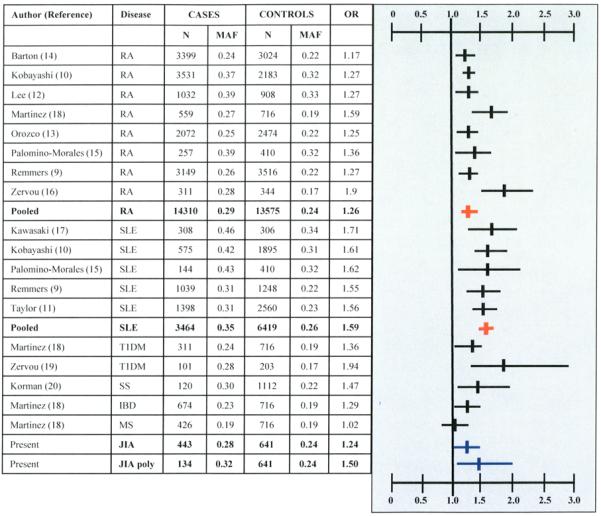
RA: rheumatoid arthritis, SLE: systemic lupus erythematosus, T1D: type 1 diabetes, SS: Sjogren’s syndrome, IBD: inflammatory bowel disease, MS: Multiple sclerosis. OR: Odds ratio (Allelic). OR (Vertical line) and 95% confidence intervals (horizontal lines) are shown. Red color indicates pooled analyses of RA and SLE studies. Blue color indicates JIA association.
DISCUSSION
JIA is a collection of chronic arthropathies in children that shares phenotypic features with several autoimmune disorders(1). Relatives of children with JIA have an increased prevalence of autoimmune disorders, and individuals with JIA also appear to have an increased prevalence of other autoimmune disorders(4, 5). These observations suggest that clinically distinct autoimmune phenotypes could share common genetic susceptibility factors. This hypothesis is supported by genetic studies. Meta-analyses of genome-wide scans of immune mediated disorders demonstrated nonrandom clustering of susceptibility loci between different human diseases(21). The identification of CTLA4 (22) and PTPN22 (23) variants underlying susceptibility to clinically distinct autoimmune disorders also supports this hypothesis.
We tested the hypothesis that variants associated with autoimmune disorders will also be associated with JIA. Our results confirmed that variants in the TNFAIP3 and STAT4 loci associated with RA and SLE also demonstrate associations with JIA. In addition, we found evidence for an association between JIA and a variant on chromosome 12 previously shown to be associated with T1D (24). To our knowledge these variants have not previously been tested for association with JIA. While the size of our JIA cohort is modest, it is still larger than ~90% of published case-control comparisons of non-HLA variants in JIA(3). Our combined case and control cohort of almost 1100 subjects has sufficient power to detect modest associations. We observed that the associations between TNFAIP3 and STAT4 variants and JIA were similar in magnitude and direction to the association between these variants and RA. While some of the subtypes did not show statistically significant associations with TNFAIP3 and STAT4 variants, it should be noted that in general the associations were in the same direction, suggesting that these cohorts might be underpowered to detect significant associations. These results suggest that TNFAIP3 and STAT4 variants might also be involved in susceptibility to multiple autoimmune phenotypes, similar to PTPN22 and CTLA4.
We did not perform a correction for multiple testing. Since our hypothesis was that different autoimmune phenotypes share common susceptibility factors, we treated JIA as a combined phenotype for the primary analysis. The loci investigated in this study were chosen based on prior findings of association in other autoimmune phenotypes, and they have an increased prior probability of association. Given that the genetic basis of JIA is not in doubt, each gene that is potentially involved in the etiology of the trait represents an individual null hypothesis to assess.(25, 26) For these reasons, we believe that a standard multiple comparisons correction would be too conservative. Nevertheless, the association with TNFAIP3 variant rs10499194 and JIA was significant after correcting for the 7 loci investigated. Ultimately, the best validation of these results will be replication in an independent cohort.
TNFAIP3 (also known as A20) is an important regulator of NF-κB signaling through ubiquitin modifications of adaptor proteins downstream of TNF-α and toll-like receptors(27, 28). Validated associations between RA and variants in the TNFAIP3 region have been reported (6, 7, 29). The TNFAIP3 SNPs investigated in our study have been shown to tag haplotypes that confer either susceptibility to or protection from RA(7). Our results confirmed that the same susceptibility and protective haplotypes are also associated with JIA. Interestingly, TNFAIP3 SNP rs6920220 is also associated with susceptibility to SLE (30).
The STAT4 gene encodes a transcription factor expressed on lymphocytes, macrophages, and dendritic cells. STAT4 is essential for IL-12 signaling and induces Th1 differentiation and production of interferon-γ, a pro-inflammatory cytokine(31). Since STAT4 variants were first described to be associated with both RA and SLE by Remmers et al. (9), several authors have confirmed associations between STAT4 variants and various autoimmune diseases(10-13, 17, 18, 20). Our demonstration of an association between the same STAT4 variant (rs7574865) and JIA adds to this literature. Both RF-positive and RF-negative polyarticular JIA subtypes demonstrated an association with this variant. Together these observations strongly support a role for STAT4 in autoimmunity.
We also found an association between JIA and a variant in the C12ORF30 region on chromosome 12. This variant shows strong association with type 1 diabetes(24). This variant also showed an association with autoimmune phenotypes analyzed together (RA, T1D, IBD) in the association study of complex traits performed by the Wellcome Trust Case Control Consortium(6). This suggests that this region might harbor genes involved in susceptibility to autoimmunity in general. Finally, there was a marginal association between the SNP rs6679677 in RSBN1 locus and JIA, and this was pronounced in male subjects. This variant, located ~73 kb from a functional variant in PTPN22 (rs2476601), is in a haplotype block suggesting that the association might be due to linkage disequilibrium with other SNPs.
We did not find associations with other autoimmunity-associated variants in the TRAF1 or IL2RA loci. Two studies have reported associations between JIA and TRAF1 variants. The variant that we tested in our cohort, rs3761847, was described by Behrens et al. to be associated with JIA in their cohort of 67 White subjects (OR 1.45, p <0.03)(32). This is the SNP that shows the strongest associations with RA(8). Another TRAF1 variant, rs10818488, which is ~14kb from rs3761847 and in linkage disequilibrium with it, was tested for an association with JIA in 338 cases and 511 controls(33). While the whole JIA cohort did not demonstrate an association with JIA, they did find an association with RF-negative polyarticular JIA subtype (OR 1.54), as well as with extended oligoarthritis and polyarticular JIA subtypes combined (OR 1.46). Our cohort had sufficient power (>80%) to detect an OR of 1.45 described in these two studies. It is possible that the actual effect sizes of the associations are lower than observed initially, and our cohort, although larger than both, is underpowered. It should also be noted that our JIA cohort was underpowered to detect associations of these variants with modest effects, given the inclusion of heterogeneous subgroups.
Our results suggest that STAT4 and TNFAIP3 variants are similar to PTPN22 and CTLA4 in predisposing to autoimmunity in general. The identification of common genetic factors underlying clinically distinct autoimmune phenotypes supports efforts to search for other variants that predispose to autoimmunity in general.
Acknowledgments
Supported by, The National Institute of Arthritis and Musculoskeletal and Skin Diseases (K23-AR50177), The National Institute of Diabetes and Digestive and Kidney Disorders (K23-DK069513) and The National Center for Research Resources (UL1-RR025764 and C06-RR11234), The Arthritis Foundation, and The Val A Browning Charitable Foundation, and The Primary Children’s Medical Center Foundation, Salt Lake City, UT.
References
- 1.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 2.Prahalad S, Ryan MH, Shear ES, Thompson SD, Giannini EH, Glass DN. Juvenile rheumatoid arthritis: linkage to HLA demonstrated by allele sharing in affected sibpairs. Arthritis Rheum. 2000;43:2335–8. doi: 10.1002/1529-0131(200010)43:10<2335::AID-ANR22>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Prahalad S, Glass DN. A comprehensive review of the genetics of juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2008;6:11. doi: 10.1186/1546-0096-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stagi S, Giani T, Simonini G, Falcini F. Thyroid function, autoimmune thyroiditis and coeliac disease in juvenile idiopathic arthritis. Rheumatology (Oxford) 2005;44:517–20. doi: 10.1093/rheumatology/keh531. [DOI] [PubMed] [Google Scholar]
- 5.Prahalad S, Shear ES, Thompson SD, Giannini EH, Glass DN. Increased prevalence of familial autoimmunity in simplex and multiplex families with juvenile rheumatoid arthritis. Arthritis Rheum. 2002;46:1851–6. doi: 10.1002/art.10370. [DOI] [PubMed] [Google Scholar]
- 6.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007 doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Ikari K, Kaneko H, Kochi Y, Yamamoto K, Shimane K, et al. Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum. 2008;58:1940–6. doi: 10.1002/art.23494. [DOI] [PubMed] [Google Scholar]
- 11.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4:e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HS, Remmers EF, Le JM, Kastner DL, Bae SC, Gregersen PK. Association of STAT4 with rheumatoid arthritis in the Korean population. Mol Med. 2007;13:455–60. doi: 10.2119/2007-00072.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orozco G, Alizadeh BZ, Delgado-Vega AM, Gonzalez-Gay MA, Balsa A, Pascual-Salcedo D, et al. Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis Rheum. 2008;58:1974–80. doi: 10.1002/art.23549. [DOI] [PubMed] [Google Scholar]
- 14.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274–9. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomino-Morales RJ, Rojas-Villarraga A, Gonzalez CI, Ramirez G, Anaya JM, Martin J. STAT4 but not TRAF1/C5 variants influence the risk of developing rheumatoid arthritis and systemic lupus erythematosus in Colombians. Genes Immun. 2008;9:379–82. doi: 10.1038/gene.2008.30. [DOI] [PubMed] [Google Scholar]
- 16.Zervou MI, Sidiropoulos P, Petraki E, Vazgiourakis V, Krasoudaki E, Raptopoulou A, et al. Association of a TRAF1 and a STAT4 gene polymorphism with increased risk for rheumatoid arthritis in a genetically homogeneous population. Hum Immunol. 2008;69:567–71. doi: 10.1016/j.humimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, et al. Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region. Arthritis Res Ther. 2008;10:R113. doi: 10.1186/ar2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez A, Varade J, Marquez A, Cenit MC, Espino L, Perdigones N, et al. Association of the STAT4 gene with increased susceptibility for some immune-mediated diseases. Arthritis Rheum. 2008;58:2598–602. doi: 10.1002/art.23792. [DOI] [PubMed] [Google Scholar]
- 19.Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN. STAT4: A risk factor for type 1 diabetes? Hum Immunol. 2008 doi: 10.1016/j.humimm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Korman BD, Alba MI, Le JM, Alevizos I, Smith JA, Nikolov NP, et al. Variant form of STAT4 is associated with primary Sjogren’s syndrome. Genes Immun. 2008;9:267–70. doi: 10.1038/gene.2008.1. [DOI] [PubMed] [Google Scholar]
- 21.Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, et al. Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci U S A. 1998;95:9979–84. doi: 10.1073/pnas.95.17.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 23.Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–71. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehringer S, Epplen JT, Krawczak M. Genetic association studies of bronchial asthma--a need for Bonferroni correction? Hum Genet. 2000;107:197. doi: 10.1007/s004390000353. [DOI] [PubMed] [Google Scholar]
- 26.Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316:1236–8. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 28.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 29.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007 doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008 doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O’Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–56. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 32.Behrens EM, Finkel TH, Bradfield JP, Kim CE, Linton L, Casalunovo T, et al. Association of the TRAF1-C5 locus on chromosome 9 with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2206–7. doi: 10.1002/art.23603. [DOI] [PubMed] [Google Scholar]
- 33.Albers HM, Kurreeman FA, Houwing-Duistermaat JJ, Brinkman DM, Kamphuis SS, Girschick HJ, et al. The TRAF1/C5 region is a risk factor for polyarthritis in Juvenile Idiopathic Arthritis. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.089060. [DOI] [PubMed] [Google Scholar]


