Abstract
Parameter estimation for infectious disease models is important for basic understanding (e.g. to identify major transmission pathways), for forecasting emerging epidemics, and for designing control measures. Differential equation models are often used, but statistical inference for differential equations suffers from numerical challenges and poor agreement between observational data and deterministic models. Accounting for these departures via stochastic model terms requires full specification of the probabilistic dynamics, and computationally demanding estimation methods. Here, we demonstrate the utility of an alternative approach, generalized profiling, which provides robustness to violations of a deterministic model without needing to specify a complete probabilistic model. We introduce novel means for estimating the robustness parameters and for statistical inference in this framework. The methods are applied to a model for pre-vaccination measles incidence in Ontario, and we demonstrate the statistical validity of our inference through extensive simulation. The results confirm that school term versus summer drives seasonality of transmission, but we find no effects of short school breaks and the estimated basic reproductive ratio ℛ0 greatly exceeds previous estimates. The approach applies naturally to any system for which candidate differential equations are available, and avoids many challenges that have limited Monte Carlo inference for state–space models.
Keywords: differential equation model, generalized profiling, state–space model, measles
1. Introduction
Mathematical models are now widely recognized as an important tool in efforts to understand, manage and contain infectious diseases in humans, other animals and plants. For some problems, general models may yield broadly applicable solutions. For example, very simple models that distinguished between ‘core’ and ‘non-core’ groups (having high and low contact rates, respectively) led to the successful adoption of contact tracing for the control of gonorrhea in the US [1]. Recently, Wallinga et al. [2] showed that in a broad range of situations, vaccinating individuals in the group experiencing the highest force of infection would be most effective for reducing the transmission of a novel infection.
But in many cases progress requires a model tailored to a specific situation with parameters estimated as accurately as possible. Fitting models to data is important for basic epidemiology, because comparison of alternative models is a powerful tool for discriminating between different hypotheses about underlying mechanisms (e.g. [3–8]) or potential environmental drivers of disease processes (e.g. [7,9–12]). And in many cases, the relative gains from different management options depend on quantitative values of parameters or on case-specific features such as multiple transmission routes and their relative importance (e.g. [6,13–18]).
An essential requirement for predicting the course of an outbreak and the responses to possible interventions is estimation of transmission rates—who infects whom, when and at what rate? But estimating transmission is also especially challenging because transmission is usually unobserved. We may know (at best) when a particular host individual contracted the infection, and in what circumstances, but (apart from sexually transmitted diseases in humans) it is rare to know with any degree of certainty which other individual passed the infection to them. Consequently, a fundamental goal of fitting dynamic models to epidemic data is to uncover the transmission process, and derive predictive models of transmission: at what rate will hosts become infected, and what factors (in the host population and its environment) influence that rate (e.g. [7,19–22])? That problem is the topic of this paper. We present a new statistical method that can be used to fit infectious disease models, generalized profiling [23], and illustrate its performance using case-report data on pre-vaccination era measles in Ontario, Canada.
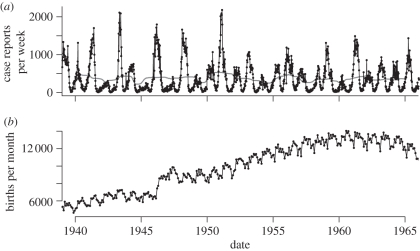
The data, plotted in figure 1, present a number of challenges for time series analysis.
— There is a substantial measurement error. Measles was contracted by essentially all children because of its high infectivity, but the number of reported cases is smaller than the number of births during the same time period by factor of ≈ 6, implying very large systematic under-reporting. In addition, an examination of a 24-month moving average of case reports indicates that there is no long-term trend in the number of case reports, even though the population increased substantially (as seen in the rising birth rate), suggesting a substantial trend in reporting rate. The low reporting rate also implies that random sampling error will be too large to ignore.
— We have data on only two processes (birth rate, and the rate of new infections) in a system with many state variables and other disease processes.
— The underlying process governing measles dynamics in Ontario, whose parameters we want to estimate, is nonlinear and stochastic, with events occurring continuously in time.
The state-of-the-art methods for this sort of fitting problem are Monte Carlo methods for state–space models, either Bayesian methods using Markov chain Monte Carlo (MCMC; e.g. [8,24,25] and references therein) or sequential Monte Carlo [7,26,27]. The method that we present here is completely different. The fitting criterion (explained in detail below) combines the least-squares trajectory matching [28–32] and gradient matching (e.g. [33–36]) fitting criteria, using basis expansion methods to reconstruct the full set of state variables. The underlying model is still a continuous time state–space model, but very few model solutions at candidate parameter values are required to converge to an excellent fit.
Figure 1.
(a) Weekly measles case reports for the province of Ontario, Canada, from 1939 through 1965. Data were digitized from unpublished, handwritten spreadsheets obtained from the Ontario Ministry of Health. The light grey line represents a moving average over the 24 months preceding and following each date. (b) Monthly live births for the province of Ontario, Canada, from 1939 through 1965. Data obtained from: Government of Canada, Dominion Bureau of Statistics, ‘vital statistics 1921–1966’. Both the weekly case reports and the monthly birth time series can be downloaded from the International Infectious Disease Data Archive (http://iidda.mcmaster.ca).
As a result, generalized profiling has some computational and other practical advantages. Most importantly, it has proven very challenging to construct MCMC proposal distributions (from which to draw new candidate parameter values) whose acceptance and ‘mixing’ rates are both high enough for the posterior distribution to be sampled well (e.g. to find all modes in the posterior) within a practical amount of time. Off-the-shelf methods have often been unsuccessful, for example New et al. [8] fitted three alternative state–space models for red grouse population cycles using WinBUGS, one of which involved grouse–parasite interactions. They observed poor mixing of the MCMC sampler for two out of the three models, and cautioned that ‘posterior densities may not accurately represent the target distribution, leading to questionable inference’. Sequential Monte Carlo methods may require careful case-specific fine-tuning to avoid ‘particle depletion’, the analogue of poor mixing (see §3.2 of Newman et al. [37]). Fitting by generalized profiling only requires nonlinear optimization. Optimization can also be computationally challenging in general, but the profiling objective function is often well-behaved because of regularization by the gradient matching component of the function (see §2.8 of Ramsay et al. [23]).
The generalized profiling method was first described by Ramsay et al. [23], and it has since been applied in several case studies. Here, in addition to challenging the method with epidemic data, we add two methodological developments: a data-based method for choosing the ‘smoothing’ parameter that sets the relative weighting of trajectory and gradient matching (previously there has been no objective way to choose the smoothing parameter), and an improved method for calculating asymptotic confidence intervals for parameter estimates.
Measles modelling is a well-ploughed field. We revisit measles because the degree of knowledge about this pathogen makes it possible to test our method in a simulation study that closely mimics the real-world data, by generating artificial data from a biologically realistic model with biologically realistic parameters. Our main ‘target’ was the pattern of seasonal variation in the transmission rate parameter, which is a key unknown in the transmission process and determines many of the most important features of the dynamics [38,39].
The results from the simulation study are encouraging in three respects: the estimated seasonal variation was very close to the actual variation in the data-generating model, the variability across replicates was similar to the width of the confidence bands on the estimate from the real data, and the computations could be completed in a reasonable amount of time on a current desktop workstation. The seasonal variability that we estimated from the real data is similar to the commonly used ‘term time’ model (high transmission when schools are in session, low when schools are out). However, there was some estimated variability within school terms, and no evidence of major decreases during short school holidays such as the Christmas break. In addition, the estimated mean transmission rate implies (with high confidence) a considerably higher value of ℛ0 than previously estimated for measles in Ontario.
2. Background
2.1. The data
In Ontario, notifications of measles cases were tabulated weekly from 1939 to 1989. Although never published, the Ontario Ministry of Health (OMoH) retained the original handwritten spreadsheets, which we digitized in 2001 for other work [40]. For the present analysis, we restricted our attention to the pre-vaccine era.
2.2. Model, measurement and parameterizations
As our model for the epidemic process, we use a version of the SEIR equations [40] with mortality assumed to be negligible in the age-group where infection occurs:
| 2.1a |
| 2.1b |
| 2.1c |
where S denotes a susceptible population (number of individuals), with recruitment rate ρ(t). Susceptibles become exposed (E, infected with the disease but not infectious) at the rate β(t)(I + v). Exposed individuals move into the infectious class I at rate σ and move out as they recover, at rate γ. We assume zero mortality in the population of interest. The recovered class (R) does not enter the equations for S, E and I, because we are assuming lifelong immunity, so there is no need to include a fourth equation (which would be Ṙ = γI ignoring mortality). The rate of infection is modified by a visiting impact v, which allows a small number of infectious individuals outside the modelled population to pass the infection to susceptibles. This has been included to remove the potential for extinction of an epidemic in the (stochastic) simulation studies below.
Our main goal is to estimate the time-varying transmission rate parameter β(t) and, in particular, to see how variation in β(t) relates to the structure of the school year. To represent this time-varying rate we used a basis expansion
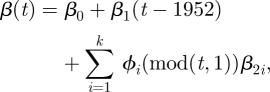 |
2.2 |
where the ϕi(t) were taken to be cyclic cubic B-splines with knots taken at intervals of 1/12 years. This number of knots is sufficient to represent both smooth seasonal variation and the sharper on/off variation of the school year versus summer holidays, and there are no apparent short-term features in the time-series of case reports (e.g., no dip in reports during or following the Christmas holiday; see figure 6 below1). The initial linear term allows for a long-term trend in the transmission rate, in addition to the annual cycle parameterized by the ϕi(t). Linear combinations of B-spline bases can represent constant functions such as β0 meaning that equation (2.2) is not identifiable as written. The β2i were therefore constrained to enforce
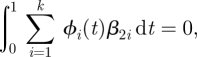 |
allowing this representation to have the effect of being a variation about an overall average for the year. The linear term was centred at 1952, the mid-point of the dates of our observations (1939 to 1965) in order to stabilize the estimate of β0 and allow its interpretation as the over-all transmission rate at the start of 1952.
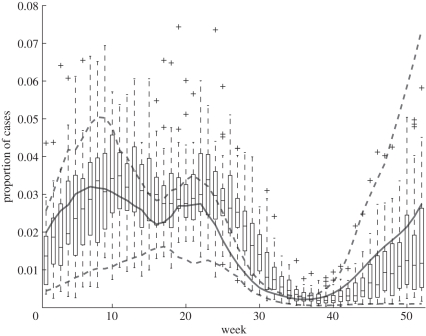
Figure 6.
Comparison of observed and model seasonal variation in measles incidence. The boxplots represent the week-by-week trend and variability in weekly case reports over the period 1940–1965, expressed as fractions of the total number of reports for the corresponding calendar year (i.e. all weekly case reports from 1940 were divided by the total number of case reports for all of 1940, and so on). The solid and dashed curves show the 25th, 50th and 75th percentiles (across years) of the same quantity in a forward solution of the deterministic model (2.1) starting from the estimated system state at the start of 1940.
In addition to disease processes, we also need to model the observation process. Only some fraction of weekly measles cases are assumed to be reported. While this can be modelled as a binomial process, the estimation methodology below only requires the specification of the expected value of observations as
for some reporting fraction p(t) and the ti occurring at 52 equal intervals across the year. The reporting ratio is also allowed to change linearly over time, represented by p(t) = p0 + p1(t − 1952), where the centring term has been used in the same manner as for the parametrization of β(t).
It will be convenient, although not necessary, to have direct observations of some state variable and we observe that equation (2.1) can be re-parameterized in terms of I*=p(t)I so that all parameters appear in the differential equation:
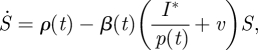 |
2.3a |
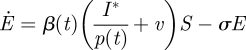 |
2.3b |
| 2.3c |
and we can set Iio = I*(ti). Note that equation (2.3c) is p(t)İ; the term ṗ(t)I is negligibly small (i.e. the mean reporting rate is nearly constant, relative to the epidemic dynamics that occur on a much shorter time scale) so for fitting purposes that term was omitted. In the remainder of this paper we drop the asterisk and use I to denote I*. It is often numerically easier to take the log of each state when solving equations such as equations (2.3) and we have followed this convention. We have also used the log of the observed data. In the simulation study below we employ a stochastic finite-population process (Gillespie algorithm, [41,42]) corresponding to equation (2.1) where the log transform also helps to make the variation in the paths more stable.
2.3. Initial parameter values
Some of the parameters in equations (2.3) were held fixed, because they are directly observable and so their values are known with effectively perfect precision relative to the uncertainties in the unobserved transmission and reporting processes. The recruitment rate ρ(t) is interpolated from the monthly birth rate data (figure 1) at a five-year lag, which approximates the age of entry into primary school. The time period between exposure and infectiousness (the latent period) is known to be around 8 days, giving a value σ = 365/8. Similarly, the mean infectious period is around 5 days, implying γ = 365/5.
Initial values also need to be specified for v, β(t) and p(t). We initialized the reporting rate to be constant (p1 = 0) and obtained an initial p0 from the ratio of total measles cases 1951–1960 to total births 1946–1955. This calculation assumes that all children are eventually infected so that the ratio of total observed cases to total births five years earlier provides an approximate proportion of reported infections. The resulting estimate is 16 per cent.
β(t) was initialized to approximate a step function providing one level at June, July and August and another during the rest of the year. Initial values are taken from Bauch & Earn [40]. v is initialized at 10−6, or about 10 infectious visitors continuously present.
3. The generalized profiling procedure
Ramsay et al. [23] describe a method for estimating parameters in ordinary differential equations such as equations (2.3) that is robust to model misspecification. In particular, it allows for small-variance stochastic forcing of a system, which would be expected to result from environmental and demographic stochasticity. The method is based on combining a collocation approach to estimating differential equations [43] and allowing deviations from differential equation models in a manner similar to the multiple shooting methods of Bock [29] and the trust region methods in Arora & Biegler [31].
To facilitate the exposition we use somewhat more general notation to write the state vector as x(t) = (S(t),E(t),I(t)) = (xS(t),xE(t),xI(t)) and refer to the components of x by xi(t) for i ∈ {S,E,I}. In this short-hand, we re-write equations (2.3) as ẋ = f(x;t,θ), where θ collects all the unknown parameters. In our model, θ = (p0,p1,v,β0,β1,β2,1,…, β2,12). Elements of f will be referenced in the same manner as elements of x.
We begin by representing x(t) in terms of a basis expansion
| 3.1 |
for Φ(t) = (ϕ1(t),…, ϕk(t)) a vector of basis functions and coefficient matrices C = (CS CE CI). Throughout we have used as Φ the cubic B-spline basis with knots at weekly intervals over 1939–1965. This basis has been chosen for its flexibility and numerical properties, but any sufficiently flexible choice of basis can be employed. Parameters are then estimated through a two-level optimization. First, for any value of the parameter vector θ, C is chosen by
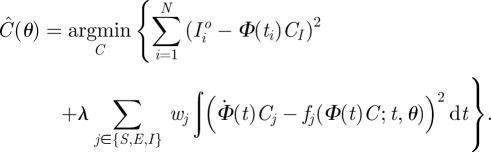 |
3.2 |
The first term on the right-hand side of equation (3.2) is the sum of squared deviations between the observed case-report data (scaled to compensate for under-reporting) and the estimated trajectory Φ(ti)CI. This is the fitting criterion for estimating parameters by least-squares trajectory matching. The second term is a weighted sum of squared deviations between the gradients (time derivatives) of the estimated trajectories for each state variable, Φ̇(t)Ci, and the corresponding gradients generated by the model, fi(Φ(t)C;t,θ). This is the fitting criterion for estimating parameters by gradient matching. In contrast with previous approaches, the gradient matching term includes unmeasured state variables, because the complete system state is estimated along with model parameters (a property shared by Monte Carlo methods for state–space models). The parameter λ determines the relative importance given to trajectory-matching and gradient-matching components of the fitting criterion.
The combined fitting criterion (3.2) means that the estimated trajectories only approximately satisfy equations (2.3). This contrasts with trajectory matching, in which estimated trajectories are solutions to the deterministic equations (2.3). In this way, equation (3.2) allows the data to impose deviations from the model if appropriate. This allows parameter estimates to be robust to departures from the model, either through imperfect specification of the functional forms of process rates, or stochastic departures from the deterministic paths predicted by the differential equations (for example, under suitable technical assumptions, the methods in Brunel [36] can be extended to show that gradient matching remains valid when the dynamics are stochastic with f(x;t,θ) being the expected value of ẋ given x(t)).
Here, equation (3.2) is expressed in similar terms to penalized objective functions for a nonparametric regression. Replacing the second term in equation (3.2) with λ∫(d2Φ(t)C/dt2)2dt corresponds exactly to standard smoothing spline regression models, as studied for example in Wahba [44]. In that situation, the value of λ controls the smoothness of the resulting regression function. Higher values of λ lead to smoother function estimates, so the optimal value of λ becomes small as the sample size increases. In our application, we think of equation (3.2) as allowing the data to impose deviations from the differential equation model, if appropriate. We thus default to thinking of λ as large, rather than small, so that estimated rates of change in state variables are always close to what the model predicts, given the estimated system state.
The weights wi allow us to place more or less emphasis on each of equations (2.3a)–(2.3c). We have placed relative weight 100 on the equation for S. This has been chosen to roughly mimic the expected size of departures from equations (2.3) associated with demographic stochasticity. The model is a ‘flow-through’ system, with all newborns passing through the S, E and I stages in turn. Therefore, the long-term average inflow and outflow rates are the same for each compartment, and roughly equal (on average) to the birth rate. We, therefore, expect approximately equal temporal variability in the rates of transitions between states S, E and I, so roughly equal values of the weights wi would be appropriate for fitting to untransformed state variables and data. Our fitting was done on a log-scale, however, and this transformation reduces the standard deviations in the rates of change in inverse proportion to the total population in each state. To undo this effect of the transformation, we should use weights with wi2 proportional to total population in each disease state. The factor of 100 is somewhat smaller than the value that this argument suggests, to allow for unmeasured migration processes. The resulting parameter estimates were insensitive to this choice; reversing the relative weighting of the variables resulted in less than a 5 per cent change in any parameter estimate. Note that while only I is measured, all state variables appear in the second term and so the minimization problem is feasible in principle.
In practice, the integral in the second term of equation (3.2) cannot be computed analytically and instead it is approximated by a numerical quadrature rule. We have used a rule that places one quadrature point tq at the midpoint between knots—i.e. halfway through each week. This allows the basis expansion to exactly solve equations (2.3) at those points, making the second term in equation (3.2) zero if need be. The resulting objective is written as
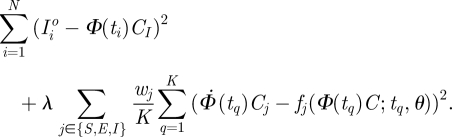 |
3.3 |
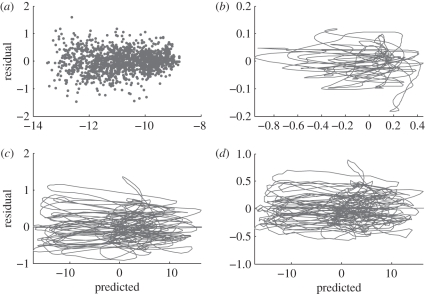
This is now a sum of squares criterion which can be minimized by a Newton–Raphson algorithm. This minimization is not guaranteed to avoid local minima and it is therefore important to find useful initialization values for C. A motivating factor in the transformation from equations (2.1) to (2.3) is that CI can be initialized by smoothing the observed Iio. From the starting values for CS, CE can be obtained by conducting the minimization with CI held fixed. Even then, the minimum found should be examined for clear lack of agreement using diagnostic plots such as those at the bottom of figure 4. When the equations cannot be written in such a way as to make some of the states directly observable, initial values for C can be obtained from a solution of equation (2.1) at the starting parameters.
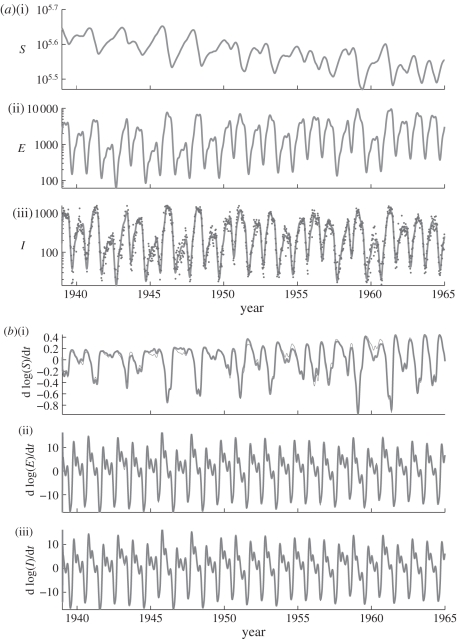
Figure 4.
(a(i–iii), Fit to data) show a comparison of estimated trajectories (lines) and observed data (points). Note that observations are available only for the I component of the state vector. (b(i–iii), Fit to model) show the derivative of the estimated trajectory (dark) compared with the derivative calculated from the right-hand side of equations (2.3) (light).
Using the definition (3.2), Ĉ is represented as a function of θ. We think of Ĉ(θ) as defining, for each θ, an estimated system trajectory in terms of the basis expansion (3.1). For choosing θ it is then natural to apply a sum of squares criterion, as would be used in nonlinear regression (e.g. [45]):
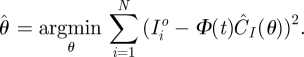 |
3.4 |
This sum-of-squares criteria can be minimized by a Gauss–Newton algorithm with gradients of ĈI(θ) obtained through the implicit function theorem.
The problems (3.2) and (3.4) can be generalized in a number of ways. Sums of squares while convenient to work with numerically are not strictly necessary and a log likelihood of the observations given the states could be used instead, as could alternative forms of the penalty. It is also not necessary to have a parameter-free comparison between a state variable and observations. However, as noted above, the ability to obtain approximate trajectories for some state variables via smoothing is helpful in providing starting values for equation (3.2). While we have instantiated equation (3.2) as an unconstrained problem, constraints between the states corresponding to conservation laws for example can be incorporated. Linear constraints, on state variables, in particular, can be translated into linear constraints on C to be enforced when minimizing (3.2). Nonlinear constraints can be enforced at each point tq used in equation (3.3). When deciding to impose such constraints a choice must be made between robustness to model violations and the maintenance of conservation laws.
Hooker [46] demonstrated that in the limit λ → ∞, the generalized profiling method is equivalent to fitting both parameters and initial conditions to the data by ordinary least squares. However, for finite values of λ, the method provides robustness to model misspecification and to disturbances of the dynamical system. It also results in optimization surfaces for both equations (3.2) and (3.4) that have fewer local maxima than attempting to fit the data by trajectory matching to solutions of equations (2.3). This leads to the strategy, when poor optima are found, of initially using a small value of λ and then increasing it in a similar manner to the use of ‘temperature’ in simulated annealing.
4. Choosing the smoothing parameter
While the methodology in Ramsay et al. [23] allows for computationally efficient and statistically robust parameter estimation, no guide was provided on how to choose the ‘smoothing parameter’ λ. As discussed above, it is natural to think of λ as being large, meaning that standard cross-validation methods that are designed to set λ → 0 are unlikely to yield appropriate values.
Ellner [47] suggested the approach of forwards cross-validation for these models. This method combines both estimates Ĉ(θ̂) and θ̂ to predict future responses. Specifically, let x(t,θ, x0) denote the solution, at time t, of equations (2.3) with x(0) = x0. Then we define the forward prediction error
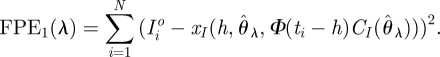 |
4.1 |
The terms in FPE1 are the (squared) errors made in predicting the observation at time ti by starting from the estimated trajectory at time ti − h, and solving the differential equation model from time ti − h to time t.
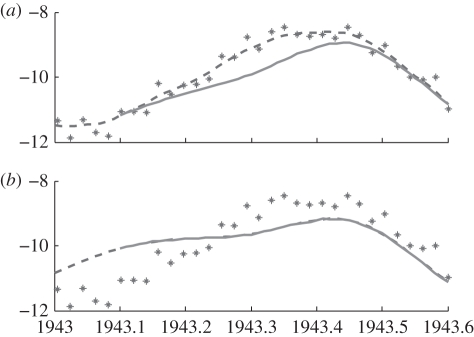
Forwards cross-validation is illustrated graphically in figure 2. The dots are the data, and the solid line is the estimated trajectory obtained from the generalized profiling approach for an overly small (figure 2a) and an overly large (figure 2b) value of λ. The dotted curve is a solution of the model with the corresponding parameter estimates θ̂(λ), starting from the estimated trajectory at t = 1943. In figure 2a, because λ is very small, the generalized profiling criterion emphasizes trajectory matching, so the estimated trajectory is close to the data. However, the estimated trajectory is not very close to being a solution of the model, so the model solution diverges from the trajectory (and from the data), generating large forward prediction errors. In figure 2b, λ is large, so the estimated trajectory is very close to being a solution of the model. However, the estimated trajectory is far from the data, so again the forward prediction errors are large. A good choice of λ will lie somewhere between these extremes.
Figure 2.
An example of forwards cross-validation from a particular point, in this case t = 1943. Dashed lines represent the estimated trajectory. Solid lines show the solution to equations (2.3) starting at the estimated system state at t = 1943. Discrepancy between the data (stars) and the solid lines represent forwards prediction error. (a) With λ = 1 the estimated trajectory is close to the data, but the solution to equations (2.3) diverges quickly from the estimated trajectory. (b) With λ = 106 the solution to equations (2.3) remain near the estimated trajectory, but the estimated trajectory does not follow the data closely.
Because of the computational cost of solving the differential equations, it will frequently be useful to use a small set of K starting observation times tik, k = 1,…,K and to solve equations (2.3) over a longer time interval. A single solution can then be compared with the next L observations. These solutions may overlap so that some observations are included more than once. For example, taking every second starting point t0, t2,…, tN−2, we could solve equations (2.3) for the next two observations. If ti+1–ti = h, this gives us
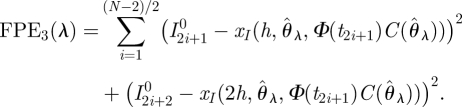 |
4.2 |
For our data, we took starting times tik at the beginning of each quarter and solved equations (2.3) for the following year, or 52 observations. We have chosen starting points at observation times and assumed a constant interval between observations for the sake of notational convenience; clearly, neither choice is strictly necessary. In practice, forwards prediction error can be minimized by a grid search, which is typically best carried out on a logarithmic scale.
Morton [48] suggested a similar approach for choosing smoothing parameters in nonparametric regression, based on predicting forward from an estimate of both the curve and its derivative. Hooker & Ellner [49] demonstrated that this approach will not, in general, yield consistent estimators of non-parametric functions. However, the strategy of minimizing FPEL(λ) can be justified from the point of view of fitting deterministic equations such as equations (2.1) in a way such that consistent estimates are obtained when the equations are correct but the estimates are robust to model violations either from stochastic effects or errors in the functional forms [49]. In particular, generalized profiling does not involve specifying (and then fitting) a stochastic model for the system dynamics—only the deterministic ‘skeleton’ is specified, and the observation process. The properties of generalized profiling for estimating process noise in an explicitly specified stochastic system have not been studied. The choice of L and of the tik allow the user to choose a level of robustness in parameter estimation that optimizes for a desired range of extrapolation forwards in time.
5. Confidence intervals
To estimate the variability of parameter estimates resulting from the equations above, we use a modification of the confidence intervals suggested by Ramsay et al. [23]. An initial covariance estimate is given by
| 5.1 |
where J is a matrix with rows
Here, σ̂2 is the residual variance. This formulation is derived from asymptotic results in nonlinear regression and is based on the assumption that the errors εi = Iio − Φ(ti)CI(θ) are independent. Because the smoothing process above can create dependence among the residuals at finite samples, we instead introduce a correction based on a Newey–West covariance estimate to allow for dependence induced by the estimate CI(θ). We derive this estimate by a modification of asymptotic results for maximum-likelihood estimates (e.g. [50]). Observing that
a Taylor expansion results in
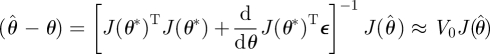 |
from which we obtain a covariance estimate
In non-linear regression, the rows of J are considered independent, resulting in (5.1). In order to account for potential serial dependence, we employ a Newey–West estimate [51] for the covariance of J,
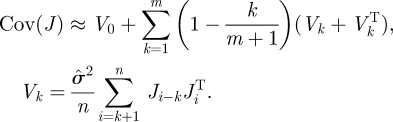 |
Effectively, these estimates allow for local dependence between the εi. Guided by the asymptotic theory [51], the number of terms m was chosen as the maximum of 5 and n1/4; empirically, the resulting confidence intervals are insensitive to the choice of m.
6. Results
The statistical methods described above were implemented in Matlab, using the software tools provided by Hooker [52]. Matlab code to run this analysis is available in the electronic supplementary material. The same functionality can be found in the CollocInfer package [53] for the R statistical programming language.
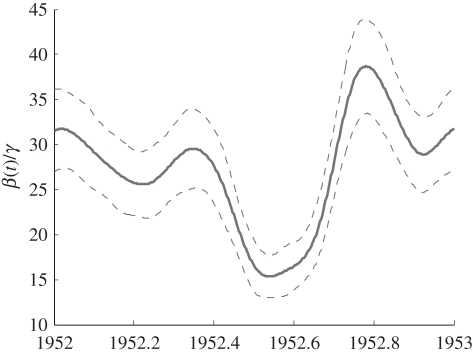
The estimated seasonal pattern of variation in the transmission parameter β(t)/γ is plotted in figure 3 along with pointwise confidence intervals. A distinct reduction in the transmission rate is evident over the summer holidays. The pronounced peak around the start of the school year may be owing to a higher rate of mixing, frailty effects or possibly an artefact to do with the influx of new susceptibles (which was not modelled here). He et al. [27] included a pulse of recruitment at the start of the school year, but assumed that contact rate was a step-function taking one value during school terms and another during holidays. As noted above, the slight decrease towards the end of the year and subsequent small peak may be artefacts owing to delayed reporting in pre-Christmas period. We estimate that the over-all level of β(t)/γ to be β̂0/γ = 27.0 ± 2.91 in 1953 and increased at rate β̂1/γ = 0.244 ± 0.115 per year. As expected, we estimated a substantial trend in the reporting rate (p̂0 = 26.9 ± 1.9% and p̂1 =− 0.0097 ± 0.002% per year). The flow of visitors into the infectious pool was estimated as essentially zero, but the objective function was effectively flat as a function of visiting rate at the point estimate of the parameters, suggesting that the data contain very little information about this parameter; this was also observed by He et al. [27] for large populations. For the purposes of numerical stability, v was therefore removed from the estimate of covariance used to compute confidence intervals.
Figure 3.
Estimated seasonal variation in the transmission rate (plotted for the year 1952; other years will be shifted slightly upwards or downwards owing to the long-term linear trend β̂1). Solid lines show the generalized profiling estimate, while the dashed lines show pointwise 95% CI.
A further advantage of the methods outlined above is that they provide readily accessible diagnostics for assessing model fit, based on the two components of the objective function: how well the estimated trajectories match the data, and how well the estimated gradients match the model. Figure 4a(i–iii) show the comparison between the data and the estimated trajectories. Figure 4b(i–iii) show the time-derivative of the estimated trajectories and the value of the right-hand side of equations (2.3) at each time point. These plots may be inspected for systematic departures from the corresponding trend. We observe that the observed data (only available for I) appear to be distributed around the estimated trajectory without obvious systematic departures. Similarly, the derivative of the estimated trajectory shows close agreement with that predicted by equations (2.3), particularly in the E and I variables. Departures from the predicted derivative of the S trajectory are evident, but do not show any systematic trends.
This analysis can be augmented by residual plots (figure 5). Residuals correspond to figure 4, either the departures of observations from their predicted trends, or the difference between the derivative of an estimated trajectory and the corresponding prediction from equations (2.3). These are plotted against predicted quantities so that any systematic relationship can be made clearer and the model potentially revised to account for them. In this case, the plots of figure 5 confirm that there are no systematic patterns of error in the trajectories or gradients, so we can interpret the discrepancies as stochastic departures of the real system from equations (2.3).
Figure 5.
Residual plots for the estimated trajectory of (a) I(t) and for the estimated gradients of (b) S, (c) E and (d) I, all on log scale. For I(t), the predicted values are the estimated trajectory, and the residuals are the departures of the weekly case report data from the estimated trajectory, i.e. the trajectory-matching component of the fitting criterion (3.2). For the gradients, predicted values are the model gradient at the estimated trajectory (fi(Φ(t)Ĉ;t,θ̂); see equation (3.2)) and the residuals are Φ̇(t)Ĉi − fi(Φ(t)Ĉ;t,θ̂), the gradient-matching component of the fitting criterion (3.2).
The estimated trend in the number of susceptibles (figure 4a(i–iii)), is owing to the estimated linear trend β̂1 in the transmission parameter β(t). In both cases, the rate of change is about 1 per cent per year (1% per year decrease in S, 1% per year increase in the mean of β(t)). However, there is very high uncertainty regarding the trend in β (e.g. a tenfold smaller trend is within two standard errors of the point estimate), so we cannot be confident that the estimated trends in S and β1 are real.
In figure 6, we look more closely at how well the fitted model captures the seasonal variation in disease incidence, by replotting the data as a function of week (within the year) and comparing this with model solutions. Because of the high year-to-year variability, the case reports for each week from 1940 to 1965 were expressed as fractions of the total number of reports for that calendar year, for both the data (box plots in figure 6) and a forward solution of the deterministic model (2.1) starting from the estimated system state at the start of 1940 (solid and dashed curves). Whereas the estimated trajectories (shown in figure 4) are ‘pulled’ towards the data by the trajectory matching component of the fitting criterion (3.2), forward solutions of equation (2.1) are free to go wherever they are pushed by the estimated seasonal variation in β(t), so this comparison is a genuine test of the fit. Forward solutions of the deterministic model follow fairly well the observed seasonal pattern of incidence. The main discrepancy is that the decrease and increase in incidence at the start and end, respectively, of the summer school holiday occur somewhat faster in model solutions than in the data. Otherwise, with the estimated seasonality of transmission rate (figure 3) the deterministic model appears to capture all of the qualitative properties of the observed seasonal pattern. This phenomenon was also described in He et al. [27], where it was explained as the effect of averaging over large spatial scales—local epidemics, each with faster onset and decline but at varying times, become blurred when averaged. This represents a failure of the mixing assumptions behind equations (2.1) and, as noted in He et al. [27], can result in poor estimation of latent periods.
7. A simulation study
Our specific goal in this paper was to show how the transmission rate β(t) could be reconstructed from a real measles time series using generalized profiling. While we have achieved that goal, it is ultimately impossible to know whether real processes that we have not modelled have influenced the success of our approach. In addition, any real incidence time series represents a single realization of a (very large) stochastic process, and we do not know how our results might vary for other realizations of the same process. Therefore, to validate the generalized profiling method for epidemiological time series, we conducted a simulation study based on a finite population model for an SEIR process. For the simulations, there is no uncertainty about the processes that generated the data, and we can examine the variability of our results across many realizations of the identical process.
Specifically, our Gillespie algorithm simulations employed a population of N = 107 individuals subject to a constant visitor impact of v = 10−6 individuals over the same 26 year time period that we studied the Ontario measles data. As in §2.3, we set σ = 365/8, γ = 365/5 and set ρ(t) using the published Ontario birth data. We set β(t) and p(t) equal to the functions that we estimated by generalized profiling (§6). Observations were generated by binomial samples with probability p(t) from transitions E → I over the previous week. Two hundred and fifty replicated histories were created, each requiring about 10 CPU hours on a current desktop workstation (one Pentium D 3 GHz processor). For each replicate,the parameters in the model were estimated using our methods. Each estimation, including a search over λ to minimize forward prediction error, required less than 2 CPU hours on the same workstation, and could be made considerably shorter by more careful initialization of the optimization and reducing the number of λ values considered.
We are interested in both the bias of the estimates and their variability. Two potential sources of bias should be distinguished: bias owing to the use of the generalized profiling objective function, and the bias associated with the use of a finite collocation basis to approximate system trajectories. The latter of these can be assessed by choosing a very large value of λ, and estimating parameters from error-free deterministic solutions to equations (2.3). Because a large value of λ places almost all emphasis on the second term in equation (3.2), this is equivalent to defining Ĉ(θ) as the coefficients that give the best possible approximation by the basis expansion (3.1) of exact solutions to the deterministic model (see [46]). Minimizing equation (3.4) is then a straightforward nonlinear least squares criterion. Since the data being fit in this procedure come from exact solutions of the differential equation without noise, the parameters will be estimated perfectly except for bias due to approximation of solutions by a finite basis expansion (3.1). The discrepancy between the original parameters and these can therefore be labelled collocation bias. Additional discrepancy between the parameters estimated by the method just described, and those estimated by generalized profiling on the Gillespie simulation data, are estimation bias resulting from the objective function.
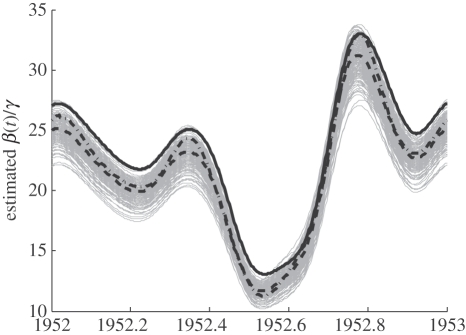
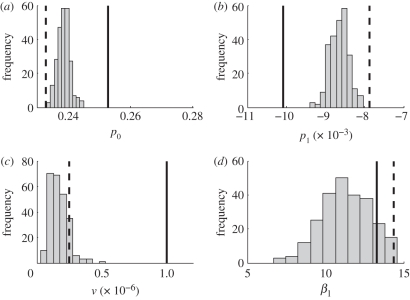
Figures 7 and 8 present estimates of the parameters on simulated data. In figure 7, the collocation bias is given by the difference between the solid and dashed-dot lines, while the estimation bias is the difference between the dashed line and the dashed-dot line. Both biases are small relative to the true magnitude of β(t), and the collocation bias is dominant, indicating that the bias could be reduced by using a larger number of terms on the basis expansion for trajectories. The spread of β(t) estimates from different stochastic simulations corresponds well to the confidence intervals in figure 3. There was collocation bias present in most of the other parameters (figure 8), and the trend in reporting rate shows some additional bias resulting from the profiling objective function. The collocation bias was highest for the visiting rate parameter, but (as noted above) this parameter was largely not identifiable.
Figure 7.
Estimates of β(t) from simulated data. Solid line: the β(t) function used in generating the simulated data. Grey lines: estimates for 250 stochastic simulations of the epidemic process (Gillespie algorithm). Dashed line: mean estimate from the stochastic simulations. Dashed-dotted line: estimate with a very large λ from an exact solution to equations (2.3).
Figure 8.
Estimated parameter values from 250 stochastic simulations of the epidemic process (Gillespie algorithm). (a) reporting rate in 1952. (b) Yearly change in reporting rate. (c) Estimated visiting rate. (d) Linear year-to-year trend in β(t). Solid vertical lines give the parameter values used in generating the simulated data. Dashed lines give values estimated with a very large λ for an exact solution to equations (2.3).
8. Discussion
Parameter estimation for continuous-time epidemic models from typically available data, such as a time series of case reports, is complicated by the nonlinear stochastic nature of the epidemic process, and by sampling variability and reporting bias in the observation process. Until very recently, these difficulties have been dealt with most effectively either by using a discrete-time approximation to the model, e.g. the TSIR model [20–22,24], or through the use of quasilikelihood indirect inference methods [54].
We have presented a method for fitting differential equation models that can deal with these challenges, without having to specify and parameterize a model for the stochastic deviations between the deterministic model and real-world outcomes. Like Monte Carlo methods (Bayesian MCMC and sequential Monte Carlo), our method involves estimating the trajectories of all state variables, measured and unmeasured, along with estimating the model parameters that are our main interest. But unlike Monte Carlo methods, this is accomplished by numerical optimization (producing a point estimate and confidence region for the coefficients specifying the trajectories), rather than by repeated sampling of possible trajectories to approximate the joint posterior distribution of parameters and trajectories. A naive MCMC approach to fitting the model in our simulation studies would have been impossible because of the computation time: ≈10 h simply to generate one proposal for state trajectories, so it would have taken several months to fit each artificial dataset. For this reason, successful applications of methods based on Monte Carlo simulations for epidemic models have employed alternative, more tractable, models of stochastic dynamics (e.g. [26–27]). Our methods avoid the computational cost of generating many proposals, and also the difficult problem of specifying a proposal distribution for trajectories and parameters that efficiently explores the full posterior distribution. Instead, inference can be based on asymptotic variance–covariance matrices for parameters and reconstructed trajectories. An important direction for further research is to compare the performance of generalized profiling with Monte Carlo methods.
This study is the first empirical application of forward cross-validation (FCV) for choosing the smoothing parameter λ in the generalized profiling method; theoretical properties of FCV are presented elsewhere [49]. FCV brings generalized profiling one step closer to being fully data-driven, and thus objective in the sense that results are not affected by ‘tuning parameters’ that a user can set at will. However, the complexity of the seasonal variation model for β(t) (the number of spline knots) was specified a priori based on the expected time scales of possible variation, as in other recent studies [20–22,24–27]. We have supported our choice in two ways: increasing the number of knots to verify that relevant features were not missed, and providing confidence bands (figure 3) that indicate which features are well supported by the data. However, a fully data-driven method for choosing the number of knots for β(t) would be advantageous.
Our results for measles in Ontario provide general support for the ‘term time’ model of seasonal variation, in which the dominant factor is assumed to be differences in contact rate between school terms and school holidays. However, our estimate of β(t) differs from the term time model in two respects. First, we did not detect any major decreases in transmission during short school holidays comparable in size to the decrease during the summer holiday, even with a weekly basis for estimating transmission seasonality. Second, our estimate includes some variation during the school year, in particular, the confidence bands in figure 3 imply that transmission is highest at the start of each school year and then declines, possibly with a secondary peak shortly after the new year. As we noted above, this within-term trend may be owing to the ‘staggered’ recruitment of individuals into the susceptible pool (a new cohort at the start of each school year), which our model omits. In addition to estimating the seasonal trend, our estimate of β(t) leads to an estimate of ℛ0, roughly given by the mean of β(t)/γ. Here, our results are somewhat surprising. The published estimate is ℛ0 ≈ 11–12 based on age-structured case reports from London, Ontario in 1912–1913. References [40,55,56] assumed this value in their analysis of incidence patterns in childhood diseases. Our estimate is considerably higher than that (figure 3), but it is within the range of values estimated for developed world cities prior to vaccination (Bauch & Earn [40], table 4 in appendix). Similarly, He et al. [27] estimated values of ℛ0 for measles in Britain that were roughly double previous estimates based on serological and age-dependent incidence data. Their model, like ours, makes a number of simplifying assumptions (homogeneous mixing, neglect of age structure, etc.) so these estimates should be interpreted cautiously until they are supported by more extensive studies using more realistic models.
Throughout this paper, we have kept the mean latent period 1/σ and the mean infectious period 1/γ fixed at their clinically observed values. Instead they could be estimated along with β(t) within the generalized profiling method. Estimating σ and γ from the Ontario measles data yielded unrealistically large values, reducing both disease stages from days to hours and compensating for this by substantially increasing β(t). This phenomenon was not found when these parameters were estimated from Gillespie process simulations of the model. In He et al. [27], two model errors were suggested to lead to different biases in the estimation of disease stage durations—unmodelled demographic stochasticity leading to shorter estimates, and spatial aggregation of data leading to longer estimates. Given these alternatives, our results suggest that the demographic stochasticity is a stronger influence than spatial aggregation in these data.
The potential for further model mis-specification suggests the investigation of the role of generalized profiling in model selection. Diagnostic plots such as those in figures 4 and 5 are clearly one way of demonstrating lack of fit, as is poor estimation of known constants. More generally, the comparison between alternative process models, for example, could be carried out by a number of statistics; minimum FPE(λ), the minimizing value of equation (3.4) or the size of the second term in equation (3.2) are obvious candidates. The properties of model selection based on these statistics have not been theoretically studied. Other questions of interest that have not received extensive treatment is to examine the uncertainty of the qualitative behaviour of equations (2.1); is it possible to determine from real data that a particular system is stable, cyclic or unstable?
Future work will apply this method to other disease time series and address biological issues. For example, we have weekly case reports for all reportable infectious diseases in Ontario over the period examined in this paper. If we consider childhood diseases with the same mean age of infection then the relevant contact pattern should be identical in all cases. If the estimated transmission rate variation is different for different diseases then we might infer that seasonal effects other than contact rate variation influence the seasonality of transmission differently for different diseases (e.g. one can imagine that humidity or temperature changes affect some pathogens more than others).
We have applied our methods to a recurrent infectious disease, but inferring transmission rate variation from case report data is also important for real-time estimation in the context of disease invasions, where one is often interested in understanding how various control measures are impacting transmission (or, in principle, in detecting evolution of transmissibility of the causative agent). In this situation, it is typical now to have daily case reports so methods have recently been developed that exploit daily data to estimate β(t) without relying on a specific epidemiological model such as the SEIR framework [57,58]. With daily case data, and an estimate of the distribution of the serial interval (the time from acquiring to transmitting the infection), one can associate reported cases with transmission events in the past, and consequently estimate the daily reproductive ratio ℛ(t) (or, equivalently, β(t) ≃ ℛ(t)/Tinf, where Tinf is the mean infectious period). The extent to which these methods can be adapted usefully for weekly time series of recurrent epidemics remains to be seen. Conversely, generalized profiling can be applied to daily time series, and this would provide a useful comparison for results obtained with the Wallinga–Teunis method [57], and other methods to estimate ℛ(t) for disease invasions (e.g. [59–61]).
Acknowledgements
This research was supported by NSF grant DEB-0813734 (S.P.E. and G.J.H.), grants from the James S. McDonnell Foundation (S.P.E. and D.J.D.E.), by the Cornell University Agricultural Experiment Station federal formula funds Project No. 150446 (G.J.H.), and CIHR and NSERC (D.J.D.E.).
Footnotes
When we used twice as many knots, the estimated β(t) contained only one additional feature: a hint of a brief decrease in the month prior to the Christmas holiday, which has been observed in other childhood diseases and interpreted as resulting from a delay in reporting caused by slower mail delivery. As this is not a property of the actual epidemic dynamics, we consider 1/12 year knot intervals to be sufficient for representing the actual β(t) process.
References
- 1.Hethcote H. W., Yorke J. A. 1984. Gonorrhea transmission dynamics and control. New York, NY: Springer [Google Scholar]
- 2.Wallinga J., van Boven M., Lipsitch M. 2010. Optimizing infectious disease interventions during an emerging epidemic. Proc. Natl Acad. Sci. USA 107, 923–928 10.1073/pnas.0908491107 (doi:10.1073/pnas.0908491107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koelle K., Pascual M. 2004. Disentangling extrinsic from intrinsic factors in disease dynamics: a nonlinear time series approach with an application to cholera. Am. Nat. 163, 901–913 10.1086/420798 (doi:10.1086/420798) [DOI] [PubMed] [Google Scholar]
- 4.Webb C. T., Brooks C. P., Gage K. L., Antolin M. F. 2006. Classic flea-borne transmission does not drive plague epizootics in prairie dogs. Proc. Natl Acad. Sci. USA 103, 6236–6241 10.1073/pnas.0510090103 (doi:10.1073/pnas.0510090103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wearing H. J., Rohani P. 2006. Ecological and immunological determinants of dengue epidemics. Proc. Natl Acad. Sci. USA 103, 11 802–11 807 10.1073/pnas.0602960103 (doi:10.1073/pnas.0602960103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller M. W., Hobbs N. T., Tavener S. J. 2006. Dynamics of prion disease transmission in mule deer. Ecol. Appl. 16, 2208–2214 10.1890/1051-0761(2006)016[2208:DOPDTI]2.0.CO;2 (doi:10.1890/1051-0761(2006)016[2208:DOPDTI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 7.King A. A., Ionides E. L., Pascual M., Bouma M. J. 2008. Inapparent infections and cholera dynamics. Nature 454, 877–880 10.1038/nature07084 (doi:10.1038/nature07084) [DOI] [PubMed] [Google Scholar]
- 8.New L. F., Matthiopoulos J., Redpath S., Buckland S. T. 2009. Fitting models of multiple hypotheses to partial population data: investigating the causes of cycles in red grouse. Am. Nat. 174, 399–412 10.1086/603625 (doi:10.1086/603625) [DOI] [PubMed] [Google Scholar]
- 9.Pascual M., Rodo X., Ellner S. P., Colwell R., Bouma M. J. 2000. Cholera dynamics and El Niño-southern oscillation. Science 289, 1766–1769 10.1126/science.289.5485.1766 (doi:10.1126/science.289.5485.1766) [DOI] [PubMed] [Google Scholar]
- 10.Koelle K., Rodo X., Pascual M., Yunus M., Mostafa G. 2005. Refractory periods and climate forcing in cholera dynamics. Nature 436 696–700 10.1038/nature03820 (doi:10.1038/nature03820) [DOI] [PubMed] [Google Scholar]
- 11.Ferrari M. J., Grais R. F., Bharti N., Conlan A. J. K., Bjornstad O. N., Wolfson L. J., Guerin P. J., Djibo A., Grenfell B. T. 2008. The dynamics of measles in sub-saharan Africa. Nature 451, 679–684 10.1038/nature06509 (doi:10.1038/nature06509) [DOI] [PubMed] [Google Scholar]
- 12.Shaman J., Pitzer V. E., Viboud C., Grenfell B. T., Lipsitch M. 2010. Absolute humidity and the seasonal onset of influenza in the continental united states. PLoS Biol. 8 10.1371/journal.pbio.1000316 (doi:10.1371/journal.pbio.1000316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blower S. M., Gershengorn H. B., Grant R. M. 2000. A tale of two futures: HIV and antiretroviral therapy in San Francisco. Science 287, 650–654 10.1126/science.287.5453.650 (doi:10.1126/science.287.5453.650) [DOI] [PubMed] [Google Scholar]
- 14.Keeling M. J. 2005. Models of foot-and-mouth disease. Proc. R. Soc. B 272, 1195–1202 10.1098/rspb.2004.3046 (doi:10.1098/rspb.2004.3046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longini I. M., Halloran M. E. 2005. Strategy for distribution of influenza vaccine to high-risk groups and children. Am. J. Epidemiol. 161, 303–306 10.1093/aje/kwi053 (doi:10.1093/aje/kwi053) [DOI] [PubMed] [Google Scholar]
- 16.Ferguson N. M., Cummings D. A. T., Fraser C., Cajka J. C., Cooley P. C., Burke D. S. 2006. Strategies for mitigating an influenza pandemic. Nature 442, 448–452 10.1038/nature04795 (doi:10.1038/nature04795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truscott J. E., Ferguson N. M. 2009. Control of scrapie in the UK sheep population. Epidemiol. Infect. 137, 775–786 10.1017/S0950268808001064 (doi:10.1017/S0950268808001064) [DOI] [PubMed] [Google Scholar]
- 18.Medlock J., Galvani A. P. 2009. Optimizing influenza vaccine distribution. Science 325, 1705–1708 10.1126/science.1175570 (doi:10.1126/science.1175570) [DOI] [PubMed] [Google Scholar]
- 19.Fine P., Clarkson J. 1982. Measles in England and Wales—1: an analysis of factors underlying seasonal patterns. Int. J. Epidemiol. 11, 5–14 10.1093/ije/11.1.5 (doi:10.1093/ije/11.1.5) [DOI] [PubMed] [Google Scholar]
- 20.Finkenstaedt B. F., Grenfell B. T. 2000. Time series modelling of childhood diseases: a dynamical systems approach. J. R. Statist. Soc. C 49, 187–205 10.1111/1467-9876.00187 (doi:10.1111/1467-9876.00187) [DOI] [Google Scholar]
- 21.Xia Y., Gog J. R., Grenfell B. T. 2005. Semiparametric estimation of the duration of immunity from infectious disease time series: influenza as a case-study. J. R. Statist. Soc. C 54, 659–672 10.1111/j.1467-9876.2005.05383.x (doi:10.1111/j.1467-9876.2005.05383.x) [DOI] [Google Scholar]
- 22.Metcalf C. J. E., Bjornstad O. N., Grenfell B. T., Andreasen V. 2009. Seasonality and comparative dynamics of six childhood infections in pre-vaccination Copenhagen. Proc. R. Soc. B 276, 4111–4118 10.1098/rspb.2009.1058 (doi:10.1098/rspb.2009.1058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsay J., Hooker G., Campbell D., Cao J. 2007. Parameter estimation for differential equations: a generalized smoothing approach. J. R. Statist. Soc. B 69, 741–770 10.1111/j.1467-9868.2007.00610.x (doi:10.1111/j.1467-9868.2007.00610.x) [DOI] [Google Scholar]
- 24.Morton A., Finkenstaedt B. 2005. Discrete time modelling of disease incidence time series by using Markov Chain Monte Carlo methods. J. R. Statist. Soc. C 54, 575–594 10.1111/j.1467-9876.2005.05366.x (doi:10.1111/j.1467-9876.2005.05366.x) [DOI] [Google Scholar]
- 25.Cauchemez S., Ferguson N. M. 2008. Likelihood-based estimation of continuous-time epidemic models from time-series data: application to measles transmission in London. J. R. Soc. Interface 5, 885–897 10.1098/rsif.2007.1292 (doi:10.1098/rsif.2007.1292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ionides E. L., Breto C., King A. A. 2006. Inference for nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 103, 18 438–18 443 10.1073/pnas.0603181103 (doi:10.1073/pnas.0603181103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He D. H., Ionides E. L., King A. A. 2010. Plug-and-play inference for disease dynamics: measles in large and small populations as a case study. J. R. Soc. Interface 7, 271–283 10.1098/rsif.2009.0151 (doi:10.1098/rsif.2009.0151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banks H. T., Burns J. A., Cliff E. M. 1981. Parameter estimation and identification for systems with delays. SIAM J. Control Optim. 19, 791–828 10.1137/0319051 (doi:10.1137/0319051) [DOI] [Google Scholar]
- 29.Bock H. G. 1983. Recent advances in parameter identification techniques for ODE. In Numerical treatment of inverse problems in differential and integral equations (eds Deuflhard P., Harrier E.), pp. 95–121 Basel, Switzerland: Birkhäuser [Google Scholar]
- 30.Biegler L., Damiano J. J., Blau G. E. 1986. Nonlinear parameter estimation: a case study comparison. AIChE J. 32, 29–45 10.1002/aic.690320105 (doi:10.1002/aic.690320105) [DOI] [Google Scholar]
- 31.Arora N., Biegler L. T. 2004. A trust region SQP algorithm for equality constrained parameter estimation with simple parametric bounds. Comput. Optim. Appl. 28, 51–86 10.1023/B:COAP.0000018879.40214.11 (doi:10.1023/B:COAP.0000018879.40214.11) [DOI] [Google Scholar]
- 32.Ciupe M. S., Bivort B. L., Bortz D. M., Nelson P. W. 2006. Estimating kinetic parameters from HIV primary infection data through the eyes of three different mathematical models. Math. Biosci. 200, 1–27 10.1016/j.mbs.2005.12.006 (doi:10.1016/j.mbs.2005.12.006) [DOI] [PubMed] [Google Scholar]
- 33.Swartz J., Bremermann H. 1975. Discussion of parameter estimation in biological modelling: algorithms for estimation and evaluation of the estimates. J. Math. Biol. 1, 241–257 10.1007/BF01273746 (doi:10.1007/BF01273746) [DOI] [PubMed] [Google Scholar]
- 34.Varah J. 1982. A spline least squares method for numerical parameter estimation in differential equations. SIAM J. Scient. Statist. Comput. 3, 28–46 10.1137/0903003 (doi:10.1137/0903003) [DOI] [Google Scholar]
- 35.Ellner S. P., Seifu Y., Smith R. H. 2002. Fitting population dynamic models to time-series data by gradient matching. Ecology 83, 2256–2270 10.1890/0012-9658(2002)083[2256:FPDMTT]2.0.CO;2 (doi:10.1890/0012-9658(2002)083[2256:FPDMTT]2.0.CO;2) [DOI] [Google Scholar]
- 36.Brunel J.-B. 2008. Parameter estimation of ODE's via nonparametric estimators. Electron. J. Statist. 2, 1242–1267 10.1214/07-EJS132 (doi:10.1214/07-EJS132) [DOI] [Google Scholar]
- 37.Newman K. B., Fernández C., Thomas L., Buckland S. T. 2009. Monte carlo inference for state–space models of wild animal populations. Biometrics 65, 572–583 10.1111/j.1541-0420.2008.01073.x (doi:10.1111/j.1541-0420.2008.01073.x) [DOI] [PubMed] [Google Scholar]
- 38.Earn D. J. D., Rohani P., Bolker B. M., Grenfell B. T. 2000. A simple model for complex dynamical transitions in epidemics. Science 287, 667–670 10.1126/science.287.5453.667 (doi:10.1126/science.287.5453.667) [DOI] [PubMed] [Google Scholar]
- 39.Earn D. J. D. 2009. Mathematical epidemiology of infectious diseases. In Mathematical biology, vol. 14 (eds Lewis M. A., Chaplain M. A. J., Keener J. P., Maini P. K.), IAS/Park City Mathematics Series, pp 151–186 Providence, RI: American Mathematical Society [Google Scholar]
- 40.Bauch C. T., Earn D. J. D. 2003. Transients and attractors in epidemics. Proc. R. Soc. Lond. B 270, 1573–1578 10.1098/rspb.2003.2410 (doi:10.1098/rspb.2003.2410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurtz T. G. 1980. Relationships between stochastic and deterministic population models. Lecture Notes in Biomathematics 38, 449–467 [Google Scholar]
- 42.Gillespie D. T. 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 10.1016/0021-9991(76)90041-3 (doi:10.1016/0021-9991(76)90041-3) [DOI] [Google Scholar]
- 43.Deuflhard P., Bornemann F. 2000. Scientific computing with ordinary differential equations. New York, NY: Springer [Google Scholar]
- 44.Wahba G. 1990. Spline models for observational data. SIAM CBMS-NSF Regional Conference Series in Applied Mathematics. Philadelphia, PA: SIAM [Google Scholar]
- 45.Bates D. M., Watts D. B. 1988. Nonlinear regression analysis and its applications. New York, NY: Wiley [Google Scholar]
- 46.Hooker G. 2007. Theorems and calculations for smoothing-based profiled estimation of differential equations. Technical Report BU-1671-M, Department of Biological Statistics and Computational Biology, Cornell University, Ithaca, NY [Google Scholar]
- 47.Ellner S. P. 2007. Commentary on ‘parameter estimation in differential equations: a generalized smoothing approach’. J. R. Statist. Soc. B 16, 741–796 [Google Scholar]
- 48.Morton R., Kang E. L., Henderson B. L. 2009. Smoothing splines for trend estimation and prediction in time series. Environmetrics 20, 249–259 10.1002/env.925 (doi:10.1002/env.925) [DOI] [Google Scholar]
- 49.Hooker G., Ellner S. P. 2010 On forwards prediction error. Technical report BU-1679-M, Department of Biological Statistics and Computational Biology, Cornell University, Ithaca, NY. [Google Scholar]
- 50.Lehmann E. L., Casella G. 1997. Theory of point estimation. New York, NY: Springer [Google Scholar]
- 51.Newey W. K., West K. D. 1987. A simple, positive semi-definite, heteroskdasticity and autocorrelation consistent covariance matrix. Econometrica 55, 703–708 10.2307/1913610 (doi:10.2307/1913610) [DOI] [Google Scholar]
- 52.Hooker G. 2006. Matlab functions for the profiled estimation of differential equations. Department of Biological Statistics and Computational Biology, Cornell University, Ithaca, NY [Google Scholar]
- 53.Hooker G., Xiao L., Ramsay J. 2010. CollocInfer: collocation inference for dynamic systems. R package version 0.1.0 [Google Scholar]
- 54.Gouriéroux C., Monfort A. 1996. Simulation-based econometric methods. CORE Lectures. Oxford, UK: Oxford University Press [Google Scholar]
- 55.Anderson R. M., May R. M. 1991. Infectious diseases of humans: dynamics and control. Oxford, UK: Oxford University Press [Google Scholar]
- 56.Anderson R. M., May R. M. 1985. Age-related changes in the rate of disease transmission: implications for the design of vaccination programmes. J. Hygiene 94, 365–436 10.1017/S002217240006160X (doi:10.1017/S002217240006160X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallinga J., Teunis P. 2004. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 160, 509–516 10.1093/aje/kwh255 (doi:10.1093/aje/kwh255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldstein E., Dushoff J., Ma J., Plotkin J., Earn D. J. D., Lipsitch M. 2009. Reconstructing influenza incidence by deconvolution of daily mortality times series. Proc. Natl Acad. Sci. USA 106, 21 825–21 829 10.1073/pnas0902958106 (doi:10.1073/pnas0902958106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pourbohloul B., et al. 2009. Initial human transmission dynamics of a novel swine-origin influenza A (H1N1) virus (S-OIV) in North America, 2009. Influenza and Other Respiratory Viruses 3, 215–222 10.1111/j.1750-2659.2009.00100.x (doi:10.1111/j.1750-2659.2009.00100.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraser C., et al. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324, 1557–1561 10.1126/science.1176062 (doi:10.1126/science.1176062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y., Sugimoto J. D., Halloran M. E., Basta N. E., Chao D. L., Matrajt L., Potter G., Kenah E., Longini I. M. 2009. The transmissibility and control of pandemic influenza A (H1N1) virus. Science 326, 729–733 10.1126/science.1177373 (doi:10.1126/science.1177373) [DOI] [PMC free article] [PubMed] [Google Scholar]