Abstract
The lethal factor of Bacillus anthracis is a major factor for lethality of anthrax infection by this bacterium. With the aid of the protective antigen, lethal factor gains excess to the cell cytosol where it manifests toxicity as a metalloprotease. For better understanding of its specificity, we have determined its residue preferences of nineteen amino acids in six subsites (from P3 to P3’) as relative kcat/Km values (specificity constants). These results showed that lethal factor has a broad specificity with preference toward hydrophobic residues, but not charged or branched residues. The most preferred residues in these six subsites are, from P1 to P3’, Trp, Leu, Met, Tyr, Pro, and Leu. The result of residue preference was used to design new substrates with superior hydrolytic characteristics and inhibitors with high potency. For better use of the new findings for inhibitor design, we have modeled the most preferred residues in the active site of lethal factor. The observed interactions provide new insights to future inhibitor designs.
Keywords: Bacillus anthracis, lethal factor, specificity, Inhibitor
1. Introduction
Anthrax is a disease caused by the infection of Bacillus anthracis, a gram-positive spore-forming bacterium usually found in the soil [1]. The infectious spores of the bacterium can enter human body through the gastrointestinal tracts (ingestion), skin (cutaneous), and lungs (inhalation) and produce distinct clinical symptoms [2]. Inhalational anthrax is the most dangerous form to human and is usually fatal. There has been an increasing interest in the development of treatment for Anthrax especially in view that B. anthracis spores have been used as a bio-weapon. Therefore, the understanding of the lethal mechanism of anthrax is of basic scientific importance.
Once B. anthracis spores are inhaled into the host, they rapidly germinate and proliferate in the circulation system. Vegetative B. anthracis secretes three plasmid-encoded toxin proteins named protective antigen (PA), lethal factor (LF) and edema factor (EF). They work together to cause most of the pathological consequences in the host. PA can combine with LF to form lethal toxin (LeTx) or with EF to form edema toxin (EdTx). These complexes gain entrance to cells through receptor binding to PA, a process ultimately delivers LF and EF to the cytosol. The pathological activities of these toxins are manifested in the cytosol by their enzymic activities. EF is an adenylate cyclase which causes increased level of cAMP in the cells. LF is a metalloproteinase and is by far the most toxic component of the B. anthracis infection [3,4]. The administration of low dosage of LT is lethal to experimental animals [1]. In human inhalation anthrax, the elimination of bacteria by antibiotics was frequently insufficient to rescue the patients [5]. Such clinical failure was thought to have caused by the presence of active LF in the cells. The best established cellular targets of LF are the members of MAP kinase kinase (MAPKK) family and the inactivation of these enzymes may account for some of the toxicity of LF [6–8]. However, other protein substrates of LF have also been proposed [9]. These observations suggest that LF is a potential therapeutic target of anthrax for the development of small molecular inhibitor drugs and the full understanding of LF specificity would be beneficial to this end.
The catalytic unit of LF which performs substrate recognition and hydrolysis is formed by three of the four domains in LF. The catalytic active site comprises a bound Zn atom and three histidine side chains. From the crystal structure of substrate peptide bound to LF [10], the binding cleft is large enough to accommodate several amino acid residues (subsites). Preliminary specificity of the subsites has been derived from the alignment of sequences around the LF cleavage sites of MAPKK enzymes [11]. The lack of clear consensus residues in the subsites (Table 1) suggests that LF has a broad amino acid preference in nearly all the subsites, an assumption supported by kinetic data on synthetic peptide substrates [12]. Although peptide inhibitors of LF based on cleavage site sequence of MAPKKs have shown good potency [13], they are too large in molecular size to be useful in clinical settings. Detailed knowledge on LF subsite specificity would provide insights for the design of small potent inhibitors with pharmacological potentials. Here we report the residue preferences in six subsites of LF (from P3 to P3’) determined as kinetic parameter, relative kcat/Km. The protein-substrate interactions was also studied by molecular modeling of binding modes of the most preferred residues in these subsites.
Table 1.
Comparison of the most preferred amino acids with those of LF biological substrates
| P3 | P2 | P1 | P1’ | P2’ | P3’ | |
|---|---|---|---|---|---|---|
| MAPKK-1 | P | T | P | I | Q | L |
| MAPKK-2 | V | L | P | A | L | T |
| MAPKK-3 | D | L | R | I | S | C |
| MAPKK-4 | A | L | K | L | N | F |
| MAPKK-6 | G | L | K | I | P | K |
| MAPKK-7 | T | L | Q | L | P | S |
| W(100) | L(100) | M(100) | Y(100) | P(100) | L(100) | |
| Most preferred amino acid | F(29) | F(68) | A(70) | L(66) | A(72) | I(81) |
| Y(20) | Y(42) | P(68) | I(39) | L(69) | M(76) |
LF cleaves it substrates between P1 and P1’ (scissile bond). The positions on the left side of the scissile bond are named P1, P2, P3 and so on; and those on the right sides are named P1’, P2’, P3 and so on. For each subsite, the amino acid with the highest initial velocity was assigned a number of 100, and others are the percentage of their initial velocity versus the highest one.
2. Materials and methods
2.1. Design of the substrate mixtures
Peptide mixtures were designed and synthesized based on a peptide template as RGKKKVLR* ILLN (in which the star denotes the cleaving site) which was known to be cleaved by LF. For characterization of each of the six subsites studied, a peptide mixture composed of 19 equal molar peptides which were differed only by one amino acid at a single subsite was designed and synthesized in an appropriate cycle of solid-phase peptide synthesis (Synpep, Dublin, CA). Because limiting the number of peptides in a mixture facilitated their identification [14], the 19 peptides were grouped into four sets of substrate mixtures according to their molecular weights of all the amino acid studied. Thus, 24 substrate mixtures in total were required for characterization of all the six subsites. A substrate with known kcat/Km will be added to each mixture to work as an internal standard.
2.2. Initial rate determination
Substrate mixtures were dissolved in DMSO at 10 mM as stocks and were further diluted to 10 µM in 20 mM HEPES, pH 7.4, 0.1 mg ml−1 BSA. Reactions were initiated at 37 °C by the addition of 10 nM LF. Aliquots were removed at different time points and reactions were stopped by the addition of formic acid (10% of reaction system in volume). Quantitative analysis was conducted by ESI-LC MS. The system was composed of an Agilent 1100 HPLC, a Clipeus 1 × 50 mm 5 mm C-18 column, and a Bruker Microtof ESI-LC MS. The HPLC buffers used were: A – 99.5% H2O, 0.5% Formic Acid, and B – 99.5% ACCN, 0.5% Formic Acid. Separations were conducted using a 5% to 50% B gradient over 4 minutes at a flow rate of 200 ml/min. Ion detection was accomplished using the time of flight instrument in positive reflector mode with ion detection between 200 and 2000 m/z through an ESI interface. Data were analyzed by Quant Analysis software equipped with the ESI mass spectrometer to obtain peak areas of the substrates and their corresponding products in a given reaction.
Each of the libraries was incubated with LF and the hydrolysis was analyzed in ESI-LC MS. To work out the hydrolytical conditions and monitor procedures, a mode peptide with the sequence of RGKKKVLRILLN was incubated with LF and analyzed. In order to compare their hydrolysis rates, relative product formation was calculated as a ratio of the peak area of a product to the sum of the areas of both the product and its corresponding substrate. Data were plotted in terms of relative product formation as a function of reaction incubation time. Initial velocity was obtained from nonlinear regression analysis representing the initial 15% formation of product. The relative initial velocity of different substrate is equal to the relative kcat/Km.
2.3. Determination of kinetic parameters
The kinetic experiments for investigating Km and kcat for synthesized LF fluorogenic substrates were performed in a buffer of 0.1 M HEPES, pH 7.4 at 37 °C. Reactions were performed in black flat-bottomed 96-well plates by mixing LF (10 nM) with varying concentrations of the fluorogenic substrates. The hydrolysis of substrate was monitored by continuously measuring the increase of fluorescence intensity using a TECAN 200, a fluorescence microplate reader. An excitation wavelength of 325 nm and an emission wavelength of 393 nm were used to monitor the changes of fluorescence intensity. The reaction rate of substrate hydrolysis for each substrate concentration was obtained as the initial velocity which was calculated as the ratio of initial 15% product formation to the reaction time. The final reaction curve was plotted as reaction rate as a function of substrate concentrations. The kinetic parameters, Km and Vmax, were gained by fitting the reaction curve using GraFit 5 (Erithacus Software, Horley, Surrey, U.K.), a nonlinear regression analysis software. Kcat was calculated as the ratio of Vmax to the initial LF concentration [14–15].
2.4. Investigation of inhibition constants of LF inhibitors
Both the kinetic assays for investigating inhibition constants (Ki) and data analysis were similar to 2.3 except that investigation of inhibition constants of LF inhibitors required the presence of LF inhibitor at different concentrations. The final result was plotted as different initial velocities as a function of varying concentrations of LF inhibitor. The Ki values were obtained by fitting the curve using software of GraFit 5 [16].
2.5. Molecular modeling of subsite binding
A hexapeptide containing the most preferred residues was modeled into the active site of LF using the X-ray structure of LF complexed with a MAPKK peptide (PDB access code 1pww) [12]. Firstly, the side chains of the original peptide were replaced with the optimized residues and then the substrate and surrounding LF residues (residues A297-I345, L368-Q417, P551-S776) were subjected to 20 cycles of conjugate gradient energy minimization calculations using program CNS / Crystallography & NMR System [17]. During the process, the total energy decreased from 3058.5 to 2767.7, finally, the residues were fixed and examined for their contacts in graphic models.
3. Results
3.1. Determination of substrate side-chain preference
We examined the residue preference of LF on six central subsites, S3, S2, S1, S1’, S2’, and S3’, which bind 3 residues on each side of the scissile bond. These subsites are the major determinants for substrate binding of proteases in general and the interaction of small potent inhibitors would not be expected to involve subsites outside of these six. We determined the relative kcat/Km (specificity constant) values of 19 amino acids in all six subsites using a competitive hydrolysis strategy which has been successfully used in the determination of subsite specificities of two other proteases [14,16,18,19]. The procedure involved the determination in a mass spectrometer of the initial rates of hydrolysis from a substrate mixture which differed only by the amino acids at a single subsite from a template sequence of RGKKKVLR*ILLN (where the subsites to be studied are shown in boldface and the astrisk denotes the cleaving site). Under such conditions, the relative rates are directly related to the relative kcat/Km, the specificity constant (see method section). This approach generated rapidly the specificity information but did not require the tedious determination of individual kcat and Km values.
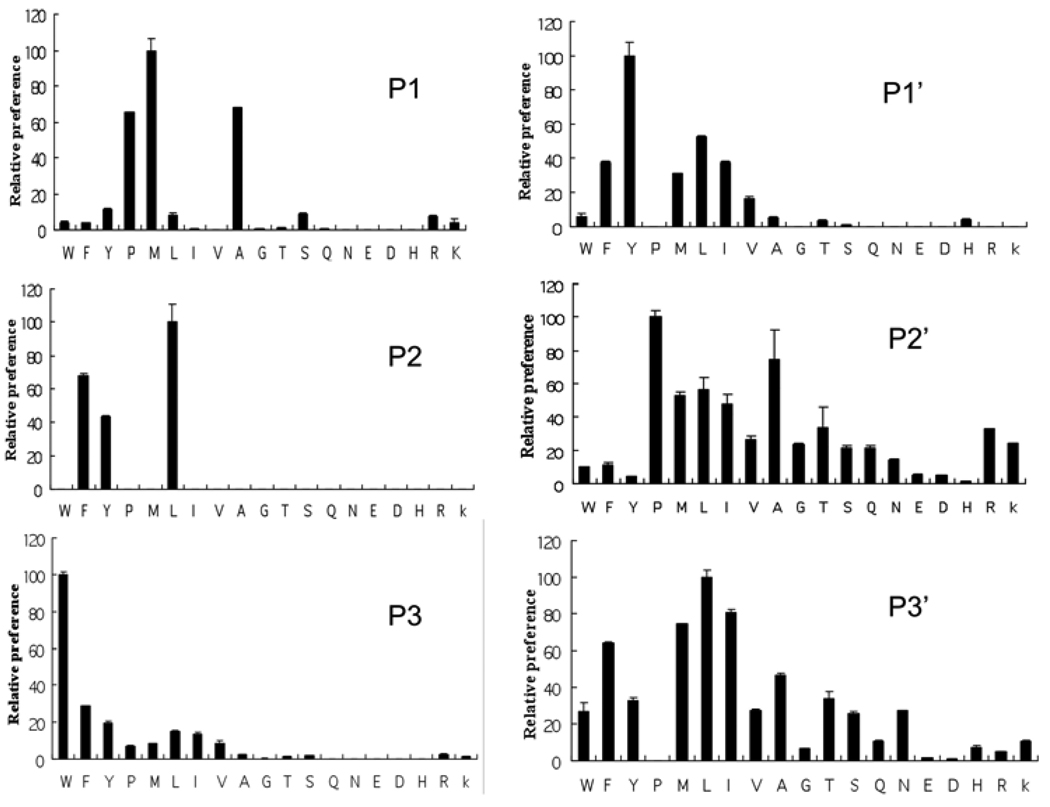
The substrate side-chain preferences for the six subsites from P3 to P3’ as expressed in relative kcat/Km, are shown in Figure 1 and numerically listed in Table 1. These results show that LF has somewhat broad specificity in all six subsites. In general, residue preferences in subsites P1, P2 and P3 are more stringent than those in subsites P1’, P2’ and P3’. Subsites P1 and P2 each prefer three residues (Met, Ala and Pro for P1 and Leu, Phe and Tyr for P2) with very low activity toward other amino acids. Subsites P3 and P1’ each prefer a single amino acid (Trp for P3 and Tyr for P1’) but have high background activities for many other residues. Subsites P2’ and P3’ showed some preferences (Pro and Ala for P2’ and Leu, Ile, Met and Phe for P3’) but can also accommodate many other residues almost as well. All the most preferred residues are hydrophobic amino acids while acidic residues (Asp and Glu) are disfavored in all subsites. Branched residues (Ile and Val) are not preferred in the P subsites but are better accommodated in the P’ subsites. Basic residues (Lys, Arg and His) are tolerated only in P2’ and P3’ subsites.
Figure 1. Amino acid preference in the six subsites of LF substrates.
The preference was shown as preference index derived from relative kcat/Km. Amino acids are in conventional single letter codes. In each subsite, the preference index for the amino acid with the highest hydrolytic rate is designated 100.
3.2. Design new LF substrate with preferred subsite residues
We used the subsite preference information in Fig. 1 to design a new LF substrate No. 3, Mca-KKWLM-YPLEK-Dnp, that incorporated six most favored residues (in boldface letters). Two franking residues at each of the N- and C-terminus were added for direct comparison with a previously reported LF substrate No. 2 [12] (Table 2). Mca and Dnp are fluorophore and quenching groups for quantitative FRET based fluorescent assay. A third substrate No. 1 (Table 2) contains the sequence from LF cleavage site of MAPKK-1. Kinetic data kcat, Km and kcat/Km for LF hydrolysis of substrates Nos. 2 and 3 were obtained by steady state kinetic method while kcat/Km was determined for substrate No. 1 by competitive hydrolysis method. We found that the kcat/Km value of the newly designed substrate No.3 (140 min−1 M−1) is about two-fold higher than that of the previously reported substrate No. 2 (76 min−1 M−1) and more than 90-fold higher than that of substrate No. 1 ( 1.5 min−1 M−1) from MAPKK1 sequence (Table 2). The kinetic data indicated that the newly designed fluorogenic substrate is the most efficient LF substrate reported so far. The high efficiency of this substrate substantiated the findings on the residue preference of LF subsites.
Table 2.
Kinetics parameters for LF-mediated hydrolysis of LF fluorogenic substrates
| Substrate | Source |
Km M |
kcat min−1 |
kcat/Km min−1 M−1 |
|---|---|---|---|---|
| 1. Mca-KKPTP-IQLN-Dnp | MAPKK-1 | ------ | ------ | 1.5a |
| 2. Mca-KKVYP-YPMEK-Dnp | Reference 11 | 58.53 | 4450 | 76 |
| 3. Mca-KKWLM-YPLEK-Dnp | New design | 16.86 | 2390 | 140 |
The kcat/Km value for this substrate was determined by competitive hydrolysis method and the individual kcat and Km were not determined.
3.3. Design new LF inhibitor with preferred subsite residues
Information on subsite specificity was used to design new inhibitor of LF. We had previously tested a LF inhibitor A (D-R)9VLR-CO-NHOH, which was designed based on the consensus P1, P2 and P3 residues from MAPPK sequences (Table 1). At the N-terminus of Inhibitor A are nine D-Arg residues as they have been shown to increase LF to legend affinity [20]. The C-terminal hydroxylamine is for chelating the active site zinc. This inhibitor has a very good potency with Ki value of 1.45 nM (Table 3). We designed and synthesized inhibitor B (D-R)9WLM-CO-NHOH using the best preferred residues in P1, P2 and P3 subsites and otherwise the same design as inhibitor A for direct comparison. As shown in Table 3, inhibitor B had a Ki value of 0.28 nM, which is about five times more potent than inhibitor A. To demonstrate the importance of subsites P1, P2 and P3, we designed inhibitor C (D-R)9LPY-CO-NHOH which contains moderately or poorly preferred residues at these subsites. Inhibitor C is about 26 times less potent than inhibitor B (Table 3). These results also substantiate the findings of LF subsite preference and confirms that the binding of sidechains P1, P2 and P3 contribute substantially the potency of the LF inhibitors.
Table 3.
Structures and potency of LF inhibitors.
| Inhibitor | Inhibitor sequence | Ki(nM) |
|---|---|---|
| A | (D-R)9VLR-CO-NHOH | 1.45 |
| B | (D-R)9WLM-CO-NHOH | 0.28 |
| C | (D-R)9LPY-CO-NHOH | 7.32 |
4. Discussion
In our current research, we successfully applied a rapid approach to determine the relative specificity at six major LF subsites. We again confirmed that the use of this approach was very powerful in yielding quickly detailed specificity information of a protease with multiple subsites. Our results showed that all the most preferred residues are hydrophobic ones (Fig. 1 and Table 1). This seems reasonable as the LF active site is a deep cleft and a hydrophobic interaction with the substrate side chains could be buried deeply inside of LF structure to form stable associations. Also, the hydrophobic side chains are mostly inside of the three-dimensional structure of globular protein substrates of LF, therefore, the utilization of these residues for subsite recognition would be expected to have significant specificity for a few cytosolic protein targeted by LF. In general, the preferences of 3 P-subsites are much more stringent than that for 3 P’-subsites. For example, subsites P1 and P2 have each 3 most preferred residues and very low preference for the remainder amino acids (Fig. 1). This is in contrast with the P2’ and P3’, which prefer many residues with activities close to one another. These results suggest that the specificity for the recognition of protein substrates by LF relies mainly on the subsites of the P side. We have confirmed the new residue preference information by the design of new substrates and inhibitors. The substrates and inhibitors incorporated the preferred residues had superior activities toward LF (Tables 2 and 3).
In the light of our new findings, we have examined the residues in 6 subsites, from P3 to P3’, of six MAPKK sequences (Table 1) for their relative preferences of LF cleavages. Interestingly, Pro (third preferred) is found at P1 in only 2 MAPKKs while the remaining 4 MAPKKs have at this subsite Arg, Lys and Gln, all have very low preferences indexes. The most high preference subsite is P2 in which 5 out of 6 MAPKKs have the highest ranking Leu residue. These results suggest that the possibility of P2 is most important among the subsites in LF substrate recognition and this line of reasoning is supported by molecular modeling to be described below. MAPKK-1 has Thr at P1 which has only a trace activity toward LF. In addition, the preference scores for MAPKK-1 in 5 subsites, from P3 to P2’, are all moderate or low, suggesting that the LF cleavage toward MAPKK-1 may be much lower than toward its other homologues. Overall, residues of various degrees of preferences appeared at many positions of these 6 MAPKKs. The lack of a consistent trend of high preference in general suggests that the specificity of LF is selected in evolution as a multifunctional protease. Logical extension from this view point is the possibility that other yet undiscovered LF substrates may be present in the cytosol of the cells which may contribute to the shaping of LF specificity.
Besides the implication for inhibitor design, we attempted to identify other possible LF substrates based on side-chain preference of LF active center. We searched a protein database and found several proteins who have the same sequences as the most preferred amino acids from P3 to P3’, such as NADH dehydrogenase subunit 1. However, after it incubated with LF even for hours, this protein was not hydrolyzed (results not shown). Thus, with known LF substrates, the relative rates determined in this study can be helpful in assessing the rate of their hydrolysis. However, the new information is not a good predictor of whether a protein is a LF substrate due to the limitation of the tertiary structures on the availability of the potential cleavage sites.
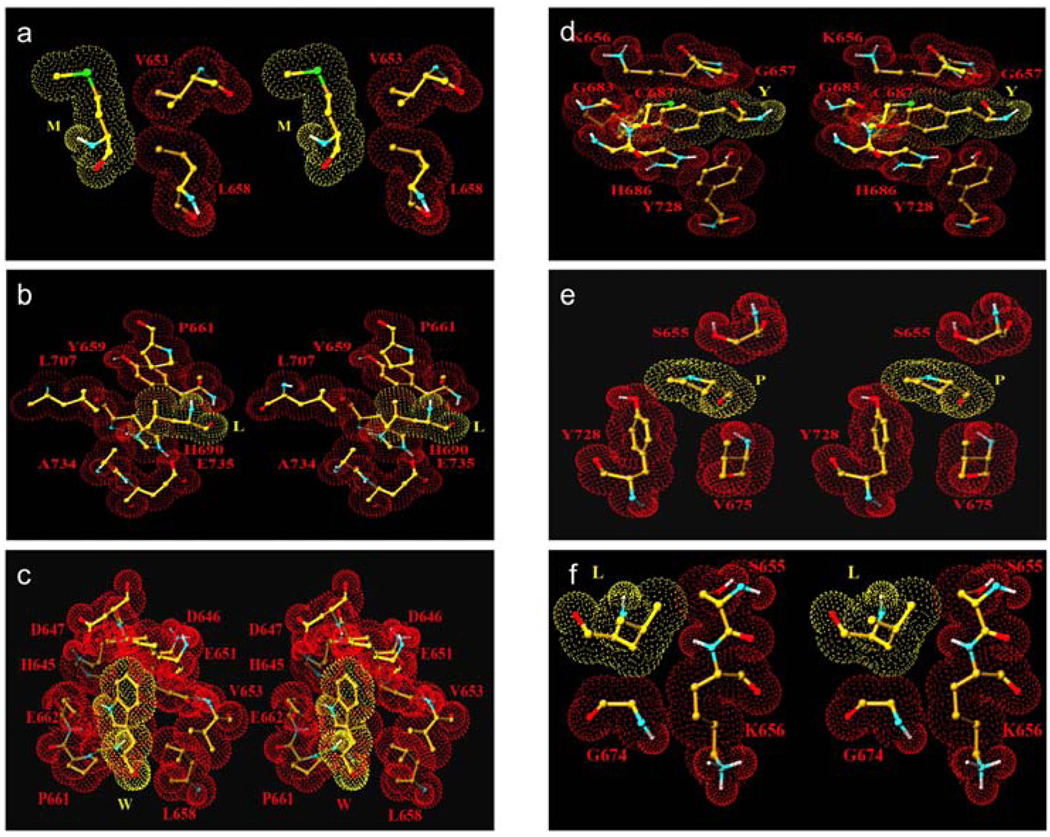
We have used molecular modeling to assess the interactions of the side chains of the six most preferred residues in the subsites of LF. We found that the interaction of Met side chain in LF subsite S1 is very limited. While the side chain of P1 Met position is extended toward the entrance of the active site cleft between side chains of H686 and K656, only the first three carbon atoms make hydrophobic interaction with the side chain of V653 of LF (Fig. 2, a). The meagerness of interactions does not indicate why Met is being preferred in this subsite. It is possible that Met is being preferred by catalytic steps of LF not directly related to its binding in the S1 subsite. Side chain of P2 Leu is located in a partial pocket formed by side chains of LF residues Y659, P661, H690, L707, A734 with ample hydrophobic interactions with side chains of Y659, P661 and H690 (Fig. 2, b). This side chain is well situated at S2, changes in side chain structure would likely result in either steric violations or reduced interactions. The model supports the above described hypothesis on the importance of P2 recognition for LF activities. For P3 Trp, the indole ring is stabilized with a polar and a hydrophobic interaction with the atoms of the enzyme. The NH1 atom of the ring interacts with the OE2 atom of the E662 while the carbon atoms of the ring interact with side chain atoms of the E651 and main chain atoms of residues H645, Q646 and D648 (Fig. 2, c). The aromatic ring of P1’ Tyr interacts with neighboring residues of the LF molecule (Fig. 2, d). Although the phenolic OH does not interact with LF, it may indeed interact with surrounding water molecules thus have a preference over Phe. The rigid Pro residue of the peptide at P2’ position is involved in weak hydrophobic interactions with the side chain atoms of residues Y728 and V675 of the lethal factor (Fig. 2, e). Additionally the carbonyl group of P2’ Pro and the backbone NH group of V675 is within a H-bond distance. The contribution of Pro to the rigidity of substrate conformation may also contribute to the preference at this subsite. The P3’ Leu is positioned at the edge of the cleft with limited hydrophobic interactions with C atom of the G674 and C and C atoms of the S655 of LF (Fig. 2, f). The position of this side chain is such that its substitution will be reasonably tolerated. This explains the lack of stringency for the preference at this position. It is clear that the interactions observed here do not always explain the side chain preference because the influence of many steps in LF catalytic mechanism to kinetic constants. However, information on subsite specificity and molecular interactions together may be taken into consideration to assist the new inhibitor designs.
Figure 2. Stereoview of the side chains of the most preferred residues interacting with LF subsite residues.
(a) Met in subsite S1; (b) Leu in subsite S2; (c) Trp in subsite S3; (d) Tyr in subsite S1’; (e) Pro in subsite S2’ and (f) Leu in subsite S3’.
Acknowledgements
This study was supported by the Molecular and Immunologic Analysis of the Pathobiology of Human Anthrax (U19 AI062629) funded by NIH. We sincerely thank Dr. Robert Turner, Oklahoma Research Facility, CoMentis, Inc, Oklahoma city, OK, USA, for the kind assistance in ESI-LC MS operation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mock M, Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 2.Shoop WL, Xiong Y, Wiltsie J, Woods A, Guo J, Pivnichny JV, Felcetto T, Michael BF, Bansal A, Cummings RT, Cunningham BR, Friedlander AM, Douglas CM, Patel SB, Wisniewski D, Scapin G, Salowe SP, Zaller DM, Chapman KT, Scolnick EM, Schmatz DM, Bartizal K, MacCoss M, Hermes JD. Anthrax lethal factor inhibition. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7958–7963. doi: 10.1073/pnas.0502159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ascenzi P, Visca P, Ippolito G, Spallarossa A, Bolognesi M, Montecucco C. Anthrax toxin: a tripartite lethal combination. FEBS Lett. 2002;531:384–388. doi: 10.1016/s0014-5793(02)03609-8. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell AJ, Abdollahi M, Bardwell L. Anthrax lethal factor-cleavage products of MAPK kinases exhibit reduced binding to their cognate MAPKs. Biochem. J. 2004;378:569–577. doi: 10.1042/BJ20031382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Y, Wiltsic J, Woods A, Guo J, Pivnichny JV, Tang W, Bansal A, Cummings RT, Cunningham BR, Friedlander AM, Douglas CM, Salowe SP, Zaller DM, Scolnick EM, Schmatz DM, Bartizal K, Hermes JD, MacCoss M, Chapman KT. The discovery of a potent and selective lethal factor inhibitor for adjunct therapy of anthrax infection. Bioorg Med Chem Lett. 2006;16:964–968. doi: 10.1016/j.bmcl.2005.10.088. [DOI] [PubMed] [Google Scholar]
- 6.Tanoue T, Nishida E. Molecular recognitions in the MAP kinase cascades. Cell Signalling. 2003;15:455–462. doi: 10.1016/s0898-6568(02)00112-2. [DOI] [PubMed] [Google Scholar]
- 7.Rainey GJ, Young JA. Antitoxins: novel strategies to target agents of bioterrorism. Nat Rev Microbiol. 2004;2:721–726. doi: 10.1038/nrmicro977. [DOI] [PubMed] [Google Scholar]
- 8.Ho DT, Bardwell AJ, Grewal S, Lverson C, Bardwell L. Interacting JNK-docking sites in MKK7 promotes binding and activation of JNK MAP kinases. J. Biol. Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turk BE. Manipulation of host signaling pathway by anthrax toxins. Biochem. J. 2007;402:405–417. doi: 10.1042/BJ20061891. [DOI] [PubMed] [Google Scholar]
- 10.Paniffer AD, Wong TY, Schwarzenbacher R, Renatus M, Petosa C, Blenkowska J. Crystal structure of the anthrax lethal factor. Nature. 2001;414:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- 11.Tonello F, Ascendzi P, Montecucco C. The metalloproteolytic activity of the anthrax lethal factor is substrate-inhibited. J. Biol. Chem. 2003;278:40075–40078. doi: 10.1074/jbc.M306466200. [DOI] [PubMed] [Google Scholar]
- 12.Turk BE, Wong TY, Schwarzenbacher R, Jarrell ET, Leppla SH, Collier RJ, Liddington RC, Cantley LC. The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Naure Struct. Mol. Biol. 2004;11:60–66. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 13.Turk BE. Discovery and development of anthrax lethal factor metalloproteinase inhibitors. Current pharmaceutical biotechnology. 2008;9:24–33. doi: 10.2174/138920108783497604. [DOI] [PubMed] [Google Scholar]
- 14.Turner RT, III, Koelsch G, Hong L, Castanheira P, Ermolieff J, Ghosh AK, Tang J. Subsite specificity of memapsin 2 (beta-secretase): implications for inhibitor design. Biochemistry. 2001;40:10001–10006. doi: 10.1021/bi015546s. [DOI] [PubMed] [Google Scholar]
- 15.Ermolieff J, Loy JA, Koelsch G, Tang J. Proteolytic activation of recombinant pro-memapsin 2 (pro-β-secretase) studied with new fluorogenic substrate. Biochemistry. 2000;39:12450–12456. doi: 10.1021/bi001494f. [DOI] [PubMed] [Google Scholar]
- 16.Turner RT, III, Lin H, Koelsch G, Ghosh AK, Tang J. Structure locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase) Biochemistry. 2005;44:105–112. doi: 10.1021/bi048106k. [DOI] [PubMed] [Google Scholar]
- 17.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Turner RT, III, Loy JA, Nguyen C, Devasamudram T, Ghosh AK, Koelsch G, Tang J. Specificity of memapsin 1 and its implications on the design of memapsin 2 (beta-secretase) inhibitor selectivity. Biochemistry. 2002;27:8742–8746. doi: 10.1021/bi025926t. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Bo H, Zhang XC, Hartsuck JA, Tang J. Predicting memapsin (beta-secretase) hydrolytic activity. Protein Sci. 2010;11:2175–2185. doi: 10.1002/pro.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonello F, Seveso M, Marin O, Mock M, Montecucco C. Screening inhibitors of anthrax lethal factor. Nature. 2002;418:386. doi: 10.1038/418386a. [DOI] [PubMed] [Google Scholar]