Abstract
The paradigm of mammary cancer induction by the mouse mammary tumor virus (MMTV) is used to illustrate the body of evidence that supports the hypothesis that mammary epithelial stem/progenitor cells represent targets for oncogenic transformation. It is argued that this is not a special case applicable only to MMTV-induced mammary cancer, because MMTV acts as an environmental mutagen producing random interruptions in the somatic DNA of infected cells by insertion of proviral DNA copies. In addition to disrupting the host genome, the proviral DNA also influences gene expression through its associated enhancer sequences over significant inter-genomic distances. Genes commonly affected by MMTV insertion in multiple individual tumors include, the Wnt, FGF, RSpo gene families as well as eIF3e and Notch4. All of these gene families are known to play essential roles in stem cell maintenance and behavior in a variety of organs. The MMTV-induced mutations accumulate in cells that are long-lived and possess the properties of stem cells, namely, self-renewal and the capacity to produce divergent epithelial progeny through asymmetric division. The evidence shows that epithelial cells with these properties are present in normal mammary glands, may be infected with MMTV, become transformed to produce epithelial hyperplasia through MMTV-induced mutagenesis and progress to frank mammary malignancy. Retroviral marking via MMTV proviral insertion demonstrates that this process progresses from a single mammary epithelial cell that possesses all of the features ascribed to tissue-specific stem cells.
Keywords: Mammary tumors, Cancer, Mouse mammary tumor virus
Introduction
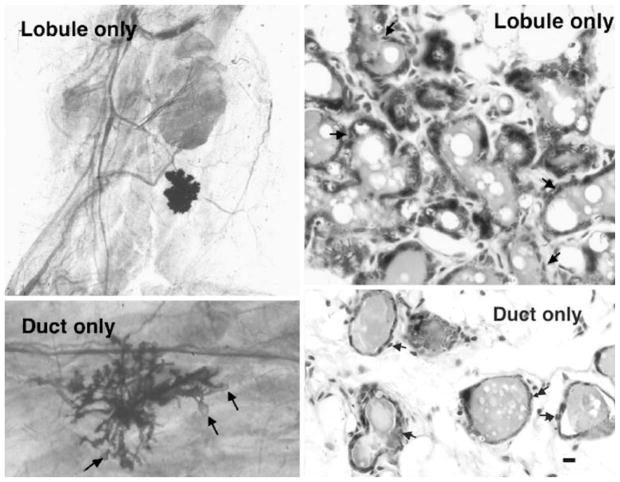
In mice, rats and humans, evidence has accumulated that a hierarchy of mammary epithelial progenitor/stem cells exists [1–3]. At present, lobule-restricted, duct-restricted and fully competent mammary stem cells are proposed to exist among the mammary epithelial population. In 1996, we demonstrated in the mouse that at least three distinct stem/progenitor epithelial cell activities were present in normal mammary glands by limiting dilution transplantation [4]. From cellular dilutions giving a nearly Poisson distribution of positive takes, three types of mammary outgrowths were recorded in full term pregnant hosts, outgrowths showing complete development, outgrowths capable of producing only mammary ducts and outgrowths producing only secretory lobular structures without ductal branching (Fig. 1). All of these epithelial structures contain epithelial cells representing diverse epithelial cell phenotypes, including luminal, myoepithelial and basal (SLC) cells. Premalignant and immortal mammary epithelial outgrowth populations, reflecting either ductal or lobular hyperplasia [5, 6] may be isolated from mouse mammary tissue, suggesting their origins began with the transformation of duct-limited or lobule-limited mammary epithelial stem/progenitors cells. These premalignant immortal populations also possess epithelial cells reflecting the full array of mammary epithelial cell types, basal, myoepithelial and luminal (shown in Fig. 2). These populations, like the MMTV-induced mammary tumors, are clonal as determined by retroviral tagging [7]. Thus, the originally transformed cell surely possessed the capacity to produce epithelial cell progeny with diverse cellular fates, suggesting a stem/progenitor cell origin for MMTV-induced hyperplasia and pregnancy independent tumors. This type of scenario occurs in mice infected with MMTV from C3H and CzechII mice. To follow up on this hypothesis, we posed the question, “Are individual mammary outgrowths from implanted epithelial fragments clonally derived?” To address this question, we transplanted individual mammary gland fragments from multiparous MMTV-infected mice to gland-free mammary fat pads and 4 weeks after implantation impregnated the hosts to generate full and complete mammary growth. We reasoned that if outgrowths clonally developed from MMTV-infected stem/progenitor epithelial cells, a specific and unique pattern of acquired proviral insertions would appear upon Southern Blot analysis of restriction enzyme-digested DNA from individual outgrowth populations. This would indicate that the majority, if not all, of the cells present in any given outgrowth were derived from a single MMTV-infected antecedent and further that this predecessor was capable of giving rise to progeny reflecting all of the epithelial cell types present in a fully functional differentiated mammary gland [8]. This result was obtained in over 60% of outgrowths from random fragments dissected from multiparous females (N=6) infected with MMTV whereas no outgrowths bearing a specific pattern of MMTV insertions was obtained when fragments were transplanted from MMTV-infected pubertal-aged females. (N=6). Outgrowths derived from MMTV-infected donors of intermediate age gave an intermediate range of retrovirus-tagged individual outgrowths. So expanding numbers of clonogenic mammary epithelial cells are infected with MMTV with increasing age and reproductive history. To confirm that individual outgrowths were comprised of single clones and not the result of the simultaneous expansion of two or more divergent clones, serial passages of random fragments from individual clonal-dominant outgrowths were carried out [9]. The results established the monoclonal nature of the tested outgrowth populations by demonstrating that all transplant generations of any given outgrowth containing MMTV insertions showed identical restriction patterns by Southern Blotting (Fig. 3). In addition to affirming the clonal nature of these populations, these experiments demonstrate the self-renewal capacity of the originally infected cell through multiple transplant generations while maintaining its multipotent properties. These results provide definitive proof that over time MMTV can infect mammary epithelial stem/progenitor cells and cause mutations in these cells through the random insertion of MMTV proviral DNA, thus, providing a basis for hypothesizing that mammary stem cells represent targets for oncogenesis in MMTV-infected mice.
Figure 1.
Lobule only and duct only outgrowths in full term pregnant transplant hosts (left panels). Both lobule-only and duct-only outgrowths comprise both luminal and myoepithelial (arrows) cells. Bar equals 10 micrometers.
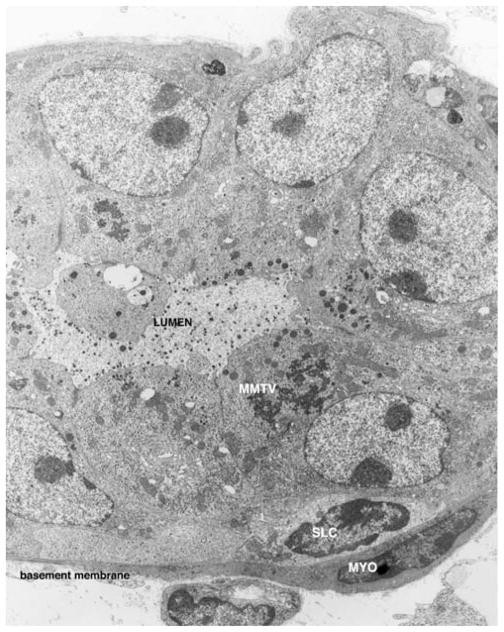
Figure 2.
This electron micrograph depicts a ultrathin section through one of the acini in an MMTV-induced alveolar hyperplasia. There is evidence of virus replication (MMTV), of secretory activity leading to secretory granule formation in the apical cytoplasm of the luminal cells and release into the lumen. An undifferentiated suprabasal cell (SLC) is present and proximal to it, a differentiated myoepithelial cell (arrow). Bar=1 μm.
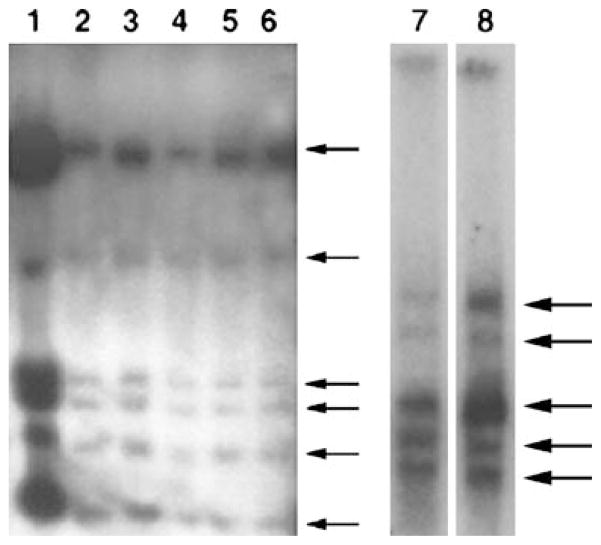
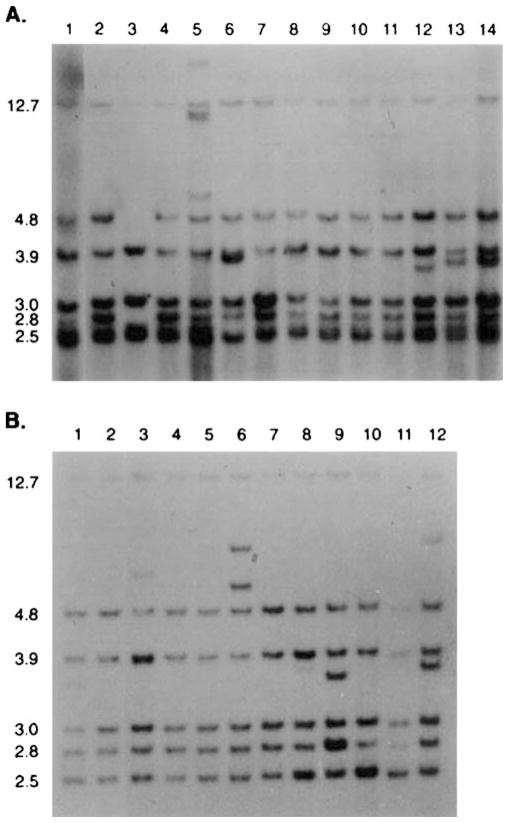
Figure 3.
Southern blot of DNA from five consecutive serial passages of a clonal-dominant normal mammary outgrowth (lanes 2–6), probed with the MMTV-LTR demonstrates that the original proviral insertion pattern is maintained in each passage. Lane 1 contains DNA from a tumor found in passage 4. In the Southern Blot (below) DNA from a lobule-limited outgrowth (lane 7) and a fully developed secretory outgrowth (lane 8) in the contralateral fat pad of the same female demonstrate the identical pattern for MMTV-host DNA restriction fragments demonstrating that each population arises from the same original MMTV-infected antecedent.
The clonal expansion of mammary stem/progenitor cells containing MMTV insertional mutations may give rise to local premalignant hyperplasia (HAN) which upon transplantation produce hyperplastic growth comprised of both luminal epithelial and myoepithelial cell types, which fills the epithelium-divested mammary fat pad. The transplanted hyperplasia does not extend beyond the limits of the fat pad; ceases growth when confronted with fat occupied by normal mammary epithelial tissue and will not grow in ectopic transplantation sites otherwise permissive for mammary tumor cell proliferation. All of these properties are shared with outgrowths of normal mammary epithelium. In contrast to normal tissue, growth senescence is never attained upon serial transplantation of the hyperplastic populations and the frequency of focal mammary tumor formation is much greater, attesting to their premalignant nature. Like the hyperplasia, the tumors that arise often comprise both myoepithelial and luminal epithelial cell phenotypes (Fig. 4). The tumors and the hyperplasia within which they develop share the identical MMTV retroviral insertions indicating their lineal relationship, often the tumors have one or more additionally acquired proviral mutations. An example of this is shown in Fig. 5A, where DNA from two separate passages of an MMTV-induced hyperplastic outgrowth is compared with tumor DNA from lesions arising stochastically within these populations at different transplant generations. The original pattern of MMTV proviral insertion is maintained in all the tumors whether they possess new insertions or not. Individual metastatic lesions from the tumors also show a lineal relation to both the originating tumor and the hyperplastic population from which they sprung. Individual metastatic lesions from the same tumor are also clonally derived and may differ from one another by the number and location of newly acquired proviral insertions (Fig. 5B). Taken together, these observations establish that individual cells among the mammary epithelium, capable of self-renewal and the production of epithelial progeny of divergent cellular phenotypes, biological attributes shared with stem cells, may be infected with MMTV acquire transforming mutations and then expand clonally to give rise to premalignant and malignant epithelial colonies. The role of epigenetic mechanisms in this progression has not been addressed except in those models where pregnancy has provided protection to malignant progression in chemical carcinogen and certain transgenic mammary tumorigenesis models [6, 10].
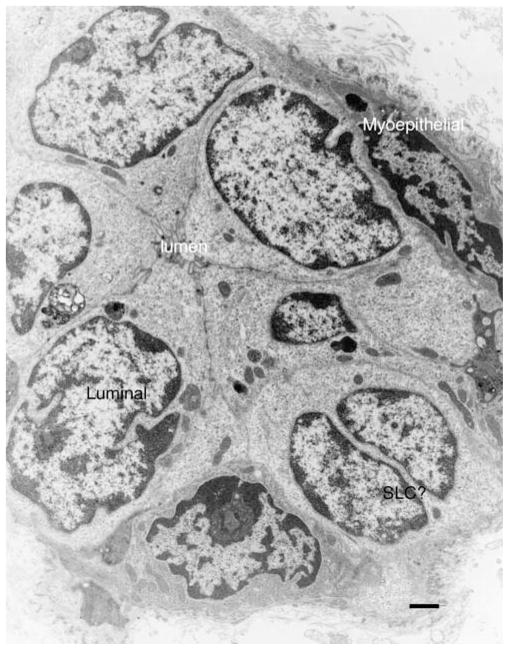
Figure 4.
An electron micrograph from an MMTV-induced primary mammary tumor shows that myoepithelial, luminal and basal (SLC) epithelial phenotypes are present. Unlike the hyperplasia, the tumor cells show very little secretory activity, only a few mitochondria, sparse rough endoplasmic reticulum development and a collapsed lumen. Bar equals 1 μm.
Figure 5.
(A) MMTV proviral restriction patterns are shown for DNA isolated from a premalignant hyperplastic mammary outgrowth (lanes 1 and 2) and from 12 independent mammary tumors (lanes 3–14) that arose focally within the hyperplastic population at various passages during its propagation in gland-free fat pads. The proviral content of the two different passages of the hyperplasia (lane 1 and 2) are present in all of the tumors. New MMTV proviral insertions were detected in the tumors shown in lanes 5, 12, 13 and 14. A loss of one of the original insertions was noted in the tumor DNA shown in lane 3. The DNA was digested with EcoR1 and probed with the MMTV-LTR sequence. EcoR1 cuts within the provirus and the LTR is represented at both ends of the provirus. Therefore each insertion produces two virus-host junction fragments detected by the LTR probe. In (B) the restriction enzymatic digestion and the probe were the same, however, DNA from a metastatic tumor (lane 1 in (B)) arising in the hyperplasia shown above in lanes 1 and 2, gave 11 independent lung metastases in the same mouse (shown in lanes 2–12). The tumor and all the metastases bear the original proviral insertions from the hyperplasia. Additional MMTV insertions were detected in the metastatic nodule DNA shown in lanes 6, 8, 9, 10 and 12. This strongly suggests that each metastatic lesion is an individual clone.
The females of the GR, BR6, Rlll, Balb/c LA, and Spretus mouse strains have a high incidence of pregnancy dependent mammary tumors or plaques that, after one or more parities, progress to a pregnancy independent tumor ([11–14] and our unpublished data). Foulds [15] and Squartini [16] described plaques as “a system of branching tubules often with bulbous ends”. Squartini et al. [13] demonstrated that differences in the manner in which the disease progressed in C3H and Rlll females was a function of the particular strain of exogenous virus. In their study BALB/c mice which have a low or no incidence of spontaneous mammary tumors [13] developed a high incidence of pregnancy-independent tumors when infected with horizontally transmitted MMTV(C3H) and pregnancy-dependent tumors which progressed to pregnancy-independence when infected with MMTV(Rlll). Even more surprising was the demonstration by Squartini that the virus could increase mammary secretory differentiation in virgin females and that the intensity of this activity was unique to the strain of MMTV present in the gland [17].
Acquisition of new MMTV insertions and fixation in the genome (sic mutation) requires that the infected cell traverse the cell cycle. Nevertheless, new viral insertions have never been observed in these clonally expanding premalignant epithelial outgrowths (or in MMTV-tagged clonally expanding normal mammary outgrowths) despite the presence of replicating MMTV displaying a full vegetative life cycle including the synthesis of reverse transcriptase, pre-pronucleocapsids, unintegrated proviral DNA and mature virions. The original retroviral insertion pattern is maintained throughout multiple transplant generations not withstanding the enormous expansion of the cellular population (Fig. 6 arrows). In addition, when one of these insertions occurs near a cellular gene, like Wnt-1, a new restriction fragment containing Wnt-1 sequence may be produced (Fig. 6, lower). This alteration is also maintained throughout malignant progression validating the clonal nature of this process. New MMTV insertions are only seen in the malignant clones arising stochastically within the premalignant outgrowth, although this also is not universally true (Fig. 6 and 5B), suggesting that epigenetic mechanisms or mutations other than those caused by proviral insertion may promote malignant progression. Thus these new MMTV insertions could represent a passenger effect and not a selected event. In 1975, Cairns [18] proposed that somatic stem cells protect themselves from mutation by selectively retaining their template DNA strands during asymmetric divisions and pass the newly synthesized strands to their offspring. Such a mechanism in the MMTV-infected cells, which generate the hyperplasia at each transplant generation, might explain the absence of new MMTV insertions, as the insertion would occur in the new DNA strand and would subsequently be transmitted to the dispensable daughter cells. Cairns [19] also postulated that original somatic stem cells die and are replaced by an immediate daughter cell which may have acquired mutation during its inception (having selectively inherited the newly synthesized DNA strands) leading to a circumstance where a stem cell with acquired mutations supplants its normal counterpart in the niche. This scenario might readily explain the appearance of the new proviral insertions observed in the DNA of the mammary tumors arising within the hyperplasia. Alternatively, local exponential, i.e. non-asymmetric, divisions of the stem/progenitor cells may result in fixation of new MMTV proviral insertions. Subsequent expansion of tumor populations from these newly mutated stem/progenitor cells will depend on the transforming nature of the newly acquired mutations.
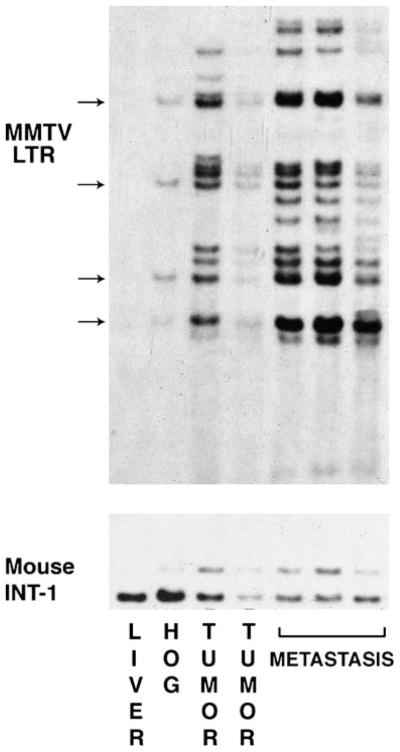
Figure 6.
A Southern Blot containing EcoR1 digested DNA from Czech mouse liver (Czech mice have no endogenous MMTV provirus), a Czech mammary hyperplastic outgrowth (HOG), two mammary tumors that developed in the contra-lateral mammary glands of the mouse bearing the hyperplastic implant and metastases from the lung of the same mouse were probed with MMTV-LTR and a probe specific for the Wnt-1 (INT-1) gene. No MMTV provirus is detected in the liver DNA, at least two insertions were evident in the HOG DNA and these were also represented in the two tumors. Additional MMTV insertions were present in the DNA from each tumor. By inspection of the proviral DNA content of the metastases, it can be determined that these most likely arose from the tumor in the 4th lane and not from the one in lane 3. In addition the metastases contain additional MMTV insertions, which may tag mutations important to metastatic progression. One of the insertions in the cell producing the hyperplastic clone occurred near the Wnt-1 gene in one allele producing a larger restriction fragment detected by the Wnt-1 probe. This rearranged Wnt-1 sequence is also found in all the tumors and metastases arising within this hyperplasia validating the clonal nature of malignant progression.
Recent evidence has confirmed that long-label-retaining epithelial (LRC) cells present among the mouse mammary epithelium selectively retention of the labeled template DNA strands while traversing the cell cycle [20, 21]. In addition, mammary transplants containing lacZ-positive lobule-limited progenitors contained long-label-retaining, lacZ-positive epithelial cells, which traversed the cell cycle and selectively retained their original DNA strands. Therefore, stem or progenitor cells and in some cases ERa and PR positive epithelial cells display the ability to selectively retain template DNA while undergoing asymmetric cell divisions. Mammary epithelial cells transformed by MMTV through insertion mutation exhibit the capacity for self-renewal, the ability to produce divergent epithelial cell progeny, and the capacity to selectively distribute newly synthesized DNA strands to their offspring while retaining their template copy, all properties of tissue stem cells. Therefore, it is likely that MMTV-induced mammary tumors arise from the mammary epithelial stem/progenitor cell subpopulation and are maintained by long-lived transformed cells capable of protecting their template DNA.
We have concluded that the existence of multipotent cells with MMTV insertions in aged multiparous mice represent substitute mammary stem cells that occupy the stem cell niche as the result of the death of the original stem cell. The fact that we observe normal complete outgrowths comprised entirely of the progeny of the affected cell, supports the conclusion that these mutated “faux” stem cells can function quite normally and in addition protect their template strands (bearing the original MMTV insertions) during asymmetric divisions (new retroviral insertions always occur during replication in the replicating DNA strand) where newly synthesized DNA copies are passed to the dispensable (differentiating and incapable of generating an independent heterogeneous clone comprised of different epithelial subtypes) progeny. Therefore upon serial transplantation of random fragments from this MMTV-marked normal clone we always find the original MMTV insertions and not new ones (because these occur randomly in the genome and are dispensed to progeny during asymmetric divisions).
Mechanisms by which MMTV Activates/Inactivates Target Genes
With few exceptions, the genes which appear to be targets for MMTV induced activation of expression are either silent in the mammary gland or are expressed at low levels. Frequently MMTV integration events occur either 5′ of the target gene in the opposite transcriptional orientation or 3′ of the target in the same transcriptional orientation [22, 23]. The activating viral integration events can occur at a significant distance (100–157 Kb) from the target gene [24, 25]. A similar phenomenon has been observed in murine leukemia virus (MLV) induced rat lymphomas in which c-Myc expression is activated by a viral integration event over 30–270 Kb distance from the gene [26]. The molecular mechanisms underlying these long-range effects of the MMTV long terminal repeat (LTR) on the target gene transcription are unclear. A complicating factor in identifying the actual target gene for a particular MMTV integration event is if it occurs within a gene that is adjacent to the target gene. Thus the expression of Wnt1/Wnt10b, Wnt3, and Fgf8 are frequently activated by MMTV integration events that have occurred within the Arf, Nsf, and Fbxw4 genes, respectively ([25]; Callahan and Smith, unpublished data). However, clearly in some cases these genes correspond to the targets. For instance, MMTV integration into one allele of the eIF3e gene (Int6) leads to the expression of a truncated eIF3e mRNA species. In each case (n=5) MMTV integrated into a eIF3e intron in the opposite transcriptional orientation of the gene. In the rearranged allele transcription was terminated at a cryptic transcription stop site in the reverse orientation of the MMTV LTR. Since the non-rearranged allele did not contain a mutation, it was concluded that the mutated allele results in the expression of a product with a dominant-negative activity. Another example of the integration of MMTV into a target gene is Notch4. In this case MMTV integrations (n=9) occurred within a 142 bp region of the gene and the viral genome was always in the same transcriptional orientation as the gene. One consequence of MMTV integration in this region was the transcription of a novel Notch4 mRNA species starting from the 3′ MMTV LTR that encodes a protein spanning the transmembrane domain to the C′ terminus of the transmembrane receptor. For Notch4, this corresponds to a gain-of-function mutation.
Core Common Integration Sites (CIS) for MMTV in Mouse Mammary Tumors Versus Low Frequency CIS
The target genes for the CIS were initially identified by recombinant cloning of restriction enzyme digested tumor DNA containing host-viral junction sequences [22, 23, 27, 28]. Recently, [24, 25] and we ([29] and Smith an Callahan, unpublished data) have used high throughput approaches involving “Splinkerette” [24, 30, 31] or inverse PCR [29, 32, 33], respectively to identify new CIS for MMTV (Table 1). A conservative definition of CIS is that two or more MMTV genomes must be integrated in the gene or near the target gene and shown to alter the target genes expression relative to other tumors having no viral integrations at that site or in normal mammary gland tissue.
Table 1.
Core MMTV CIS in mammary tumors of different MMTV infected mouse strains.
| %
|
|||
|---|---|---|---|
| Mouse Strain | Wnt | Fgf | RSpo |
| Czech (n=36)a | 23 | 42 | 17 |
| Balb/cfCz (n=51)a | 36 | 28 | 18 |
| Balb/cfSp (n=44)a | 31 | 29 | 13 |
| Balb/cfC3H (n=155)b | 41 | 39 | 4 |
| Czech HOG (n=31)a | 16 | 10 | 32 |
| Tumor (n=14)a only | 0 | 14 | 0 |
Our unpublished data
Data taken from Theodorou et al. [25]
Theodorou et al. [25] identified 33 CIS in a panel of 160 Balb/cfC3H mammary tumors. In our study the mammary tumors analyzed were from CzechII (M. musculus musculus, n=66), Balb/cf CzechII (n=77), and Balb/cf Spretus (MMTV from M. spretus, n=79) along with 31 independent CzechII HOGs and 14 HOG derived mammary tumors. Although our analysis is ongoing, the most frequently identified CIS target genes whose expression has been verified to be altered as a consequence of MMTV integration in the particular CIS and that are found in each of the four tumor panels as well as Theodorou et al. [25] tumor panel, include members of the Wnt (-1/10b, -3, and -3a), Fgf (-3, -4, and -8), and RSpo (-2 and -3) gene families. In another recent study of pregnancy independent tumors that arose from pregnancy dependent mammary tumors in Balb/c mice infected with novel strains of MMTV, RSpo3 was identified as a new CIS [11]. We refer to these genes as “Core” CIS since they occur at a high frequency (n>5) irrespective of the mouse strain or the inducing strain of virus. In addition, we have found that eIF3e/Int6 and Notch4/Int3 are also high frequency (n>5) CIS in our panel of Czech HOGs and mammary tumors, respectively [27, 28]. A second group of CIS occurs at low frequency (n<4) in mammary tumors. Theodorou et al. [25] found 25 low frequency CIS in Balb/cfC3H mammary tumors, of these CIS, 22 are unique to this strain of virus. Although our study is incomplete, it is clear that there are similar numbers of low frequency CIS which are unique to particular mouse strains and strains of MMTV. Furthermore, MMTV activation of the Core CIS appears to be an early event since in 14 out of 31 Czech mammary HOGS having a MMTV RIS contained an activated member of one of these gene families. There does appear to be specificity for particular Core CIS in these HOGs. Thus only Wnt1, Fgf3/4 and RSpo2 in addition to eIF3e/Int6 are activated in Czech mammary HOGs. Conversely, Notch4/Int3 [27, 34, 35] is a frequent CIS in CzechII and M.m jkg mammary tumors, but is a low frequency CIS in BR6 and Balb/cfC3H mammary tumors [25, 36]. Interestingly, Notch4 has never been found to be a MMTV CIS in HOGs, suggesting that activation of this gene does not lead to a premalignant lesion recognized as a hyperplastic alveolar nodule (HAN). Similarly Rspo3 has only been found in mammary tumors. This suggests that there may be earlier initiating events that collaborate with these two genes for tumor progression.
Based on the current available data ([25]; Callahan and Smith, unpublished data), the Eras (3 hits), Astn2 (2 hits), Fgf6 (3 hits), Fgf10 (4 hits), Igf2/H19 (4 hits), Lambl-1 (3 hits), Map3k8 (4 hits), Dpp10 (2 hits) and Odz1 (2 hits) genes correspond to CIS that are unique to Balb/cfC3H mammary tumors. MMTV integration at other CIS target genes, such as Notch4/Int3, eIF3e/Int6, Stmbt2, and FGFR2 appear either to depend on particular mouse strains or the strain of MMTV. In addition, there could be as many as 20 additional CIS that occur infrequently (2–3 tumors out of several hundred analyzed) and correspond to either anonymous or relatively uncharacterized genes (Callahan and Smith, unpublished data). However, based on a statistical analysis of MuLV CIS, [37, 38] it has been calculated that in a data set of 1200 integration events a particular CIS having less than 5 integrations could simply occur by chance. Therefore, the low frequency CIS would require a higher level of proof to confirm their significance to mammary tumorigenesis.
Wnt/Rspo/Fgf Core Signaling Pathways
The Wnt signaling pathway is involved in the determination of cell and tissue polarity, stimulation of cell proliferation and differentiation, and adult tissue homeostasis. The Wnt genes are members of a family of 19 genes related to the Drosophila segmented polarity gene, wingless (wg) (reviewed in [39]). Wnt1 encodes cysteine-rich secreted proteins of 41–44 kD that is palmitoylated and glycosylated associates with extracellular matrix and cell surface [40]. The primary receptor for the Wnt protein is comprised of Frizzled (Fz) a seven transmembrane receptor family (10 members) and low density lipoprotein receptor-related protein (LRP) single transmembrane receptors, LRP-5 or LRP-6. Members of the R-Spondin gene family (RSpo1–4) encode non-conventional secreted ligands for the Wnt co-receptors. RSpo proteins contain an N-terminal signal peptide, a cysteine rich region containing two furin like domains, a thrombospondin type 1 domain, and a C-terminal region comprised of positively charge amino acids [41]. RSpo proteins appear to enhance or synergize with Wnt in the “canonical” Wnt signaling pathway that leads to the stabilization of β-catenin. Stabilized β-catenin subsequently moves to the nucleus where it activates transcription factor T-cell factor (Tcf)/lymphocyte enhancing factor (Lef). Target genes include c-Myc and CyclinD1. It has been hypothesized that the mechanism by which RSpo enhances Wnt signaling relates to its ability to interfere with Dickkopf (Dkk1)/Kremen mediated internalization of LRP6 through an interaction with Kremen, resulting increased levels of LRP6 on the cell surface [42].
The non-canonical Wnt pathway does not require LRP6 or β-catenin and can be further defined as the Wnt/PCP (Planar Cell Polarity) and the Wnt/Ca+ signaling pathways [39, 43]. It is not known whether the RSpo proteins also affect non-canonical Wnt signaling pathways (reviewed in [43]). The Wnt/PCP pathway modulates organization of the cytoskeleton through activation of JNK by Fz through a cascade of interacting proteins that include: Strabismus (Stbm), Dishevelled (Dvl), Dishevelled-associated activator of morphogenesis (Daam1), and GTPase RhoA and Rho-associated kinases. The Wnt/Ca+ signaling pathway leads to the release of cytosolic Ca+ that activates calcineurin phosphatase that in turn dephosphorylates NF-AT that is followed by its transfer to the nucleus where it activated the expression of target genes.
Fibroblast growth factors (Fgf) represent a large family (28 members) of multifunctional peptide growth factors (reviewed by [44]). Members of the Fgf family vary in length, but are homologous to one another within a core of 120 amino acid residues. They play a critical role during embryonic development, including the early mammary gland development (reviewed by [45]), by regulating cellular proliferation, differentiation and migration. In the adult, Fgfs are homeostatic factors functioning in wound healing, control of the nervous system and tumor angiogenesis. Their signaling is mediated through binding to a family (five members) of high affinity receptor tyrosine kinases (RTK) designated Fgf receptors (FGFRs). FGFRs are transmembrane proteins composed of an extracellular ligand binding domain and an intracellular cytoplasmic domain containing a tyrosine kinase core. Formation of a complex comprised of FgF-Syndecan/heparin-FGFR leads to receptor autophosphorylation and activation of four intracellular signaling cascades including: the PI3K/Akt pathway, the PLC γ/Ca+/PKC pathway, and the Ras/MAPK pathway which leads to the phosphorylation of specific members of the Ets family of transcription factors that activate the expression of FGF target genes (DAG, IP3, and SOS).
The Notch Gene Family
The Notch gene family (designated 1 through 4) is related to the Drosophila Notch gene (reviewed in [46]). Members of this gene family encode transmembrane receptor proteins that are involved in cell fate decisions during development [47]. Expression of each of the murine Notch genes, their ligands (Delta-like-1, -2, and 4; Jagged-1 and -2) and target genes can be detected in mammary glands of virgin, pregnant, lactating, and involuting mice (our unpublished data). Upon the interaction of the ligand with the Notch extracellular domain, the Notch intracellular domain is released through a series of proteolytic cleavages and travels to the nucleus. In the nucleus it binds to RBP-J, a transcription repressor/activator, where it relieves the repression of target genes and activates their transcription. This constitutes the primary known or “canonical” Notch signaling pathway. The known target genes for Notch ICD/RBP-J include: the Hes and Hey gene families that encode basic helix loop helix repressor proteins, c-Myc, Narp, Notch4, and Cyclin D1.
Activation of the Notch4 locus [34, 48] was first detected in the CzechII mouse mammary tumors [27, 49]. All of the viral integration events within Notch4 occurred within one of three exons encoding amino acid residues just N-terminal to the transmembrane domain of the encoded protein. These viral integration events result in the constitutive expression of a novel 2.3 kb mRNA species from the 3′ MMTV LTR that encodes the ICD of the protein (designated Int3). This mRNA species was not detected in tumors where the locus was intact or in the normal mammary gland. In similar studies, Dievart et al [50] found Notch1 to be rearranged by MMTV in 2 out of 24 MMTV-Neu transgenic mouse mammary tumors. The viral induced rearrangement of Notch1 also led the constitutive expression of the Notch1 ICD. Notch2 and Notch3 appear not to be targets for rearrangement by MMTV integration in mouse mammary tumors.
Experiments in which the ICD of the Drosophila Notch protein are over-expressed demonstrated that this represents a gain-of-function mutation, mimicking the consequences of the interaction between the Notch protein and its ligand [51]. In transgenic mice expression of Notch-1, -3, -4(Int3) ICD from the MMTV LTR or in the case of Int3 from the whey acidic protein (Wap) promoter there are two consequences on the biology of the mammary gland. In each strain of mice the development of mammary alveolar/lobular structures is arrested and the mice cannot lactate. Secondly with a high degree of penetrance (70–100%) the mice develop mammary adenocarcinomas [52–55].
The eIF-3e (Int6) Gene
The gene corresponding to CIS Int6 encodes a 48 kDa protein that is expressed in all adult tissues which have been tested including the mammary gland and as early as day 8 of embryonic development [56, 57]. It has been highly conserved through evolution, the amino acid sequence of the mouse and human Int6 gene products are identical and related sequences are present in Drosophila, C. elegans, and Saccharomyces pombe [28, 56–58]. It has been shown to be component of the eucaryotic translation initiation factor-3 (eIF-3e) as well as an interactive participant in the COP9 signalosome and the regulatory lid of the 26S proteasome. Integration of MMTV into the eIF-3e gene has been demonstrated in two independent CzechII preneoplastic hyperplastic outgrowth lines (HOG) as well as two independent mammary tumors from unrelated CzechII mice [28]. In each case an MMTV genome integrated into an intron of the eIF-3e gene in the opposite transcriptional orientation as the gene and resulted in the expression of a truncated RNA species that terminates at a cryptic transcription termination signal in the reverse sequence of the MMTV LTR. This novel RNA species encodes a C-terminally truncated form of the protein, designated Int6sh. The non-rearranged allele of eIF-3e in these tumors was checked for the presence of a somatic mutation, but none was found.
Expression of Int6sh in stably transfected human and mouse mammary epithelial cell lines leads to cellular transformation [59, 60]. To validate that Int6sh has transforming activity in vivo, a transgenic mouse model was designed using the Wap promoter to target expression of Int6sh to differentiating alveolar epithelial cells in the mammary gland. In Wap-Int6sh mice, the contralateral mammary glands contained widespread focal alveolar hyperplasia [61]. Mammary tumors developed in 42% of Wap-Int6sh parous females at an average age of 18 months. Only 4% of Wap-Int6sh non-breeding females developed tumors by 2 years of age. The Wap promoter is active only during estrus in the mammary tissue of cycling non-pregnant mice. These observations provide direct in vivo evidence that mammary-specific expression of the Int6sh leads to persistence of alveolar hyperplasia with the accompanying increased predisposition to mammary tumorigenesis. We have speculated that in this setting MMTV integration into eIF-3e either results in the expression of a biologically activated form of eIF-3e/Int6 or that it corresponds to a dominant-negative mutation. A different scenario appears to be operative in human carcinomas. Decreased expression of eIF-3e is observed in approximately one third of all human breast, lung and colon carcinomas [62]. In small cell lung carcinomas this is associated with poor outcome and is caused by hyper-methylation of the eIF-3e transcription promoter [63].
The Effect of Genetic Background on the Frequency of MMTV CIS in Mammary Tumors in Feral and Inbred Mouse Strains
Although approximately 50 CIS have been identified in MMTV induced mammary tumors from feral and inbred strains of mice, many appear to be unique to particular mouse strains or are associated with particular strains of MMTV ([25] and our unpublished data). Members of the Wnt, Fgf, and Rspo gene families represent the Core CISs which appear to be selected independent of genetic background, including M.m.musculus [7], M.m.domesticus [7, 36], M.m.jyg [35] and M.cervicolor.popaeus [64] or strain of MMTV. In contrast, MMTV integration into eIF3e has only been detected in M.m.musculus (CzechII) mammary tumors. Although Notch4 has been reported as a CIS in mammary tumors from the inbred high incidence BR6 (2 out of 30, [36]) and Balb/cfC3H (2 out of 160, [25]) mouse strains, it is more commonly altered in mammary tumors (18–40%) from the feral CzechII and M.m.jyg mouse strains [27, 34, 35].
One consequence of inbreeding mouse strains for a high incidence of MMTV induced mammary tumors with a short latency may have been the fixation of host mutations that provide selective advantage for viral integrations that occur at particular CISs. A classic example is the C3H mouse strain that has been inbred for a high incidence of mammary tumors for ~60 years. Wnt1 is activated by MMTV in 60–80% of the C3H mammary tumors (reviewed by [36]). When this same virus is transmitted to Balb/c mice only 20% of the mammary tumors contain an MMTV activated Wnt1 gene [7, 65]. Similarly, the BR6 mouse strain was inbred for a high incidence of pregnancy dependent mammary tumors that evolve into pregnancy independent tumors after several parities. In 70% of the pregnancy independent tumors Wnt1 and Fgf3/Fgf4 were activated by MMTV integration events [36].
Early studies of MMTV CIS noted that Wnt1 and Fgf3 or Fgf4 were frequently activated by MMTV in the same tumor leading to the speculation that these activated genes collaborate to promote tumor progression. This concept was tested by infecting, either Wnt1 or Fgf3, transgenic mice with MMTV(C3H) and identify collaborating CIS. In fact in the Wnt1 transgenic mammary tumors Fgf3, Ffg4, and Fgf8 were frequently (80%, 5%, and 11%, respectively) activated by the integration of an MMTV genome in adjacent host sequences [66, 67]. Similarly, in MMTV infected Fgf3 transgenic mammary tumors Wnt1 and Wnt10b were frequently (23% and 6%, respectively) activated by viral integration [32]. The cooperation between activated Wnt1 and Fgf3 was further substantiated by the phenotype of Wnt1/Fgf3 bi-transgenic mice [68]. In virgin bi-transgenic females mammary tumor development occurs 2 months earlier than sibling Wnt1 females and Fgf3 females rarely develop mammary tumors. Interestingly, virtually all bi-transgenic male mice develop mammary tumors whereas only 15% of Wnt1 males and no Fgf3 males develop mammary tumors. Although Wnt1 and Fgf3 clearly cooperate in driving mammary tumorigenesis, together they are not sufficient to induce tumorigenesis. In the Wnt1/Fgf3 bi-transgenic mice tumor development does not begin until several months after birth and fewer than 30% of animals have tumors in more than 1 of the 10 mammary glands. Thus additional genetic or epigenetic events must occur to initiate tumorigenesis. In our unpublished data on MMTV induced mammary tumors in CzechII, Balb/cfCz, and Balb/cfSp mice, greater than 50% of the tumors have more than one CIS. In fact it is not uncommon to have as many as 4 to 6 CIS per tumor. Lowther et al. [29] observed that mammary tumors in which Rspo2 was activated by MMTV, frequently Fgf3/Fgf4, Wnt3a or Wnt10b were also activated by MMTV. As noted earlier Rspo2 appears to use the same co-receptors (Fz and LRP) as Wnt and may synergize with it in the Wnt/canonical β-catenin or one of the noncanonical pathways.
The strategy of using MMTV induced insertional mutagenesis as a means to identify genes (CIS) that will collaborate with particular transgenes to induce mammary tumors has been expanded, but with mixed results. The difficulties in large part have been due to the different source of virus (recombinant hybrid versus milk borne MMTV), time of introduction of virus into the host (milk borne at birth, intraperitoneal injection at 3–4 weeks, 5–8 weeks and 7–12 weeks), the host genetic background (Balb/c, FVB/N, and C57Bl/C3H/Balb/c mixture) and the particular transgene (Wnt1, Fgf3, ErbB2, cyclinD, and P53172H) that precludes any rational comparison of the different studies [32, 50, 67, 69–71]. It seems likely that if a particular transgene is not among the genes that are normally targets for MMTV induced mammary tumorigenesis then there is no reason a priori to expect that they would affect the latency of tumor development, but they may affect the distribution of CIS relative to those found in infected non-transgenic mammary tumors. Thus in 24 MMTV infected ErbB2 mammary tumors, 4 contained a viral activated Wnt1 and 2 additional tumor contained a viral integration in Notch1 [50]. In another study Chatterjee et al. [69] genetically crossed FVB/N mice containing a WAP-P53172H transgene with C3H/He mice and then continued to backcross on to C3H/He mice. They found that as the FVB/N genetic background was diluted into the C3H/He background the latency of MMTV infected mammary tumors increased such that N2 transgenic tumors had a median latency of 242 day whereas the viral induced non-transgenic tumors had a latency of 269 day. Using inverse PCR they found RIS at 12 known genes (CD44, Pde1b, Galgt1, GalNAc4ST2, Col5a1, Wnt1, Wnt10b, Fgf8, Cnk1, Olig1, Olig2, and Uncx4.1 and 4 uncharacterized genes (EST-KIAA0952, 3167888, AK021163, AK015267) in DNA from viral infected transgenic mammary tumors. Other than the Wnt and Fgf genes, the remainder of the genes were novel and seem likely candidates to collaborate with the WAP-P53172H transgene in mammary tumorigenesis. It may be that CIS represent “initiating” mutations in a two step tumorigenesis model where the “promoting” influences depend upon modifying genes in the host genome and/or subsequent events that lead to expansion and maintenance of the “initiated” cell, e.g. pregnancy or wound repair.
The Contribution of Somatic Mutations in MMTV Induced Mouse Mammary Tumorigenesis
Based on current available data it seems that activation of the Core CIS and eIF3e are the early events contributing to the development of preneoplastic HOGs. Some HOGs contained CIS at Wnt1 and Fgf3 or Rspo2 and Fgf4or Wnt1 but still require additional mutagenic event(s) for malignant transformation. One possibility is that the low frequency CIS are the events that drive malignant transformation. Another equally plausible possibility is that malignant progression may result from the selection of somatic mutations that collaborate with MMTV induced mutations during tumor progression. In fact analysis of several transgenic mouse mammary tumor models has revealed signature chromosomal aberrations that are unique to the particular transgene [72–74]. A similar argument can be made that particular genes known to contribute to tumor progression are targets for point mutations that compliment MMTV CIS induced mammary tumorigenesis. For instance Wnt1 transgenic mammary tumors frequently have point mutations in Ha-Ras, but not K-Ras or N-Ras [75]. We have found that Wap-Int3 transgenic mammary tumors frequently contain point mutations in Trp53 (our unpublished data).
The Relevance of MMTV Induced CIS in Mouse Mammary Tumors to Human Breast Cancer
Until a few years ago the relevance of the genetics of mouse mammary tumorigenesis to human breast cancer was an open question. For instance there are morphological, hormonal, and life style differences represented in the two biological systems. In addition, the scientific community felt that the histopathology of mouse mammary tumors did not mirror the analogous parameters of the most frequent human breast tumor, i.e. invasive ductal carcinoma. Theodorou et. [25] interrogated a microarray analysis of 295 primary breast tumors [76] for the expression of the human homologues of the MMTV CIS target genes. They found that the expression of 11(ASTN2, CENTG2, EGR3, FGFR2, GSE1, JMJDIC, IGF2, LAMB1, PDGFRB, PROS1, and RREB1) out of the 33 CIS were deregulated in 5–43% of the human breast tumors. In another approach Wood et al. [77] have surveyed by high through put nucleotide sequence analysis of 18,191 genes in 11 primary human breast tumor DNAs. Interestingly, 7 (ASTN2, DPP10, FGFR2, JMJD1C, NOTCH4, ODZ1, and RREB1) out of the 33 CIS target genes [25] were mutated (Bert Vogelstein, personal communication). In addition, they found mutations in related family members of some of the MMTV CIS genes, including WNT (2, 14, and16), FGF (13 and14), ATP2B1 (many other ATP genes), CENTG2 (CENTG1), LAMB1(LAMB4), and MAP3K8 (MAP3K6) (Bert Vogelstein, personal communication). Collectively these studies provide strong support for the conclusion that there is a significant overlap in the genetics of breast cancer in these two biological systems.
A related issue is the persistent reports of MMTV related sequences in primary human breast tumor samples and whether these contribute to the etiology of human breast cancer as an insertional mutagen [78–84]. This topic is reviewed in more detail by Susan Ross in a separate article of this issue of the Journal. Recently, the Pogo Laboratory [85] reported that cells isolated from ascites or pleural effusions of patients with metastatic breast cancer contained MMTV sequences in their DNA, expressed the MMTV Env protein, and showed β-retroviral particles by electron microscopy. In another report, Indik et al. [86] reported that they were able to infect Hs578T human mammary cells with MMTV (GR) and demonstrate that it spread through the culture. This virus in mice is associated with the induction of pregnancy dependent mammary tumors. In fact Indik et al. [86] were the first, in this tissue culture system, to provide evidence the virus had integrated into the human genome by providing the sequence of the host-viral junction fragment. The inverse PCR high throughput methodology has been available at least since Wang et al. [82] first reported MMTV sequences in human breast tumor DNA. It seems reasonable to expect that in the near future evidence will be provided that the MMTV related sequences reported in DNA from primary human breast tumors was acquired by integration of a viral genome in the host DNA or corresponds to contaminating mouse genomic DNA.
Conclusions
Based on a statistical analysis of the frequency of MMTV induced CIS the Core signaling pathways contributing to MMTV induce mammary tumors have been identified. The lack of complexity of CIS in MMTV induced tumors is striking. The Core CIS include the Wnt, RSpo, and Fgf gene families that have been shown to collaborate or synergize with one another during mammary tumorigenesis. The Notch4 and eIF3e CIS appear to be mouse strain specific. MMTV induced collaborating genes for Notch gene family and eIF3e are unknown. It seems possible that in Notch4 or eIF3e positive mammary tumors malignant progression is facilitated by other selected somatic mutations or epigenetic events. At the present time there has been no systematic effort to evaluate these possibilities. The other low frequency CIS may reflect mouse strain or virus specific CIS, the effects host genetic background that contribute to tumor progression or chance. It is striking that the human homologs of many of these genes are mutated or their expression is deregulated in primary human breast carcinomas. These findings may provide an approach to develop new prognostic indicators or targets for therapeutic intervention.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- MMTV
mouse mammary tumor virus
- HAN
hyperplastic alveolar nodule
- LRC
label retaining cells
- MLV
murine leukemia virus
- HOG
hyperplastic outgrowth
- Tcf
T-cell factor
- Lef
lymphocyte enhancing factor
- RTK
receptor tyrosine kinase
- ICD
intracellular domain
- CIS
common integration site
- RIS
retroviral integration site
Contributor Information
Robert Callahan, Email: rc54d@nih.gov, Mammary Gland Biology and Tumorigenesis Laboratory, National Cancer Institute, Building 37/Room 1118A, MSC4254, Bethesda, MD 20892, USA.
Gilbert H. Smith, Email: gs4d@nih.gov, National Cancer Institute, Building 37/Room 1112A, MSC 4254, Bethesda, MD 20892, USA
References
- 1.Kamiya K, Gould MN, Clifton KH. Quantitative studies of ductal versus alveolar differentiation from rat mammary clonogens. Proc Soc Exp Biol Med. 1998;219(3):217–25. doi: 10.3181/00379727-219-44335. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67(2):93–109. doi: 10.1023/A:1010615124301. [DOI] [PubMed] [Google Scholar]
- 3.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 4.Smith GH. Experimental mammary epithelial morphogenesis in an in vivo model: evidence for distinct cellular progenitors of the ductal and lobular phenotype. Breast Cancer Res Treat. 1996;39(1):21–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- 5.Medina D. The preneoplastic phenotype in murine mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2000;5(4):393–407. doi: 10.1023/A:1009529928422. [DOI] [PubMed] [Google Scholar]
- 6.Medina D. Mammary developmental fate and breast cancer risk. Endocr Relat Cancer. 2005;12(3):483–95. doi: 10.1677/erc.1.00804. [DOI] [PubMed] [Google Scholar]
- 7.Callahan R, Smith GH. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene. 2000;19(8):992–1001. doi: 10.1038/sj.onc.1203276. [DOI] [PubMed] [Google Scholar]
- 8.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125 (10):1921–30. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 9.Smith GH, Boulanger CA. Mammary stem cell repertoire: New insights in aging epithelial populations. Mech Ageing Dev. 2002;123:1505–19. doi: 10.1016/S0047-6374(02)00114-8. [DOI] [PubMed] [Google Scholar]
- 10.Rajkumar L, Kittrell FS, Guzman RC, Brown PH, Nandi S, Medina D. Hormone-induced protection of mammary tumorigenesis in genetically engineered mouse models. Breast Cancer Res. 2007;9(1):R12. doi: 10.1186/bcr1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gattelli A, Zimberlin MN, Meiss RP, Castilla LH, Kordon EC. Selection of early-occurring mutations dictates hormone-independent progression in mouse mammary tumor lines. J Virol. 2006;80(22):11409–15. doi: 10.1128/JVI.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee AE. Genetic and viral influences of mammary tumours in BR6 mice. Br J Cancer. 1968;22(1):77–82. doi: 10.1038/bjc.1968.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squartini F. Responsiveness and progression of mammary tumors in high-cancer-strain mice. J Natl Cancer Inst. 1962;28:911–26. [PubMed] [Google Scholar]
- 14.van Nie R, Verstraeten AA. Studies of genetic transmission of mammary tumour virus by C3Hf mice. Int J Cancer. 1975;16(6):922–31. doi: 10.1002/ijc.2910160606. [DOI] [PubMed] [Google Scholar]
- 15.Foulds L. The histologic analysis of mammary tumors of mice. J Natl Cancer Inst. 1956;17(6):701–801. [PubMed] [Google Scholar]
- 16.Squartini F. Tumours of the mammary gland. IARC Sci Publ. 1979;23:43–90. [PubMed] [Google Scholar]
- 17.Squartini F, Basolo F, Bistocchi M. Lobuloalveolar differentiation and tumorigenesis: two separate activities of mouse mammary tumor virus. Cancer Res. 1983;43(12 Pt 1):5879–82. [PubMed] [Google Scholar]
- 18.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 19.Cairns J. Somatic stem cells and the kinetics of mutagenesis and carcinogenesis. Proc Natl Acad Sci U S A. 2002;99(16):10567–70. doi: 10.1073/pnas.162369899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith GH. Label-retaining mammary epithelial cells divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–7. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 21.Booth BW, Smith GH. Estrogen receptor-alpha and progesterone receptor are expressed in label-retaining mammary epithelial cells that divide asymmetrically and retain their template DNA strands. Breast Cancer Res. 2006;8(4):R49. doi: 10.1186/bcr1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 23.Peters G, Brookes S, Smith R, Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33(2):369–77. doi: 10.1016/0092-8674(83)90418-X. [DOI] [PubMed] [Google Scholar]
- 24.Theodorou V, Boer M, Weigelt B, Jonkers J, van der Valk M, Hilkens J. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene. 2004;23(36):6047–55. doi: 10.1038/sj.onc.1207816. [DOI] [PubMed] [Google Scholar]
- 25.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39(6):759–69. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 26.Lazo PA, Lee JS, Tsichlis PN. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc Natl Acad Sci U S A. 1990;87(1):170–3. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneo-plasia. J Virol. 1995;69(3):1932–8. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowther W, Wiley K, Smith GH, Callahan R. A new common integration site, Int7, for the mouse mammary tumor virus in mouse mammary tumors identifies a gene whose product has furin-like and thrombospondin-like sequences. J Virol. 2005;79(15):10093–6. doi: 10.1128/JVI.79.15.10093-10096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32(1):153–9. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 31.Qureshi SJ, Porteous DJ, Brookes AJ. Alu-based vectorettes and splinkerettes. More efficient and comprehensive polymerase chain reaction amplification of human DNA from complex sources. Genet Anal Tech Appl. 1994;11(4):95–101. doi: 10.1016/1050-3862(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 32.Lee FS, Lane TF, Kuo A, Shackleford GM, Leder P. Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci U S A. 1995;92(6):2268–72. doi: 10.1073/pnas.92.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver J, Keerikatte V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J Virol. 1989;63(5):1924–8. doi: 10.1128/jvi.63.5.1924-1928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14(16):1883–90. doi: 10.1038/sj.onc. 1201035. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar NH, Haga S, Lehner AF, Zhao W, Imai S, Moriwaki K. Insertional mutation of int protooncogenes in the mammary tumors of a new strain of mice derived from the wild in China: normal-and tumor-tissue-specific expression of int-3 transcripts. Virology. 1994;203(1):52–62. doi: 10.1006/viro.1994.1454. [DOI] [PubMed] [Google Scholar]
- 36.Peters G. Oncogenes at viral integration sites. Cell Growth Differ. 1990;1(10):503–10. [PubMed] [Google Scholar]
- 37.Fehse B, Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2008;15(2):143–53. doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Luke BT, Burgess SM. Redefining the common insertion site. Virology. 2006;344(2):292–5. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 39.Mohinta S, Wu H, Chaurasia P, Watabe K. Wnt pathway and breast cancer. Front Biosci. 2007;12:4020–33. doi: 10.2741/2368. [DOI] [PubMed] [Google Scholar]
- 40.Smolich BD, McMahon JA, McMahon AP, Papkoff J. Wnt family proteins are secreted and associated with the cell surface. Mol Biol Cell. 1993;4(12):1267–75. doi: 10.1091/mbc.4.12.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendrickx M, Leyns L. Non-conventional frizzled ligands and Wnt receptors. Dev Growth Differ. 2008;50(4):229–43. doi: 10.1111/j.1440-169X.2008.01016.x. [DOI] [PubMed] [Google Scholar]
- 42.Binnerts ME, Kim KA, Bright JM, Patel SM, Tran K, Zhou M, et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc Natl Acad Sci U S A. 2007;104(37):14700–5. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikuchi A, Yamamoto H. Tumor formation due to abnormalities in the beta-catenin-independent pathway of Wnt signaling. Cancer Sci. 2008;99(2):202–8. doi: 10.1111/j.1349-7006.2007.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cotton LM, O’Bryan MK, Hinton BT. Cellular signaling by fibroblast growth factors (FGFs) and their receptors (FGFRs) in male reproduction. Endocr Rev. 2008;29(2):193–216. doi: 10.1210/er.2007-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dillon C, Spencer-Dene B, Dickson C. A crucial role for fibroblast growth factor signaling in embryonic mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9(2):207–15. doi: 10.1023/B:JOMG.0000037163.56461.1e. [DOI] [PubMed] [Google Scholar]
- 46.Callahan R, Egan SE. Notch signaling in mammary development and oncogenesis. J Mammary Gland Biol Neoplasia. 2004;9(2):145–63. doi: 10.1023/B:JOMG.0000037159.63644.81. [DOI] [PubMed] [Google Scholar]
- 47.Muskavitch MA. Delta-notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166(2):415–30. doi: 10.1006/dbio.1994. 1326. [DOI] [PubMed] [Google Scholar]
- 48.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary protooncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122 (7):2251–9. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 49.Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66(4):2594–9. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18(44):5973–81. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 51.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74(2):331–45. doi: 10.1016/0092-8674(93)90424-O. [DOI] [PubMed] [Google Scholar]
- 52.Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56(8):1775–85. [PubMed] [Google Scholar]
- 53.Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168(3):973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6(3):345–55. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 55.Smith GH, Gallahan D, Diella F, Jhappan C, Merlino G, Callahan R. Constitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional development. Cell Growth Differ. 1995;6(5):563–77. [PubMed] [Google Scholar]
- 56.Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272(38):23477–80. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 57.Diella F, Levi G, Callahan R. Characterization of the INT6 mammary tumor gene product. DNA Cell Biol. 1997;16(7):839–47. doi: 10.1089/dna.1997.16.839. [DOI] [PubMed] [Google Scholar]
- 58.Miyazaki S, Rasmussen S, Imatani A, Diella F, Sullivan DT, Callahan R. Characterization of the Drosophila ortholog of mouse eIF-3p48/INT-6. Gene. 1999;233(1–2):241–7. doi: 10.1016/S0378-1119(99)00130-4. [DOI] [PubMed] [Google Scholar]
- 59.Mayeur GL, Hershey JW. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e) FEBS Lett. 2002;514(1):49–54. doi: 10.1016/S0014-5793(02)02307-4. [DOI] [PubMed] [Google Scholar]
- 60.Rasmussen SB, Kordon E, Callahan R, Smith GH. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene. 2001;20(38):5291–301. doi: 10.1038/sj.onc.1204624. [DOI] [PubMed] [Google Scholar]
- 61.Mack DL, Boulanger CA, Callahan R, Smith GH. Expression of truncated Int6/eIF3e in mammary alveolar epithelium leads to persistent hyperplasia and tumorigenesis. Breast Cancer Res. 2007;9(4):R42. doi: 10.1186/bcr1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol. 2001;18(1):175–9. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- 63.Buttitta F, Martella C, Barassi F, Felicioni L, Salvatore S, Rosini S, et al. Int6 expression can predict survival in early-stage non-small cell lung cancer patients. Clin Cancer Res. 2005;11(9):3198–204. doi: 10.1158/1078-0432.CCR-04-2308. [DOI] [PubMed] [Google Scholar]
- 64.Escot C, Hogg E, Callahan R. Mammary tumorigenesis in feral Mus cervicolor popaeus. J Virol. 1986;58(2):619–25. doi: 10.1128/jvi.58.2.619-625.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marchetti A, Robbins J, Campbell G, Buttitta F, Squartini F, Bistocchi M, et al. Host genetic background effect on the frequency of mouse mammary tumor virus-induced rearrangements of the int-1 and int-2 loci in mouse mammary tumors. J Virol. 1991;65(8):4550–4. doi: 10.1128/jvi.65.8.4550-4554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapoun AM, Shackleford GM. Preferential activation of Fgf8 by proviral insertion in mammary tumors of Wnt1 transgenic mice. Oncogene. 1997;14(24):2985–9. doi: 10.1038/sj.onc.1201146. [DOI] [PubMed] [Google Scholar]
- 67.Shackleford GM, MacArthur CA, Kwan HC, Varmus HE. Mouse mammary tumor virus infection accelerates mammary carcinogenesis in Wnt-1 transgenic mice by insertional activation of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci U S A. 1993;90(2):740–4. doi: 10.1073/pnas.90.2.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwan H, Pecenka V, Tsukamoto A, Parslow TG, Guzman R, Lin TP, et al. Transgenes expressing the Wnt-1 and int-2 protooncogenes cooperate during mammary carcinogenesis in doubly transgenic mice. Mol Cell Biol. 1992;12(1):147–54. doi: 10.1128/mcb.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatterjee G, Rosner A, Han Y, Zelazny ET, Li B, Cardiff RD, et al. Acceleration of mouse mammary tumor virus-induced murine mammary tumorigenesis by a p53 172H transgene: influence of FVB background on tumor latency and identification of novel sites of proviral insertion. Am J Pathol. 2002;161(6):2241–53. doi: 10.1016/S0002-9440(10)64500-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gadd M, Pisc C, Branda J, Ionescu-Tiba V, Nikolic Z, Yang C, et al. Regulation of cyclin D1 and p16(INK4A) is critical for growth arrest during mammary involution. Cancer Res. 2001;61(24):8811–9. [PubMed] [Google Scholar]
- 71.Zelazny E, Li B, Anagnostopoulos AM, Coleman A, Perkins AS. Cooperating oncogenic events in murine mammary tumorigenesis: assessment of ErbB2, mutant p53, and mouse mammary tumor virus. Exp Mol Pathol. 2001;70(3):183–93. doi: 10.1006/exmp.2001.2357. [DOI] [PubMed] [Google Scholar]
- 72.Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21(6):890–8. doi: 10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- 73.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, et al. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63(9):2179–87. [PubMed] [Google Scholar]
- 74.Weaver ZA, McCormack SJ, Liyanage M, du Manoir S, Coleman A, Schrock E, et al. A recurring pattern of chromosomal aberrations in mammary gland tumors of MMTV-cmyc transgenic mice. Genes Chromosomes Cancer. 1999;25(3):251–60. doi: 10.1002/(SICI) 1098-2264(199907)25:3<251::AID-GCC7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 75.Podsypanina K, Li Y, Varmus HE. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2004;2:24. doi: 10.1186/1741-7015-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 77.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 78.Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6 (4):1273–8. [PubMed] [Google Scholar]
- 79.Liu B, Wang Y, Melana SM, Pelisson I, Najfeld V, Holland JF, et al. Identification of a proviral structure in human breast cancer. Cancer Res. 2001;61(4):1754–9. [PubMed] [Google Scholar]
- 80.Melana SM, Holland JF, Pogo BG. Search for mouse mammary tumor virus-like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res. 2001;7(2):283–4. [PubMed] [Google Scholar]
- 81.Wang Y, Go V, Holland JF, Melana SM, Pogo BG. Expression of mouse mammary tumor virus-like env gene sequences in human breast cancer. Clin Cancer Res. 1998;4(10):2565–8. [PubMed] [Google Scholar]
- 82.Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55(22):5173–9. [PubMed] [Google Scholar]
- 83.Wang Y, Jiang JD, Xu D, Li Y, Qu C, Holland JF, et al. A mouse mammary tumor virus-like long terminal repeat superantigen in human breast cancer. Cancer Res. 2004;64(12):4105–11. doi: 10.1158/0008-5472.CAN-03-3880. [DOI] [PubMed] [Google Scholar]
- 84.Zammarchi F, Pistello M, Piersigilli A, Murr R, Di Cristofano C, Naccarato AG, et al. MMTV-like sequences in human breast cancer: a fluorescent PCR/laser microdissection approach. J Pathol. 2006;209(4):436–44. doi: 10.1002/path.1997. [DOI] [PubMed] [Google Scholar]
- 85.Melana SM, Nepomnaschy I, Sakalian M, Abbott A, Hasa J, Holland JF, et al. Characterization of viral particles isolated from primary cultures of human breast cancer cells. Cancer Res. 2007;67(18):8960–5. doi: 10.1158/0008-5472.CAN-06-3892. [DOI] [PubMed] [Google Scholar]
- 86.Indik S, Gunzburg WH, Kulich P, Salmons B, Rouault F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology. 2007;4:73. doi: 10.1186/1742-4690-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]